- *Corresponding Author:
- K. R. Iyer
Department of Pharmaceutical Chemistry, Bombay College of Pharmacy (Autonomous), Kalina, Mumbai 400 098, India
E-mail: krishna.iyer@bcp.edu.in
| Date of Received | 14 October 2020 |
| Date of Revision | 25 November 2021 |
| Date of Acceptance | 13 March 2023 |
| Indian J Pharm Sci 2023;85(2):318-324 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
An isoniazid N-acetylation phenotyping study was carried out in 200 human volunteers. A single urine sample taken 5 h post-isoniazid oral dose of 300 mg was analyzed and urinary concentrations of isoniazid and its metabolite acetylisoniazid were determined by high-performance liquid chromatography. Acetylator phenotype was determined from the metabolic ratio of N-acetylisoniazid to isoniazid concentration in the urine sample. The cut-off value, antimode, to distinguish between slow and fast acetylators was determined by probit analysis. Of the total 194 subjects phenotyped, 70.1 % were slow acetylators, 27.6 % were intermediate acetylators and 2.7 % were fast acetylators.
Keywords
Isoniazid, acetylisoniazid, phenotyping, N-Acetyltransferase 2, Indian population
The factors responsible for the individual pharmacokinetic and pharmacodynamic variability are manifold and include drug-drug interactions, ethnicity, age, renal and liver function, concomitant diseases, nutritional status, smoking and alcohol consumption. In addition, genetic factors can also have an influence on drug efficacy and toxicity. Genetic variation in the human genome is an accepted phenomenon and can be divided into germ-line variations in the Deoxyribonucleic Acid (DNA) and somatic gene mutations. Germ- line polymorphisms are mostly Single Nucleotide Polymorphisms (SNPs) at certain points along the human genome, but can also involve additions, deletions, duplication and sequence rearrangements. Somatic mutations, as in tumors for example, explain the variety of epidemiological patterns observed in cancer. Polymorphisms in drug metabolizing enzymes can result in differences in the pharmacokinetic and pharmacodynamic profiles of therapeutic agents. Polymorphisms in drug transporters, such as P-glycoprotein (P-gp), can influence the uptake and excretion of a drug, thereby also causing inter-individual variability in the pharmacokinetics of that drug[1]. The number of polymorphisms identified in genes encoding drug-metabolizing enzymes and drug transporters is rapidly increasing and will hopefully lead to a better explanation for the observed variation in efficacy and toxicity of drugs in patients[1].
Aryl amine N-Acetyl transferases (NAT’s) are drug metabolizing enzymes capable of N-acetylation and detoxification of aryl amine and hydrazine drugs[2]. Two functional isoforms of NAT are expressed in humans, NAT1 and NAT2. Tissue expression of NAT2 is confined mainly to liver and intestine, while NAT1 is found in many tissues[2]. NAT2 substrates include procainamide, isoniazid, sulphonamides, dapsone and some carcinogenic aryl amines. Likewise, NAT1 substrates include p-amino benzoic acid, p-aminosalicylic acid, sulfamethoxazole, sulfanilamide and p-amino- benzoyl-L-glutamate (pABGlu). Several alleles of NAT2 have been reported[3]. The rate of acetylation of Isoniazid (INH) to Acetylisoniazid (AcINH) is genetically determined by acetylator phenotype as being slow or fast. Thus, individuals with deficient metabolism of drugs are referred to as slow metabolizers (or poor metabolizers) as compared with normal or fast metabolizers (extensive or rapid metabolizers). Genetically, extensive INH acetylators are either homozygous (RR) or heterozygous (Rr) for a dominant wild-type rapid acetylator allele (R), while slow acetylators are homozygous (rr) for a recessive slow acetylator allele[3,4]. The objective of the present study was to conduct an acetylation phenotyping study on a sample Indian population (200 volunteers), using INH as a probe drug.
