- *Corresponding Author:
- Khanzad Khudhur Jarjees
Department of Food Technology,
College of Agriculture, Salahaddin University,
Erbil 44002,
Iraq
E-mail: khanzad.jarjees@su.edu.krd
| This article was originally published in a special issue, “Diagnostic and Therapeutic Advances in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2021:83(5)Spl Issue “275-282” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Pseudomonas aeruginosa is one of the most common opportunistic pathogens that cause infections. The drug resistance in Pseudomonas aeruginosa may relate to the production of broad spectrum beta lactamase enzymes. This study aimed to detect multidrug resistant, extensively drug resistant and pan drug resistant isolates of Pseudomonas aeruginosa and to determine the frequency of extended spectrum beta lactamases enzymes by using a reliable test for detection blaCTX-M1, blaCTX-M2 and blaCTX-M3 genes in Pseudomonas aeruginosa. Pseudomonas aeruginosa isolated from different clinical samples, including sputum, wound, burn and urine. Identification and antibiotic susceptibility test were made by VITEK® 2 automated system. Identification confirmed by 16S ribosomal ribonucleic acid. The bacterial deoxyribonucleic acid was extracted and specific primers were used for detection blaCTX-M1, blaCTX-M2 and blaCTX-M3 genes. 62 Pseudomonas aeruginosa strains were obtained from 650 different clinical specimens (9.54 %). Of the 62 Pseudomonas aeruginosa isolates, the highest resistances were related to the antibiotics of ticarcillin, ticarcillin/clavulanic acid, piperacillin, piperacillin/tazobactam, ceftazidime and cefepime (100 %). However, the lowest resistance was observed to meropenem, imipenem (3.23 %), amikacin, ciprofloxacin (6.45 %) and gentamycin, tobramycin (19.35 %). Out of 62 bacterial isolates, 46 (74.19 %) were multidrugresistance strains, 2 (3.23 %) were extensive drug-resistance and no pan drug resistance was isolated. The gene frequency of blaCTX-M1 was 12 (19.35 %) and blaCTX-M2 5 (8.06 %), all specimens were negative for blaCTX-3 gene. This study highlights the increased prevalence of multidrug resistant Pseudomonas aeruginosa. The blaCTX-M genes provide Pseudomonas aeruginosa with an additional powerful resistance mechanism with potentially serious clinical implications, including limitation of the therapeutic options. The incidence of extended spectrum beta lactamases varies with geographic location and time.
Keywords
BlaCTX-M1, blaCTX-M2, blaCTX-M3, extended spectrum beta lactamases, Pseudomonas aeruginosa
Pseudomonas aeruginosa (P. aeruginosa) is an obligate aerobic and can persist in both community and hospital settings due to its ability to survive on minimal nutrition requirements and tolerate various physical conditions [1]. P. aeruginosa is the most frequent bacterial species associated with infections such as urinary tract infections, respiratory infections, dermatitis, soft tissue infections, gastrointestinal infections and various systemic infections, particularly in patients with severe burns [2,3].
P. aeruginosa utilizes many secreted virulence factors that have been implicated in the pathogenesis of infection. These including exotoxin A, phospholipase, alkaline protease, pili, flagella and lipopolysaccharide [4]. The important problem of this organism is the development of drug resistance mechanisms. As a matter of fact, P. aeruginosa has an outer membrane with low permeability, multidrug discharge pumps, efflux pumps, inactivation and modification of antibiotics lactamase and the outer membrane purine degradation set, which could be the reason for the resistance of these microorganisms during the treatment [5-7].
Two major classification schemes exist for categorizing beta lactamase enzymes: Ambler classes A through D, based on amino acid sequence homology and Bush- Jacoby-Medeiros groups 1 through 4, based on substrate and inhibitor profile [8]. However, a growing number of Ambler class A Extended Spectrum Beta Lactamases (ESBLs), class B Metallo-Beta-Lactamases (MBLs) and class D Extended Spectrum Oxacillinases (ES-OXAs) have been reported in P. aeruginosa clinical isolates. Class A ESBLs reported in P. aeruginosa include Pseudomonas Extended Resistance (PER), Sulfhydryl Variable (SHV), Temoniera (TEM), Vietnam Extended Spectrum Beta Lactamase (VEB), Belgium (BEL), Guiana Extended Spectrum Beta Lactamase (GES) and Cefotaximase (CTX-M) type [9,10].
