- *Corresponding Author:
- Jiayu Chen
Department of Clinical Laboratory, Shaoxing University, Shaoxing, Zhejiang, China
E-mail: limingcheng1964@163.com
| This article was originally published in a special issue, “XXXXXX” |
| Indian J Pharm Sci 2020:82(1)spl issue1;XX-XX |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Streptococcus pneumoniae is one of the leading causes of invasive infections. Traditional culture and identification methods require a long time period and are liable to be complicated by normal flora. In this study, a new method was developed based on loop-mediated isothermal amplification. A series of primers for autolysin (LytA) from S. pneumoniae were designed with a bioinformatics technique, and a loop-mediated isothermal amplification system for the detection of S. pneumoniae was set up. Comparative analysis of specificity and sensitivity from the culture and polymerase chain reaction methods in 50 samples collected from patients with respiratory infections was performed. The data demonstrated that the selected primers and reaction system in the loop-mediated isothermal amplification were respondent to Streptococcus pneumoniae and did not cross-react with other bacteria. The detection of sensitivity to the loop-mediated isothermal amplification method was 102 CFU/ml. From all of the clinical specimens tested, 9 cases (18.0 %) were confirmed to be positive for Streptococcus pneumoniae using the loop-mediated isothermal amplification method, whereas only 4 cases (8.0 %) were diagnosed using the culture method. Loop-mediated isothermal amplification technology is a specific, simple and accurate method for the efficient differentiation of S. pneumoniae in clinical specimens.
Keywords
LAM, Streptococcus pneumoniae, bioinformatics
Streptococcus pneumoniae is a common clinical pathogen, which often causes pneumonia, meningitis, otitis media and sepsis[1]. The traditional detection method of isolating the pathogen from clinical specimens is the diagnostic golden standard[2]. However, classical bacterial culture and identification of S. pneumoniae requires rich nutrients and as it grows slowly, high possibility for contamination by alpha-haemolytic streptococci that beong to normal human flora[3].
Loop-mediated isothermal amplification (LAM) is a novel isothermal nucleic acid amplification method which is conducted under the condition of isothermal (60-65°) and a short time (usually 1 h). The process is used for nucleic acid amplification and is simple, rapid, accurate and highly cost effective[4]. In contrast to conventional polymerase chain reaction (PCR), the method does not require a template of thermal denaturation, temperature cycles, electrophoresis and ultraviolet observations[5]. The reaction can achieve 109-1010 times amplification in 15 to 60 min and produces a large number of amplification products which are called magnesium pyrophosphate. This forms white precipitates which can be observed by the naked eye indicating the presence of target genes, unlike ordinary PCR methods which require gel electrophoresis to observe amplification products[6]. LAM technology is already optimized and validated in the fields of food safety, bacterial and fungal pathogens, and viral diseases[7].
In this study, a target sequence of the S. pneumoniae’s autolysin A (LytA) gene was selected as a reporter gene and primers of 6 different areas were designed by LAM software. The LAM reaction system was established to amplify S. pneumoniae strains. Simultaneously, 2 primers were designed to identify the target genes of 6 distinct areas by PCR. Although the LAM method has been established for detection of pneumococcus in cerebrospinal fluid specimens, its efficacy in diagnosing bacterial pneumoniae has not yet been reported. The aim of the study was to establish a quick, simple and economical method for the detection of S. pneumoniae for clinical applications. To our knowledge, this is the first report of applying the LAM method for detecting S. pneumoniae in clinical sputum samples. Clinical specimens were consecutively obtained from sputum (n=50) in the affiliated hospital in Jilin between 2014 and 2015. Clinical protocols to collect the samples were approved by ethical committees of Beihua University (protocol number 2015-1-10) and written informed consent was obtained from all participants in the study. Reference strains were included as internal standards in all tests, S. pneumoniae ATCC 49619, S. pyogenes ATCC 19615, S. aureus ATCC 25923, E. coli ATCC 25922, P. aeruginosa ATCC 27853 and C. albican ATCC 90029. Genomic DNA was extracted from all of the standard strains in the study by boiling method. In brief, all strains were inoculated on Columbia blood agar plates (Becton Dickinson, USA), incubated at 5 % CO2 and 37° for 18-24 h and the pure colony was isolated into 50 μl L-B broth. The broth was boiled at 95° for 15 min, then cooled for 2 min, centrifuged at 15 000 rpm for 30 s, the supernatants were removed and later used as the detection template.