Materials and Methods
INH (100 g) was a gift sample by Amsal Chem. Pvt. Ltd., Ankleswar, Gujarat. AcINH was synthesized and characterized by melting point, IR and UV spectra[5,6]. Glacial acetic acid, acetic anhydride, benzene, potassium dihydrogen phosphate, sodium dodecyl sulphate, sodium hydroxide and conc. hydrochloric acid of analytical grade were obtained from S.d. Fine Chem. Ltd. Methanol and acetonitrile of High-Performance Liquid Chromatography (HPLC) grade was obtained from Qualigens Fine Chemicals. Solonex® tablets from Macleods Pharmaceuticals Ltd. were used for the study.
Study protocol:
The study protocol was approved by the “Institution Ethics Committee of Research Society of Bombay College of Pharmacy”. The trial group comprised 200 adults (Male: n=125, Female: n=75). The age of volunteers ranged from 18-39 y. A written informed consent was obtained from all study participants. After overnight fasting, a single oral dose of INH (300 mg) as a Solonex® tablet (Macleod’s Pharmaceuticals Ltd.) was administered. The subjects were prohibited from taking any food and drink overnight and during the study, except for water. Urine samples were collected at 5 h post dose and a 50 ml aliquot was retained for analysis. Urine samples were stored at -70° until analysis.
Standard curve for determination of INH and AcINH in human urine:
Blank urine was collected following the same procedure as that of the study protocol, without intake of the tablet and was stored at -70° until analysis. Standard curve was prepared in duplicate. Blank urine was allowed to thaw to room temperature. An aliquot (400 µl) of blank urine was added to a 1 ml capacity micro centrifuge tube to which 50 µl of a suitable stock INH solution (in distilled water) and 50 µl of a suitable stock AcINH solution (in distilled water) was spiked. To the resulting solution, 500 µl acetonitrile was added and the tubes were centrifuged at 16000 x g for 30 min at 4° in a refrigerated centrifuge (Biofuge Stratos). An aliquot (500 µl) of the supernatant was diluted with 2 ml of 0.01 M KH2PO4 buffer; pH 2.5 and 20 µl of this solution was subjected to HPLC analysis. The complete protocol for preparation of standard curve in blank urine is given in Table 1.
| Final Concentration (uM) | 50 μl sub stock solution (mM) | Volume of blank urine (μl) | Volume of ACN (μl) | After centrifugation | ||||
|---|---|---|---|---|---|---|---|---|
| INH | AcINH | INH | AcINH | Volume of supernatant taken (ul) | Volume of buffer (ml) | Total volume (ml) | ||
| Blank | Blank | - | - | 500 | 500 | 500 | 2 | 2.5 |
| 0.7 | 0.451 | 7 | 4.51 | 400 | 500 | 500 | 2 | 2.5 |
| 1.4 | 0.902 | 14 | 9.02 | 400 | 500 | 500 | 2 | 2.5 |
| 4.2 | 2.71 | 42 | 27.06 | 400 | 500 | 500 | 2 | 2.5 |
| 7 | 4.51 | 70 | 45.1 | 400 | 500 | 500 | 2 | 2.5 |
| 9.8 | 6.31 | 98 | 63.14 | 400 | 500 | 500 | 2 | 2.5 |
| 14 | 9.02 | 140 | 90.2 | 400 | 500 | 500 | 2 | 2.5 |
Table 1: Protocol for Preparation of Standard Curve in Blank Urine
Determination of INH and AcINH in sample human urine:
Sample urine was collected by following the procedure dictated by the study protocol and then stored at -70° until analysis. A 500 µl aliquot of sample was taken and to this 500 µl aliquot of acetonitrile was added. The mixture was then processed as indicated for standard curve samples. Urine samples of 15-20 volunteers were analyzed in batches and a standard curve was included with each run. All samples of the 194 volunteers were analyzed in duplicate and the mean values of the estimates have been reported.