The CTX-M type ESBLs developed from TEM and SHV and can be divided into five subgroups according to their amino acid sequence similarities, including CTX-M-I, CTX-M-II, CTX-M-III, CTX-M-IV and CTX-M-V [11,12]. The most common group of ESBLs not belonging to the TEM or SHV families was termed CTX-M to highlight their greater activity against cefotaxime than against ceftazidime [13]. Some of them, such as CTX-M-15 and CTX-M-19, also hydrolyze ceftazidime efficiently [14], which may complicate their phenotypic recognition [15]. CTX-M ESBLs are the most commonly isolated ESBLs in many parts of the world, particularly Europe [8,16].
Resistance to ESBLs in P. aeruginosa is associated in most cases with the overproduction of cephalosporinases Ampicillin Class C (AmpC) [1]. ESBLs is one of the main leading causes of resistance to beta lactam antibiotics among Gram-negative bacteria [17]. This enzyme is plasmid-encoded beta lactamase that mediate resistance to penicillin, first, second and third generation cephalosporin, such as cefotaxim, ceftriaxone and ceftazidime [17,18]. TEM, SHV and CTX-M are the major genetic groups of ESBLs amongst clinically Gramnegative bacteria [18,19].
Aminoglycosides are used in the treatment of a broad range of life-threatening infections. The activity of aminoglycosides depends on binding to a highly conserved motif of 16S Ribosomal Ribonucleic Acid (rRNA). Aminoglycosides resistance mechanisms include decreased outer membrane permeability, active efflux and amino acid substitutions in ribosomal proteins. In contrast, the most common resistance mechanism is enzymatic, leading to a modification of the drug. Methylation of 16S rRNA has recently been demonstrated to be another mechanism of resistance encountered in Gram-negative organisms, corresponding to a modification of the antibiotic target. Methylases interfere in the binding of these antibiotics to their site of action. These 16S rRNA methylases confer a high level of resistance to clinically useful aminoglycosides such as amikacin, gentamicin and tobramycin [20,21]. The corresponding genes are associated with transposon structures located on transferable plasmids, enhancing their horizontal spread. Isolates producing 16S rRNA methylases are multidrug-resistant, particularly to broad spectrum beta lactams through the production of ESBLs or MBLs [21,22].
European Centre for Disease Control (ECDC) and Centre for Disease Control and Prevention (CDC), Atlanta have proposed standardized definitions for the Multidrug Resistant (MDR), Extensively Drug Resistant (XDR) and Pan Drug Resistant (PDR) bacteria. MDR was defined as acquired nonsusceptibility to at least one agent in three or more antimicrobial categories as per guidelines. XDR was defined as non-susceptibility to at least one agent in all but two or fewer antimicrobial categories and PDR was defined as non-susceptibility to all agents in all antimicrobial categories [23,24]. The main objective of this study was to detect MDR, XDR and PDR bacteria and to investigate the prevalence of blaCTX-M1, blaCTX-M2 and blaCTX-M3 genes producing ESBL among P. aeruginosa isolated from different clinical samples.
Materials and Methods
Collection and identification of samples:
650 different clinical samples including sputum, wound, burn and urine were collected from the patients admitted to Erbil hospitals and health centers from May 2019 to November 2019. Gram stains were used to determine the form of microscopic organisms. To determine the phenotypic identity, oxidase and catalase tests were used. The Gram-Negative card for the VITEK® 2 compact system is completely automated and provides identification and antibiotic susceptibility test. For the final determination, the molecular detection method of Polymerase Chain Reaction (PCR) was used with the gene of 16S rRNA [25].