Clinical sputum specimens were processed by adding 0.25 % trypsin into each sputum specimen, which had a volume of 1-3 times that of the trypsin. Samples were then incubated with shaking at 37° for 1 h, centrifuged at 2000 rpm for 3 min and then 1 ml of supernatant was transferred into a centrifuge tube before being centrifuged at 15 000 rpm for 3 min. The precipitate was resuspended in 100-150 μl distilled water, incubated at 56° in a water bath for 1 h and then at 95° for 15 min before transferring to an ice bath for 2 min. The samples were then centrifuged at 15 000 rpm for 30 s and the supernatants were saved for LAM and conventional PCR analysis. A series of primers were designed for detection of S. pneumoniae lytA, with the Primer Explore V4 (Eiken Chemical Co., Japan) software. A comparison of the sequence to exclude strains was performed using the BLAST alignment tool (NCBI, Maryland, USA). All primers were synthesized by the Shengon Bioengineering Company (Table 1). The LAM reaction system included 1.6 M of primers FIP and BIP, 0.2 M primers F3 and B3, 8 μ Bst DNA polymerase, 20 mM tris-HCl, 10 mM KCl, 10 mM (NH4)2SO4, 6 mM MgSO4, 0.1 % triton X-100, 0.8 Triton X-100 M hydrochloric acid theophylline and 5 μl of template, which was made up to a final volume of 50 μl with double distilled water. The reaction conditions were 63° for 60 min and 63° for 2 min. Products were observed for turbidity by the naked eye and 2 % agarose electrophoresis containing ethyl bromide, respectively. To evaluate the specificity of the LAM primers, all reference strains included as internal standards were tested. To ascertain the detection limit of the LAM method, serial ten-fold dilutions of S. pneumoniae genomic DNA were amplified. Briefly, the strain of S. pneumoniae, ATCC 49619, was prepared in the pure culture colony. Bacterial broth was distilled in sterile water and counted. Based on the preliminary count, serial ten-fold dilution of colony was performed. The final colony forming units were 106, 105, 104, 103, 102 and 101 CFU/ml. All bacterial DNA templates were extracted using a DNA extraction kit (Qiagen, Valencia, CA) according to the manufacture’s protocol. LAM was performed as previously described. To evaluate the LAM assay’s efficacy in diagnosing bacterial pneumoniae, we compared the detection performance of the LAM method with those of conventional PCR and traditional culture method by testing fifty samples of clinical sputum. The conventional PCR reaction system included 0.2 mM dNTP, 0.2 U Taq polymerase, 0.4 μM forward and reverse primers, 1.5 mM Mg2+ and 5 μl template of DNA extracted directly from clinical sputum and made up to a final volume of 50 μl with double distilled water. The reaction conditions were set as 94° for 3 min, 94° for 30 sec, 57° for 30 sec, 72° for 1 min, for a total of 30 cycles and 72° for 10 min. The PCR product was separated on 2 % agarose gel by electrophoresis and observed by gel imaging system.
Table 1: IGO Nucleotide Primer Sequences Used For Lam and PCR
| Primers | Sequence (5' to 3' ) |
|---|---|
| LAM-FIP | CCGCCAGTGATAATCCGCTTCAGGCAAGTACACGCACACT' |
| LAM-BIP | TCTCGCACATTGTTGGGAACGGTCCCAGGCACCATTATCAAC |
| LAM-F3 | AGTCGGCGTGCAACCATA |
| LAM-B3 | GCATAGGTCTCAGCATTCCA |
| LAM-FLP | AGGATCCTGCGAGCGTAG |
| LAM-BLP | AAGAAGGCGTACTCGACCTG |
| PCR | F-GCTTGTTCCCGGTGCCACAC |
| R-GGAGCGCGATGCGTCAGGAC |
FIP: a forward inner primer; BIP: a reverse inner primer; F3 and B3: two outer primers; FLP and BLP: two loop primers
LAM was performed as described above. Clinical sputum specimens were streaked on Columbia blood agar culture medium and incubated in 5 % CO2 at 37° for 3 d and checked daily for growth. Bacterial isolation and identification relies on the standard microbiological criteria. All tests were performed in duplicate. Data are expressed as the mean±SD, calculated from at least three independent experiments. Statistical comparisons were performed using the two-tailed Student’s t-test. Statistical analysis was carried out using SPSS 16.0 (USA) software package with the level of statistical significance designated as p<0.05.