HPLC conditions:
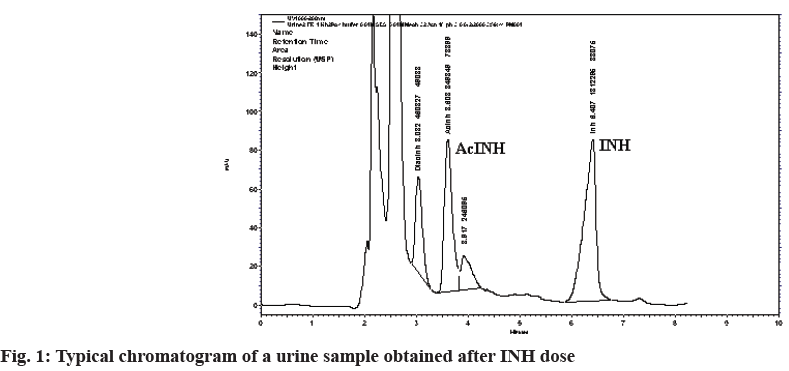
HPLC was performed on a Thermo Finnigan system, which consisted of a System controller (Spectra System P2000), a variable wavelength detector (Spectra System UV1000) and data processor (Chromquest 4.1 software). Stationary phase was a reverse phase BDS Hypersil C18 column [4.6 mm i.d×250 mm, particle size 5 µm, Thermo Electron Corporation]. The mobile phase composition was 50:32:18 (v/v) 0.01 M KH2PO4 buffer (pH 2.5), methanol and acetonitrile, respectively, to which sodium dodecyl sulphate was added to yield a final concentration of 0.01 M. Analysis was performed at ambient temperature at a detector wavelength of 266 nm and flow rate of 1 ml/min. AcINH and INH eluted after approximately 3.6 min and 6.3 min, respectively.
Pilot study:
A pilot study was carried out on three volunteers, to determine the stability of urine samples at -70°. Urine samples were found to be stable for a period of 4 w, post collection when stored at -70°.
Determination of acetylator phenotype status:
The acetylator phenotype status of individuals was determined from their Metabolic Ratio (MR). MR was defined as the molar AcINH concentration to molar INH concentration ratio. The value of antimode, cut off point, to distinguish between slow and fast acetylators was derived by multiple regression analysis using the Probit plot i.e. probit analysis[7]. Using the Hardy-Weinberg law the frequencies of allele controlling fast acetylation (p), frequencies of allele controlling slow acetylation (q), frequencies of homozygous slow acetylators (q2), heterozygous intermediate acetylators (2pq) and homozygous fast acetylators (p2) were determined[8].
Results and Discussion
INH is a pivotal agent in the treatment of tuberculosis in combination with other drugs or alone as a prophylactic agent. INH is also used as a test drug for determination of the acetylator phenotype. The genetically polymorphic NAT2 is responsible for INH metabolism. The hereditary variability of INH metabolism may have clinical relevance in both drug efficacy and toxicity. Following oral administration, INH is rapidly and completely absorbed. It is metabolized mainly in the liver through acetylation to form AcINH The HPLC method was developed and evaluated for interday precision, linearity, Limits Of Detection (LOD) and Lower Limit Of Quantitation (LLOQ). Briefly, the interday precision values (% RSD) for INH at concentrations 14.0, 9.8, 7.0, 4.2, 1.4 and 0.7 mM were 6.1 %, 9.8 %, 10.1 %, 10.6 %, 11.3 % and 19.9 % respectively. The interday precision values (% RSD) for AcINH at concentrations 9.02, 6.31, 4.51, 2.71, 0.902 and 0.451 mM were 8.12 %, 6.65 %, 5.86 %, 5.20 %, 11.24 % and 19.67 % respectively. The calibration curves of INH and AcINH in urine were linear in the range of 0.7–14.0 mM and 0.451–9.02 mM, respectively, with a correlation coefficient (r2) of 0.9967±0.002 for INH and 0.9972±0.0023 for AcINH (equations for a typical standard curve run). The LOD for INH and AcINH was found to be 0.35 mM and 0.23 mM respectively. The LLOQ for INH and AcINH was 0.70 mM and 0.451 mM respectively. The results of 194 subjects out of a total of 200 subjects whose urine sample was collected are reported, since 6 samples could not be analyzed due to interfering peaks in the HPLC chromatograms. A representative chromatogram of the urine sample obtained from a human volunteer is shown in fig. 1 indicating the resolution of INH and AcINH from most of the urine components.