Antibiotic susceptibility test:
Antibiotic susceptibility test is done using VITEK® 2 automated system (bio Merieux) for the following antimicrobial ticarcillin, ticarcillin/clavulanic acid, piperacillin, piperacillin/tazobactam, ceftazidime, cefepime, imipenem, meropenem, amikacin, gentamicin, tobramycin and ciprofloxacin. MDR, XDR and PDR strains were detected according to guidelines proposed by ECDC and CDC [23,24].
Identification of species based on 16S rRNA gene:
The extraction of bacteria's genome was performed using an extraction kit (Invitek Stratec Business, Canada). Then, PCR test was done on the extracted gene for the 16S rRNA gene. In this study, one pair of primer with the sequence of F: GGGGGATCTTCGGACCTCA and R: TCCTTAGAGTGCCCACCCG were used together in a PCR reaction to identify the species [25]. The total of 25 μl PCR master mix reaction volume was performed for identification (16S rRNA) that containing 4 μl of genomic DNA, 12.5 μl of 2X GoTaq® Green Master Mix (Promega, USA) and 1.5 μl was added for each of the forward and reversed primer and completed with 5.5 μl nuclease free water and the thermal cycle with following steps were repeated for 30 times as shown in Table 1.
| Test Procedure | Temperature (°) | Time (s) |
|---|---|---|
| Initial denaturation | 95 | 300 |
| Denaturation | 95 | 30 |
| Annealing | 58 | 45 |
| Extension | 72 | 60 |
| Final extension | 72 | 300 |
Table 1: Steps and Thermal Cycle of The 16s Rrna For Identification of Pseudomonas
Identification of the genes of blaCTX-M1, blaCTX-M2 and blaCTX-M3 in P. aeruginosa:
P. aeruginosa were nightly cultured in Luria-Bertani liquid medium at 37° for 24 h and DNA was extracted from the bacteria of P. aeruginosa using the kit (Invitek Stratec Business kit, Canada). Then, they were used on the genes of blaCTX-M1, blaCTX-M2 and blaCTX-M3 with the specific oligonucleotide primers which were simultaneously used for the process of PCR as shown in Table 2 [25,26]. Thermal cycler device was accurately set up for doing PCR process based on the mentioned resources in Table 3 [25,27,28]. The PCR products were separated in a 2 % agarose gel and stained with Safe dye (GenetBio, Korea). The gel was visualized under an Ultraviolet (UV) transilluminator (Synegene, UK).
| Primer name | Amplicon size bp | Nucleotide sequence | References |
|---|---|---|---|
| CTX-M1 | 499 | F-GACGATGTCATCGGCTGAGC R-AGCCGCCGACGCTAATACA |
[28] |
| CTX-M2 | 351 | F-GCGACCAGGTTAACTACAATCC R-CGGTAGTATTGCCCTTAAGCC |
[28] |
| CTX-M3 | 307 | F-CGCTTTGCCATGTGCAGCACC R-GCTCAGTACGATCGAGCC |
[28] |
Table 2: Primers Used For Detection Of CTX-M1, CTX-M2 and CTX-M3 Genes
| Test Procedure | Temperature (°) | Time (s) |
|---|---|---|
| Initial denaturation | 94 | 180 |
| Denaturation | 94 | 60 |
| Annealing | 55 | 30 |
| Extension | 72 | 60 |
| Final extension | 72 | 600 |
Table 3: Steps and Thermal Cycle of The Genes of CTX-M1 and CTX-M2
Results and Discussion
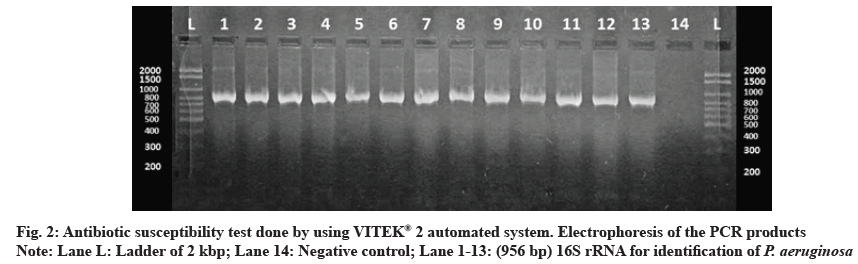
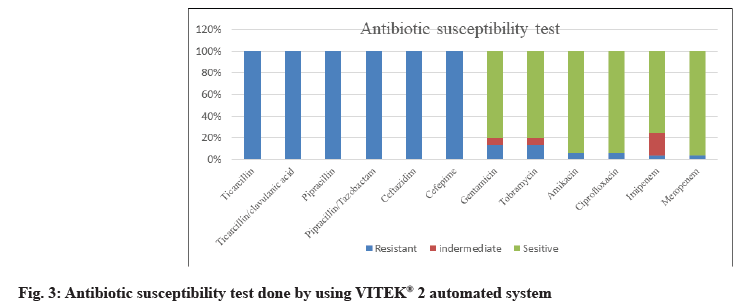