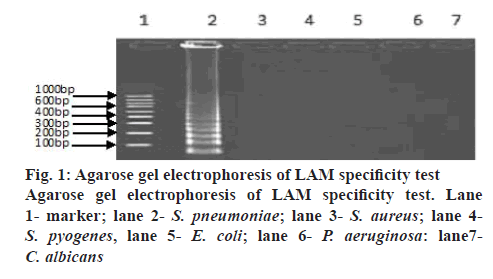
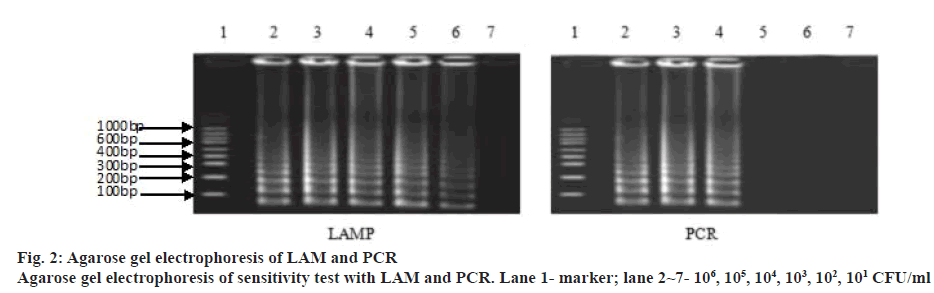
A LAM protocol was established and successfully performed to amplify the target sequence. A typical ladder-like pattern as the characteristic of the LAM was visible on agarose gel electrophoresis (figure 1). Additionally, amplification tubes in the test showed positive results as white opacity. To evaluate the species-specificity for the LAM primers, 10 strains of S. pneumoniae and 6 non-S. pneumoniae strains were tested using similar concentration of genomic DNA in each assay mixture. All S. pneumoniae strains yielded a positive LAM reaction after 30 min. In contrast, non- S. pneumoniae strains were negative after 60 min of incubation. The minimal bacteria concentration was 102 CFU/ml for S. pneumoniae detection by LAM method, whereas, the latter was 104 CFU/ml in S. pneumoniae detection by PCR method showing that the sensitivity of the LAM method was higher than the PCR method (figure 2). Fifty clinical samples of sputum were tested using LAM, PCR and a traditional culture method. Among 9 LAM-positive specimens, 8 cases (16.0 %) were confirmed to be positive as S. pneumoniae by the PCR and 1 case was PCR negative, whereas, only 4 (8.0 %) were S. pneumoniae culture positive (Table 2). In addition, 41 LAM-negative specimens were also negative by PCR and culture methods. All 4 S. pneumoniae culture positive specimens were positive by both LAM and PCR assays. There was a statistically significant difference between the LAM and PCR methods (p<0.01). The diagnosis of S. pneumoniae infection relies on a traditional culture method, which is regarded as the golden standard worldwide. However, the detection rate of S. pneumoniae with this method is low because it presents a positive result in children who harbour α-hemolytic streptococci or closely related pneumoniae strains. In fact, accurate diagnosis of S. pneumoniae presents a significant challenge because of the limited utility of routine microbiology laboratory tests and the injudicious use of antimicrobial therapy in China. As a consequence, people are trying to develop rapid and accurate methods to detect S. pneumoniae[7,8].
Table 2: Comparison of Three Methods for Testing 50 Cases of Clinical Specimens
| Detection method | Positive | Negative | Rate of positive |
|---|---|---|---|
| Traditional culture method | 4 | 46 | 8 |
| PCR method | 8*∆∆ | 42 | 16 |
| LAM method | 9∆ | 41 | 18 |
*One case positive is oral Streptococcus by PCR method. Compared LAM method ∆ with PCR method ∆∆ there was a difference (p<0.01)
Molecular methods possess inherently valuable merits due to their enhanced sensitivity and specificity in the clinic. The most functional gene in S. pneumoniae is lytA that encodes autolysin[9,10]. Moreover, the specificity level for the gene has been evaluated and reported at 100 % compared to other genes such as ply and psaA which are expressed at levels of 98 % and 81 % respectively. In another study using the real time PCR technique, the presence of the lytA gene was observed in all collected specimens from the clinic[4,11].
In the study, a LAM protocol was successfully performed to amplify the target sequence based on the lytA gene. In order to validate the LAM protocol, 50 sputum specimens were tested using the LAM, PCR and traditional culture methods. The results showed that the detection rate of the 50 clinical specimens was 8.0 % using the traditional method, 18 % using the LAM and 16 % using the PCR method. Five cases were detected as positive specimens by the LAM method but were detected as negative by the traditional culture method. It was assumed that patients were administered antibacterial drugs before collecting samples, which inhibited the growth of the bacteria and lead to negative culture results. The LAM method allows direct detection of S. pneumoniae DNA and is unaffected by antimicrobial treatment. From a clinical viewpoint, the high sensitivity and specificity of LAM will be valuable to diagnose or exclude the presence of S. pneumoniae infection, even in the clinical samples containing the small amount of bacteria, similar to the reports previously described[3,12,13].