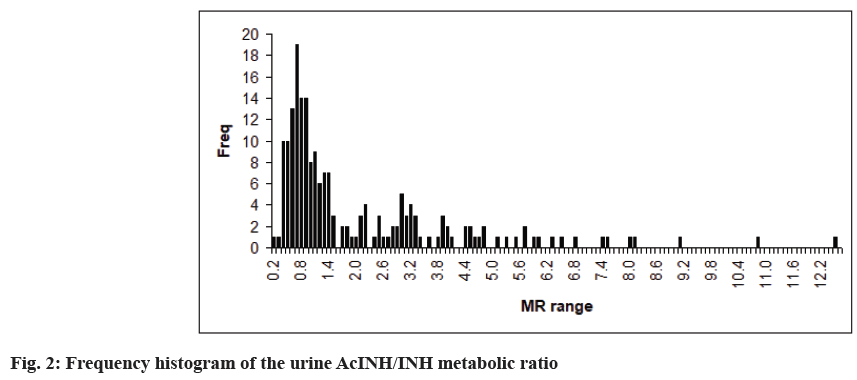
A frequency histogram was obtained by plotting MR range vs. frequency (number of individuals) of that range and is shown in fig. 2. A test for deviation from Gaussian distribution was determined by using Kolmogorov Smirnov (KS) test. Since the Gaussian distribution is also called “normal distribution”, the test is also referred to as the normality test. The KS statistic quantifies the discrepancy between the distribution of the data and an ideal Gaussian distribution. The KS test for normality was done using Graph Pad Prism 4.0. Interpretation of the results of normality depends on the p-value calculated by the test and on the sample size. The requirement for passing the normality test is a large p-value irrespective of the sample size. In contrast, a small p-value (less than 0.05) is indicative of non- Gaussian distribution. Data obtained from this study was subjected to the KS test and yielded a p value lesser than 0.0001, indicating that the data is not normally distributed.
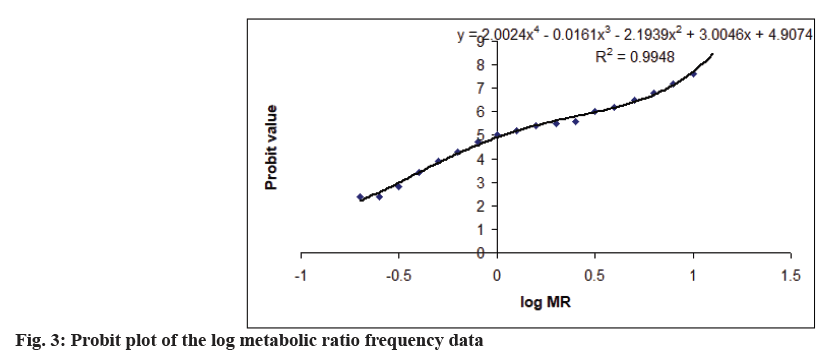
The cut-off value, the antimode, to distinguish between slow and fast acetylators was then determined by multiple regression analysis using the Probit plot i.e. Probit analysis. The probit plot obtained is shown in fig. 3. The antimode value obtained was obtained by solving the quadratic equation obtained from the probit plot and was about 2.4. Thus, individuals with MR<2.4 were categorized as slow acetylators. Of the 194 subjects, 136 subjects (70.1 %) were found to be slow acetylators.