62 P. aeruginosa strains were obtained from 650 different clinical specimens (9.54 %). All isolates were identified phenotypically, as shown in fig. 1. Using the VITEK® 2 automated system, then identification was confirmed by 16S rRNA gene according to the expected size of (956 bp) for identification of P. aeruginosa as shown in fig. 2. Of the 62 P. aeruginosa isolates, the highest resistances were related to the antibiotics of ticarcillin, ticarcillin/clavulanic acid, piperacillin, piperacillin/tazobactam, ceftazidime and cefepime (100 %). However, the lowest resistance was observed to meropenem and imipenem 2 (3.23 %), amikacin and ciprofloxacin 4 (6.45 %) and gentamycin tobramycin 12 (19.35 %) as shown in fig. 3.
Out of 62 bacterial isolates, 46 (74.19 %) were multidrugresistance strains, 2 (3.23 %) were extensive drugresistance and no PDR was isolated, as shown in Table 4. The gene frequency of blaCTX-M1 was 12/62 (19.35 %) and blaCTX-M2 5/62 (8.06 %) as shown in fig. 4, all specimens were negative for blaCTX-M3 gene. This study revealed that among P. aeruginosa isolates exhibiting broad spectrum beta lactam resistance, there was a prevalence of blaCTX-M1 and blaCTX-M2.
| Antimicrobial resistance | P. aeruginosa (62) |
|---|---|
| MDR | 46 (74.19 %) |
| XDR | 2 (3.23 %) |
| PDR | 0 |
Table 4: Distribution of Multidrug-Resistance Strains, Extensive Drug-Resistance and Pan Drug Resistance in Isolated P. Aeruginosa
P. aeruginosa has recently emerged as a major cause of healthcare-associated infections, especially in immunocompromised people and burn patients [29,30]. The treatment of P. aeruginosa infections is increasingly complicated due to both intrinsic and acquired resistance to the most commonly prescribed antibiotics in hospital settings [1,31].
Inappropriate empirical therapy has been associated with increased mortality in P. aeruginosa infections; delays in starting appropriate therapy may increase length of hospital stay and infection persistence. With an increased prevalence of ESBL-producing pathogens, such isolates' fast and accurate identification becomes increasingly clinically important. Despite a variety of available methods, the identification of ESBLs by conventional phenotypic methods remains difficult in practice due to the large number of beta lactamase variants, the association of ESBLs with overproduced or plasmidborne cephalosporinases AmpC or MBL [32] and the change of outer-membrane permeability [33]. Hence, molecular methods that have the potential to disentangle this complex resistance etiology and translate it into clinically useful information have the potential to improve the current diagnostic practice [34]. ESBL-producing isolates were MDR and showed relatively higher resistance rates to most antibiotics tested [35,36].
Of the 62 P. aeruginosa isolates, the highest resistances were related to the antibiotics of ticarcillin, ticarcillin/ clavulanic acid, piperacillin, piperacillin/tazobactam, ceftazidime and cefepime (100 %). However, the lowest resistance was observed to meropenem and imipenem 2 (3.23 %), amikacin and ciprofloxacin 4 (6.45 %) and gentamycin tobramycin 12 (19.35 %). This result is in accordance with other reports, Asghar and Ahmed [37]; they reported that carbapenems are often used as a last resort treatment for Pseudomonas infections. Resistance to imipenem was shown to be 35.4 %. This difference in the distribution of aminoglycoside genes may be attributed to differences in aminoglycoside prescription patterns, selection of bacterial populations or geographical differences in the occurrence of aminoglycoside resistance genes.