A case also occurred in which a strain was detected as positive by the PCR method but negative using the LAM method. The strain was confirmed to be an oral streptococcus detected by traditional culture method. Due to the poor specificity of the PCR primer amplification of typical and atypical LytA genes, the PCR method cannot distinguish between S. pneumoniae and oral streptococcus, besides the typical LytA gene in S. pneumoniae, there are some of the other atypical LytA genets which lead to a false positive reaction.
In summary, a LAM method was successfully developed as a valuable diagnostic tool to identify S. pneumoniae from clinical specimens without using a culture method. The data demonstrated that the selected primers and reaction system in LAM were specific to S. pneumoniae. LAM technology is specific, simple and accurate in differentiating S. pneumoniae from clinical specimens without the need for large laboratory equipment
Financial interest and scholarship:
This study was a part of National Natural Science Foundation of China project (81402979). This research was financially supported by the Health Commission of Jilin Province of China (200112108), the Jilin and Zhejiang Science and Technology Development Program (20140307008YY and 2014C33155).
The study was partially presented in the International Conference on Advanced Nano-Technology and Biomedical Material (ANTBM2014). The authors declare that they have no competing interests. The authors alone are responsible for the content and writing of the paper.
References
- Rozenbaum MH, Pechlivanoglou P, Van Der Werf TS, Lo-Ten-Foe JR, Postma MJ, Hak E. The role of Streptococcus pneumoniae in community-acquired pneumonia among adults in Europe: a meta-analysis. Eur J Clin Microbiol Infect Dis 2013;32:305-16.
- Johansson N, Kalin M, Tiveljung-Lindell A, Giske CG, Hedlund J. Etiology of community-acquired pneumonia: increased microbiological yield with new diagnostic methods. Clin Infect Dis 2010;50:202-9
- Seki M, Yamashita Y, Torigoe H, Tsuda H, Sato S, Maeno M. Loop-mediated isothemal amplification method targeting LytA gene for detection of Streptococcus pneumoniae. J Clin Microbiol 2005;43:1581-6.
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. J Nucleic Acids Res 2000;28:e63.
- Llull D, Lopez R, Garcia E. Characteristic signatures of the lytA gene provide a basis for rapid and reliable diagnosis of Streptococcus pneumoniae infections. J Clin Microbiol 2006;44:1250-6.
- Kim DW, Kilgore PE, Kim EJ, Kim SA, Anh DD, Seki M. Loop-mediated isothermal amplification assay to detect Haemophilus influenzae type b in cerebrospinal fluid. J Clin Microbiol 2011;49:3621-6.
- Nandakumar R, Whiting J, Fouad AF. Identification of selected respiratory pathogens in endodontic infections. Oral Surg Oral Med 2008;106:145-51.
- Sherwin RL, Gray S, Alexander R, McGovern PC, Graepel J, Pride MW, et al. Distribution of 13-valent pneumococcal conjugate vaccine Streptococcus pneumoniae serotypes in US adults aged 50 years with community-acquired pneumonia. J Infect Dis 2013;208:1813-20.
- Sanz JC, Ríos E, Rodríguez-Avial I, Ramos B, Marín M, Cercenado E. Identification of Streptococcus pneumoniae lytA, plyA and psaA genes in pleural fluid by multiplex real-time PCR. Enferm Infecc Microbiol Clin 2018;36:428-30.
- Jourdain S, Drèze PA, Vandeven J, Verhaegen J, Van Melderen L, Smeesters PR. Sequential multiplex PCR assay for determining capsular serotypes of colonizing S. pneumoniae. BMC Infect Dis 2011;11:100.
- Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol 2006;44:124-31.
- Hajia M, Farzanehkhah M, Hajihashemi B, Dolatyar A, Imani M, Saburian R, et al. Real-time assay as tool for detection lytA gene in Streptococcus pneumoniae isolates. Cell J 2014;16:141-6.
- Kim DW, Kilgore PE, Kim EJ, Kim SA, Anh DD, Dong BQ, et al. The enhanced pneumococcal LAMP assay: a clinical tool for the diagnosis of meningitis due to Streptococcus pneumonia. PLoS One 2012;7:e42954.