Application of the Hardy-Weinberg law indicated that the frequencies of the allele controlling the fast acetylators (p), which is the wild type allele, was 0.163 and for the allele controlling the slow acetylators (q), which is the mutant allele, was 0.837. The frequencies of homozygous fast acetylators (p2), heterozygous intermediate acetylators (2pq) and homozygous slow acetylators (q2) are thus 0.027, 0.276 and 0.701 respectively, i.e. analysis of the sample population indicated that 2.7 % are fast acetylators, 27.3 % are intermediate acetylators and 70.1 % are slow acetylators. The sample population was further divided into subgroups categorized as natives of Maharashtra, natives of Gujarat, natives of North India, natives of South India, males and females and the distribution of slow and fast acetylators for each subgroup was determined. The results obtained are summarized in Table 2 and Table 3.
| p | q | p2 | 2pq | q2 | |
|---|---|---|---|---|---|
| Entire population | 0.163 | 0.837 | 0.027 | 0.276 | 0.701 |
| Maharashtrians | 0.171 | 0.829 | 0.029 | 0.284 | 0.687 |
| Gujaratis | 0.204 | 0.796 | 0.042 | 0.325 | 0.633 |
| South Indians | 0.139 | 0.861 | 0.019 | 0.24 | 0.741 |
| North Indians | 0.2 | 0.8 | 0.04 | 0.32 | 0.64 |
| Males | 0.131 | 0.869 | 0.017 | 0.228 | 0.756 |
| Females | 0.208 | 0.792 | 0.043 | 0.33 | 0.627 |
Table 2: Frequencies of Allele Controlling Acetylation
| % Slow | % Intermediate | % Fast | |
|---|---|---|---|
| Sample population | 70.1 | 27.6 | 2.7 |
| Maharashtrians | 68.7 | 28.4 | 2.9 |
| Gujaratis | 63.3 | 32.5 | 4.2 |
| South Indians | 74.1 | 24 | 1.9 |
| North Indians | 64 | 32 | 4 |
| Males | 75.6 | 22.8 | 1.7 |
| Females | 62.7 | 33 | 4.3 |
Table 3: Percentages of Slow, Intermediate and Fast Acetylators in the Sample Population and Each Subgroup
One of the well described and genetically determined polymorphic drug metabolisms is the NAT2 acetylation polymorphism. At least seven different point mutation sites have been described for NAT2. Rapid acetylators are homozygous or heterozygous for the dominant NAT2*4 wild type allele, whereas slow acetylators carry different combinations of the multiple recessive alleles. There are marked differences in genotypes and allele frequencies of the NAT2 gene in different populations. Certain NAT2 traits may contribute to the occurrence of adverse drug effects and, moreover, are susceptibility factors for certain malignancies such as bladder or lung cancer. In addition, NAT2 acetylation polymorphism has been linked to susceptibility to certain autoimmune diseases such as Systemic Lupus Erythematosus (SLE) and scleroderma[9]. The extent of N-acetylation of a number of drugs like isoniazid, procainamide, dapsone, sulfamethazine, sulfapyridine, phenelzine, thyroxamine, nitrazepam, caffeine, clonazepam, dapsone, and other drugs has been shown to be under the control of a polymorphic trait that determines the acetylator phenotype[10]. The determination of NAT2 genotype or NAT2 phenotype has been proposed as a way to predict adverse reactions in patients with tuberculosis.
During the past four decades, a considerable amount of information has accumulated on the distribution of acetylator phenotypes among populations of different ethnic and geographic origin. The proportions of rapid and slow acetylators vary considerably between different populations in relation in ethnicity and geographic location. The therapeutic and epidemiologic implications of INH acetylation phenotype have become more apparent. The acetylation polymorphism has important consequences for both drug therapy and xenobiotic toxicity. It has been shown that in patients treated with INH and rifampicin, the incidence of jaundice and possibly raised serum alanine aminotransferase, are higher in subjects with the slow INH acetylator phenotype. Earlier studies have suggested more frequent occurrence of phenytoin toxicity in patients concurrently being treated with INH. Furthermore, associations between acetylator phenotype and diseases such as cancer of the bladder, Gilbert’s disease, leprosy and early development of thyrotoxicosis have also been reported. Slow acetylators are also reportedly at higher risk of INH adverse effects such as peripheral neuritis and hepatic toxicity. Phenotyping the acetylator status of patients for appropriate individualization of dosage regimen appears worthwhile[11].