The highest gene frequencies of ESBLs are related to blaCTX-M1 (19.35 %) and blaCTX-M2 (8.06 %). These explain the high resistance of P. aeruginosa to the third and fourth generation of cephalosporin.
Different results have been reported in other countries for the prevalence of the enzymes of ESBL related to CTX-M. Lin et al. [38] reported that amikacin had the highest susceptibility rate, whereas aztreonam had the lowest in Taiwan. Amikacin 91.2 %, ciprofloxacin 71.9 %, imipenem 70.2 %, cefepime 59.6 %, ceftazidime 56.1 %, piperacillin/tazobactam 52.6 % and aztreonam 17.5 %, and blaCTX-M genes were not detected in this study. In another study in Iran Bokaeian et al. [39], showed that ciprofloxacin (95.7 %) and piperacillin (92.2 %) were the most effective antipseudomonal agents and blaCTX-M1 genes were not detected.
Our results consistent with the findings of other studies in Saudi Arabia [40] that reported about (25.9 %) of P. aeruginosa isolates were confirmed as ESBL producers and the frequencies of blaCTX-M genes were 10.7 %. In a similar study in a Chinese hospital, the prevalence of ESBLs-producing P. aeruginosa isolates was (87.5 %) and (12.5 %) didn’t produce ESBLs. All ESBLsproducing isolates showed nearly identical antimicrobial susceptibility profiles. In another study in India blaCTXM amplified in (10.71 %) isolates [41]. In Bandar-Abbas, Iran, the frequencies of blaCTX-M genes were 23.95 %. They reported that the prevalence of different ESBL genes varies in different geographic locations. The blaCTX-M genotypes are prevalent in Asian countries [42]. CTX-M-type ESBLs, within some cases evidence of their horizontal transfer from Enterobacteriaceae to P. aeruginosa, are very rarely identified in P. aeruginosa. The genotypic prevalence of ESBLs for blaCTX-M-1, blaCTX-M-2 genes was 14.3 % and 9.6 %, respectively [43]. In Tabriz, Iran, the highest gene frequencies of ESBLs are related to the genes of CTX-M1 (27.27 %), CTX-M2 (23.63 %) and CTX-M3 (9.09 %), respectively [25]. A single CTX-M-1-producing P. aeruginosa isolate has been reported from The Netherlands in 2006 [44] and CTX-M-2 positive P. aeruginosa have been identified in South America [9,45,46]. Recently, CTX-M-3 was identified from several P. aeruginosa isolates from China [47].
Jafari [48] reported that in Marand city, East Azarbijan of the 85 P. aeruginosa, the highest resistance was to amikacin antibiotics (82.35 %), nalidixic acid (80 %) and ceftazidime (72.94 %), and the highest susceptibility was observed to ampicillin (38.82 %) and cefotaxime (38.82 %). The highest gene frequencies of ESBLs are related to the genes of CTX-M1 (34.12 %), CTX-M2 (28.24 %) and CTX-M3 (12.94 %), respectively.
The multidrug resistant phenotype in P. aeruginosa could be mediated by several mechanisms, including multidrug efflux systems, enzyme production, outer membrane protein (porin) loss and target mutations. These resistant organisms are clinically important because they result in increased morbidity and mortality. In addition, ESBL producing bacteria is frequently resistant to many antibiotics classes, resulting in difficult-to-treat infections. Currently, carbapenems are regarded as the drugs of choice for the treatment of infections caused by ESBL-producing organisms. The development and spread of ESBLs are most likely caused by the overuse of expanded spectrum cephalosporins in the hospital setting. Proper infection control practices and barriers are essential to prevent the spreading and outbreaks of ESBL-producing bacteria.
The difference in result for gene frequencies and antibiotic resistance may be due to either the different common colons present in a certain region or different antibiotic treatment patterns [49].
This study highlights the increased prevalence of multidrug resistant P. aeruginosa. The CTX-M ESBLs provide P. aeruginosa with an additional powerful resistance mechanism with potentially serious clinical implications, including limitation of the therapeutic options. The incidence of ESBLs varies with geographic location and time. ESBL producing strains are usually found in those areas of hospitals where antibiotic use is frequent and the patient’s condition is critical. This study revealed that among P. aeruginosa isolates exhibiting extended spectrum beta lactam resistance, there was a prevalence of blaCTX-M genes. The complete identification of ESBL and the restriction of beta lactam antibiotics can maintain the efficacy of beta lactam antibiotics as much as possible.
Conflict of interests:
The authors declared no conflict of interests.
References
- Lister PD, Wolter DJ, Hanson ND. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin Microbiol Rev 2009;22:582–610.
- Peerayeh SN, Mahabadi RP, Toupkanlou SP, Siadat SD. Diversity of β-lactamases produced by imipenem resistant, Pseudomonas aeruginosa isolates from the bloodstream. Burns 2014;40(7):1360-4.
- Milczewska J, Wołkowicz T, Zacharczuk K, Kwiatkowska M, Sands D. Cross-infections with Pseudomonas aeruginosa in patients with cystic fibrosis attending the Warsaw Centre. Dev Period Med 2015(1):60-5.
- Salyers AA, Whitt DD, Whitt DD. Bacterial pathogenesis: a molecular approach. Washington, DC: ASM press; 1994.
- Hancock RE, Speert DP. Antibiotic resistance in Pseudomonas aeruginosa: mechanisms and impact on treatment. Drug Resist Updat 2000;3(4):247-55.
- Papp-Wallace KM, Endimiani A, Taracila MA, Bonomo RA. Carbapenems: past, present and future. Antimicrob Agents Chemother 2011;55(11):4943-60.
- Kanani M, Khazaei S, Madani H, Melikian Zadeh A. Drug resistance of Pseudomonas aeruginosa to ceftazidime and imipenem in Imam Reza (AS) Kermanshah 89-85 y. J Lorestan Univ Med Sci 2013;15(4):52-60.
- Drawz SM, Bonomo RA. Three decades of β-lactamase inhibitors. Clin Microbiol Rev 2010;23(1):160-201.
- Picão RC, Poirel L, Gales AC, Nordmann P. Diversity of β-lactamases produced by ceftazidime-resistant Pseudomonas aeruginosa isolates causing bloodstream infections in Brazil. Antimicrob Agents Chemother 2009;53:3908-13.
- Amoudizaj FF, Aghayi E, Matin MG, Soltani N, Mala P. Antibiotic Resistance Pattern and Frequency of PER-1, SHV-1 and AMPC Type B-Lactamase Genes in Pseudomonas aeruginosa Isolated from Clinical Samples. Open Microbiol J 2020;13:308-12.
- Pitout JDD, Hossain A, Hanson ND. Phenotypic and molecular detection of CTX-M-β-lactamases produced by Escherichia coli and Klebsiella spp. J Clin Microbiol 2004;42:5715-21.
- Roschanski N, Fischer J, Guerra B, Roesler U. Development of a multiplex real-time PCR for the rapid detection of the predominant beta-lactamase genes CTX-M, SHV, TEM and CIT-type AmpCs in Enterobacteriaceae. PLoS One 2014;9:e100956.
- Bonnet R. Growing Group of Extended-Spectrum β-Lactamases: The CTX-M Enzymes. Antimicrob Agents Chemother 2004;48(1):1-14.
- Poirel L, Naas T, Le Thomas I, Karim A, Bingen E, Nordmann P. CTX-M-type extended-spectrum β-lactamase that hydrolyzes ceftazidime through a single amino acid substitution in the omega loop. Antimicrob Agents Chemother 2001;45(12):3355-61.
- Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J Antimicrob Chemother 2005;56(1):52-9.
- Zahra T, Rezvan M, Ahmad K. Detection and characterization of multidrug resistance and Extended-Spectrum-Beta-Lactamase-producing (ESBLS) Pseudomonas aeruginosa isolates in teaching hospital. African J Microbiol Res 2011;5:3223-8.