Several methods that use different test molecules such as caffeine, sulfamethazine, sulfadimidine, dapsone, procainamide, clonazepam, INH etc. are available for assessment of the acetylation capacity in an individual. The advantages of INH for determining acetylator phenotype are that no serious side effects are likely during the phenotyping test. Thus, INH has been used extensively and in this study as a test probe.
It is evident from the results that over 60 % of the entire population, each subgroup (Maharashtrians, Gujaratis, North Indians and South Indians), males and females exhibit a slow acetylator phenotype. Our results are comparable to the results obtained by studies carried out earlier in an Indian subpopulation. However, our study does show a slightly high percentage of slow acetylators as compared to the previous study carried out in a South Indian population (Table 3). Table 4 indicates our results in the light of other reported NAT phenotyping studies carried out in different populations. Our studies also indicate that overall, the Indian population has a distribution of NAT phenotypes similar to that observed in Caucasian and European populations[12-18].
| Population | Probe drug | % Slow |
|---|---|---|
| Saudi Arabs[3] | INH | 94.9 |
| Swedish[11] | INH | 70 |
| Iranian[2] | INH | 59.6 |
| Afghani[2] | INH | 59.2 |
| Bangladeshi[12] | Sulfadimidine | 20.5 |
| South Indian[13] | Sulfamethazine | 51 |
| South Indian[13] | Dapsone | 59 |
| Japanese[13] | Dapsone | 6.6 |
| Finnish Lapland[14] | ||
| Saame Lapps | INH | 27 |
| Nellim Skolts | INH | 40 |
| Japanese[15] | Procainamide | 7 |
| Amerindians[16] | ||
| Cunas | INH | 24 |
| Teribes | INH | 29 |
| Chinese[17] | Dapsone | 13 |
| Eskimos[18] | INH | 5 |
| Present Study | ||
| Total Population | INH | 70.1 |
| Maharashtrians | INH | 68.7 |
| Gujaratis | INH | 63.3 |
| South Indians | INH | 74.1 |
| North Indians | INH | 64 |
| Males | INH | 75.6 |
| Females | INH | 62.7 |
Table 4: Comparison of Present and Reported NAT Phenotyping Data
Acknowledgements:
The authors wish to thank AICTE (File No. 8019/ RDII/R&D/PHA (213) 2000-01) and “DST-FIST Program”-[SR/FST/LS1-163/2003] for providing the research grants for this work. The authors wish to acknowledge Dr. Nithya J. Gogtay and Dr. Sanish Davis of the KEM Hospital, Mumbai for help with the probit analysis of the data, and Prashant Tannu and Deepali Desle for help in the manuscript preparation.
Conflict of interests:
Authors declared there is no conflict of interest.
References
- Bosch TM, Meijerman I, Beijnen JH, Schellens JH. Genetic polymorphisms of drug-metabolising enzymes and drug transporters in the chemotherapeutic treatment of cancer. Clin Pharmacokinet 2006;45:253-85.
[Crossref] [Google Scholar] [PubMed]
- Hassanzadeh K, Khashayarmanesh Z, Panahi M, Esmaielie DM, Mokrie LS. N-acetylation Phenotyping using Isoniazid in an Iranian and an Afghani Population. Indian J Pharm Sci 1999;61(5):297.
- Matar KM, Mayet AY, Ayoola EA, Bawazir SA, Al-Faleh FZ, Al-Wazzan A. Isoniazid acetylation phenotyping in Saudi Arabs. J Clin Pharm Ther 2004;29(5):443-7.
[Crossref] [Google Scholar] [PubMed]
- Olson W, Miceli J, Weber W. Dose-dependent changes in sulfamethazine kinetics in rapid and slow isoniazid acetylators. Clin Pharmacol Ther 1978;23(2):204-11.