- Rawat D, Nair D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J Glob Infect Dis 2010;2(3):263.
- Paterson DL, Bonomo RA. Extended-spectrum β-lactamases: a clinical update. Clin Microbiol Rev 2005;18(4):657-86.
- Bradford PA. Extended-spectrum β-lactamases in the 21st century: characterization, epidemiology and detection of this important resistance threat. Clin Microbiol Rev 2001;14(4):933-51.
- Doi Y, Arakawa Y. 16S ribosomal RNA methylation: emerging resistance mechanism against aminoglycosides. Clin Infect Dis 2007;45(1):88-94.
- Doi Y, Wachino JI, Arakawa Y. Nomenclature of plasmid-mediated 16S rRNA methylases responsible for panaminoglycoside resistance. Antimicrob Agents Chemother 2008;52(6):2287-8.
- Bueno MF, Francisco GR, O'Hara JA, de Oliveira Garcia D, Doi Y. Coproduction of 16S rRNA methyltransferase RmtD or RmtG with KPC-2 and CTX-M group extended-spectrum β-lactamases in Klebsiella pneumoniae. Antimicrob Agents Chemother 2013;57(5):2397-400.
- Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012;18(3):268-81.
- Pattnaik D, Panda SS, Singh N, Sahoo S, Mohapatra I, Jena J. Multidrug resistant, extensively drug resistant and pan drug resistant Gram-negative bacteria at a tertiary care centre in Bhubaneswar. Int J Community Med Public Health 2019;6:567-72.
- Sales A, Fathi R, Mobaiyen H, Bonab FR, Kondlaji KB. Molecular Study of the Prevalence of CTX-M1, CTX-M2, CTXM3 in Pseudomonas aeruginosa Isolated from Clinical Samples in Tabriz Town, Iran. Electronic J Biol 2017;13(3):253-9.
- Khan SA, Nawaz MS, Khan AA, Hopper SL, Jones RA, Cerniglia CE. Molecular characterization of multidrug-resistant Enterococcus spp. from poultry and dairy farms: detection of virulence and vancomycin resistance gene markers by PCR. Molecular and cellular probes 2005;19(1):27-34.
- Owlia P, Bameri Z, Chitsaz M. Detection of CTX-M-β Lactamases form Isolated Klebsiella pneumoniae. Iran J Pathol 2010;5:137-42.
- Chaudhari V, Gunjal S, Mehta M. Antibiotic resistance patterns of Pseudomonas aeruginosa in a tertiary care hospital in Central India. Int J Med Sci Public Health 2013;2(2):386-9.
- Sydnor ER, Perl TM. Hospital epidemiology and infection control in acute-care settings. Clin Microbiol Rev 2011;24(1):141-73.
- Anvarinejad M, Japoni A, Rafaatpour N, Mardaneh J, Abbasi P, Shahidi MA, et al. Burn patients infected with metallo-beta-lactamase-producing Pseudomonas aeruginosa: Multidrug-resistant strains. Arch Trauma Res 2014;3(2):e18182.
- Dalhoff A. Global fluoroquinolone resistance epidemiology and implictions for clinical use. Interdiscip Perspect Infect Dis 2012;2012.
- Drieux L, Brossier F, Sougakoff W, Jarlier V. Phenotypic detection of extended‐spectrum β‐lactamase production in Enterobacteriaceae: review and bench guide. Clin Microbiol Infect 2008;14:90-103.
- Martínez‐Martínez L. Extended‐spectrum β‐lactamases and the permeability barrier. Clin Microbiol Infect 2008;14:82-9.
- Leinberger DM, Grimm V, Rubtsova M, Weile J, Schröppel K, Wichelhaus TA, et al. Integrated detection of extended-spectrum-beta-lactam resistance by DNA microarray-based genotyping of TEM, SHV and CTX-M genes. J Clin Microbiol 2010;48(2):460-71.
- Fallah F, Borhan RS, Hashemi A. Detection of bla (IMP) and bla (VIM) metallo-β-lactamases genes among Pseudomonas aeruginosa strains. Int J Burn Trauma 2013;3(2):122-4.
- Vala MH, Hallajzadeh M, Hashemi A, Goudarzi H, Tarhani M, Tabrizi MS, et al. Detection of Ambler class A, B and D ß-lactamases among Pseudomonas aeruginosa and Acinetobacter baumannii clinical isolates from burn patients. Ann Burns Fire Disasters 2014;27(1):8-13.
- Asghar AH, Ahmed OB. Prevalence of aminoglycoside resistance genes in Pseudomonas aeruginosa isolated from a tertiary care hospital in Makkah, KSA. Clin Pract 2018;15:1-7.
- Lin SP, Liu MF, Lin CF, Shi ZY. Phenotypic detection and polymerase chain reaction screening of extended-spectrum β-lactamases produced by Pseudomonas aeruginosa isolates. J Microbiol Immunol Infect 2012;45(3):200-7.
- Bokaeian M, Zahedani SS, Bajgiran MS, Moghaddam AA. Frequency of PER, VEB, SHV, TEM and CTX-M genes in resistant strains of Pseudomonas aeruginosa producing extended spectrum β-lactamases. Jundishapur J Microbiol 2015;8(1):e13783.
- Ahmed OB, Asghar AH, Bahwerth FS. Prevalence of ESBL genes of Pseudomonas aeruginosa strains isolated from Makkah Hospitals, Saudi Arabia. Euro J Biol Med Sci Res 2015;3(6):12-8.
- Bahrami M, Mmohammadi-Sichani M, Karbasizadeh V. Prevalence of SHV, TEM, CTX-M and OXA-48 β-Lactamase Genes in Clinical Isolates of Pseudomonas aeruginosa in Bandar-Abbas, Iran. Avicenna J Clin Microbiol Infect 2018;5(4):86-90.
- Minhas N, Sharma PC. Molecular characterization of Pseudomonas aeruginosa isolates recovered from human patients in Himachal Pradesh (India) for selective genes: extended spectrum beta-lactamase (ESBL), ampicillin class c (AmpC) and metallo beta-lactamase (MBL) genes. Int J Pharm Sci Res 2016;7:4905.
- Chen Z, Niu H, Chen G, Li M, Li M, Zhou Y. Prevalence of ESBLs-producing Pseudomonas aeruginosa isolates from different wards in a Chinese teaching hospital. Int J Clin Exp Med 2015;8:19400-5.
- Al Naiemi N, Duim B, Bart A. A CTX-M extended-spectrum β-lactamase in Pseudomonas aeruginosa and Stenotrophomonas maltophilia. J Med Microbiol 2006;55:1607-8.
- Celenza G, Pellegrini C, Caccamo M, Segatore B, Amicosante G, Perilli M. Spread of blaCTX-M-type and blaPER-2 β-lactamase genes in clinical isolates from Bolivian hospitals. J Antimicrob Chemother 2006;57:975-8.
- Polotto M, Casella T, de Lucca Oliveira MG, Rúbio FG, Nogueira ML, de Almeida MTG, et al. Detection of Pseudomonas aeruginosa harboring bla CTX-M-2, bla GES-1 and bla GES-5,bla IMP-1 and bla SPM-1 causing infections in Brazilian tertiary-care hospital. BMC Infect Dis 2012;12:176.
- Qing Y, Cao KY, Fang ZL, Huang YM, Zhang XF, Tian GB, et al. Outbreak of PER-1 and diversity of β-lactamases among ceftazidime-resistant Pseudomonas aeruginosa clinical isolates. J Med Microbiol 2014;63:386-92.
- Jafari-Sales A, Shadi-Dizaji A. Molecular analysis of CTX-M genes among ESBL producing in Pseudomonas aeruginosa isolated from clinical samples by Multiplex-PCR. Hozan J Environ Sci 2018;2:17-29.
- Rao P, McCaughan J, McCalmont M, Goldsmith CE, Hall V, Millar BC, et al. Comparison of antibiotic susceptibility patterns in Pseudomonas aeruginosa isolated from adult patients with Cystic Fibrosis (CF) with invasive Pseudomonas aeruginosa from non-CF patients. J Cyst Fibros 2012;11:349-52.