[Crossref] [Google Scholar] [PubMed]
- Fox HH, Gibas JT. Synthetic tuberculostats. VIII. Acyl derivatives of isonicotinyl hydrazine. J Org Chem 1953;18(10):1375-9.
- Yale HL, Losee K, Martins J, Holsing M, Perry FM, Bernstein J. Chemotherapy of Experimental Tuberculosis. VIII. The Synthesis of Acid Hydrazides, their Derivatives and Related Compounds1,2. J Am Chem Soc 1953;75(8):1933-42.
- Panchabhai TS, Noronha SF, Davis S, Shinde VM, Kshirsagar NA, Gogtay NJ. Evaluation of the activity of CYP2C19 in Gujrati and Marwadi subjects living in Mumbai (Bombay). BMC Clin Pharmacol 2006;6(1):1-5.
[Crossref] [Google Scholar] [PubMed]
- Haining RL, Yu. A. Cytochrome P450 pharmacogenetics. In: Lee JS, Obach RS, Fisher MB, editors. Drug metabolizing enzymes-Cytochrome P450 and other enzymes in drug diseases and development. Netherlands and New York: FontisMedia S.A. and Marcel Dekker Inc; 2003. p. 375-420.
- Aynacioglu SA, Bozkurt A, Nacak M, Kortunay S, Tunc R, Dincel A, et al. N-acetyltransferase polymorphism in patients with Behçet's disease. Eur J Clin Pharmacol 2001;57:659-62.
[Crossref] [Google Scholar] [PubMed]
- Miller ME, Garland WA, Min BH, Ludwick BT, Ballard RH, Levy RH. Clonazepam acetylation in fast and slow acetylators. Clin Pharmacol Ther 1981;30(3):343-7.
[Crossref] [Google Scholar] [PubMed]
- Smith CA, Wadelius M, Gough AC, Harrison DJ, Wolf CR, Rane A. A simplified assay for the arylamine N-acetyltransferase 2 polymorphism validated by phenotyping with isoniazid. J Med Genet 1997;34(9):758-60.
[Crossref] [Google Scholar] [PubMed]
- Zaid RB, Nargis M, Neelotpol S, Hannan JM, Islam S, Akhter R, et al. Acetylation phenotype status in a Bangladeshi population and its comparison with that of other Asian population data. Biopharm Drug Dispos 2004;25(6):237-41.
[Crossref] [Google Scholar] [PubMed]
- Peters JH, Gordon GR, Karat AB. Polymorphic acetylation of the antibacterials, sulfamethazine and dapsone, in South Indian subjects. Am J Trop Med Hyg 1975;24(4):641-8.
[Crossref] [Google Scholar] [PubMed]
- Ellard GA, Gammon PT, Tiitinen H. Determination of the acetylator phenotype from the ratio of the urinary excretion of acetylisoniazid to acid-labile isoniazid: A study in Finnish Lapland. Tubercle 1973;54(3):201-10.
[Crossref] [Google Scholar] [PubMed]
- Okumura K, Kita T, Chikazawa S, Komada F, Iwakawa S, Tanigawara Y. Genotyping of N-acetylation polymorphism and correlation with procainamide metabolism. Clin Pharmacol Ther 1997;61(5):509-17.
[Crossref] [Google Scholar] [PubMed]
- Inaba T, Arias TD. On phenotyping with isoniazid: The use of urinary acetylation ratio and the uniqueness of antimodes. Study of two Amerindian populations. Clin Pharmacol Ther 1987;42(5):493-7.
[Crossref] [Google Scholar] [PubMed]
- Horai Y, Zhou HH, Zhang LM, Ishizaki T. N-acetylation phenotyping with dapsone in a mainland Chinese population. Br J Clin Pharmacol 1988;25(1):81-7.
[Crossref] [Google Scholar] [PubMed]
- Armstrong AR, Peart HE. A comparison between the behavior of Eskimos and non-Eskimos to the administration of isoniazid. Am Rev Respir Dis 1960;81(4):588-94.
[Crossref] [Google Scholar] [PubMed]