- *Corresponding Author:
- G. Wang
Research & Development Department
Mab-Venture Biopharma, Shanghai 201319, China
E-mail: george_wang@mab-venture.com
| Date of Received | 20 February 2020 |
| Date of Revised | 22 February 2021 |
| Date of Acceptance | 25 March 2021 |
| Indian J Pharm Sci 2021;83(2):284-296 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Although the physical/chemical stability and potential interactions of trastuzumab and pertuzumab in a single infusion bag before co-administration have been evaluated preliminarily, it is not easy to clarify which monoclonal antibodies changes when the admixtures were analyzed as a whole, especially for in vitro potency evaluation. In this study, take admixtures of pertuzumab and trastuzumab biosimilar products as samples, an in vitro potency assay system was developed that can monitor the changes of potency of each monoclonal antibodies as well as their admixtures. Development of the assay system included 3 steps, the first step is to develop protein quantification assay for each monoclonal antibodies by specific antigen based enzyme-linked immunosorbent assay, then a specified cell based potency assay that could be used to measure the biological potency of each monoclonal antibodies and their admixtures was developed. After that, the enzyme-linked immunosorbent assay based protein quantification assay and the specified cell based potency assay were qualified for accuracy and precision. Meanwhile, admixtures containing different amount ratios of pertuzumab and trastuzumab biosimilar products were prepared and measured by the well qualified assays. Finally, the potency test data generated from admixtures of different amount ratio were summarized as an analysis table. The analysis table plus protein quantification results were then used as the basic tool for further in vitro potency evaluation of unknown admixtures. Using this system, the change of potency of each monoclonal antibodies in admixtures could be monitored. The in vitro potency assay system was then qualified in evaluating forced degraded samples. This study represents a good example on thorough biological potency evaluation for monoclonal antibodies admixtures.
Keywords
Antibody admixtures, potency, human epidermal growth factor receptor 2, pertuzumab, trastuzumab
Monoclonal antibody (mAb) derived biologics grow steadily and rapidly during the last 30 y, since the clinical approvals of rituximab and trastuzumab in the mid- 1990s [1]. MAbs play an important part in anti-cancer therapy including solid or haematological malignancies because of their high selectivity and specificity. However, mAbs alone are seldom used as the first-line choice for cancer therapy, their therapeutic utility and widespread use are limited by the development of patient resistance or the biomarker’s expression level [2]. Combinational use of two or more monoclonal antibodies in order to gain synergistic effect is a promising strategy to overcome the hurdle [3]. One typical example is the synergistic use of pertuzumab, trastuzumab, small molecule docetaxel [4] or vinorelbine [5] that have become an alternative first-line treatment regimen for breast cancer. This synergistic use of trastuzumab, pertuzumab and chemotherapy was also compared with that of trastuzumab emtansine plus pertuzumab in a phase 3 KRISTINE trail, resulted in significantly more patients achieving a pathological complete response [6].
Pertuzumab and trastuzumab are recombinant humanized monoclonal antibodies targeting the extracellular subdomain II and IV of human epidermal growth factor receptor 2 (HER2) respectively [7,8]. Both mAbs alone are active against HER2-overexpressing metastatic breast cancer through different mechanisms. The in vitro study finds that combination of pertuzumab and trastuzumab are more potent than the single one in inducing receptor degradation [9] and Antibody- dependent cellular cytotoxicity (ADCC) [10]. Their synergistic effect was also confirmed in HER2-positive breast xenografts that reveals strongly enhanced inhibition of growth [11]. The later clinical trial further established the therapeutic benefits of pertuzumab and trastuzumab admixture. In clinical trial CLEOPATRA and VELVET Cohort 1, pertuzumab and trastuzumab were administered separately followed by docetaxel or vinorelbine respectively, the results showed no unexpected toxicities with much better efficacy than trastuzumab plus docetaxel plus placebo for the treatment of first-line patients with HER2 positive metastatic breast cancer [4]. In clinical trial VELVET Cohort 2, pertuzumab and trastuzumab were co- administered in a single bag, followed by vinorelbine. This study also ended up with similar results compared to VELVET Cohort 1, which support the safety and efficacy of pertuzumab and trastuzumab co-infusion in application [5].
Co-infusion of pertuzumab and trastuzumab could save medical care related time and resources, but also might cause incompatibility [12]. So the stability and compatibility of these 2 mAbs together in one formulation are needed to be investigated, because agitation [13], high temperature [14] or light exposure [15] might make unwanted modifications, like oxidation, deamidation, aggregation and so on. In fact, to support co-administration of pertuzumab and trastuzumab in VELVET study, a comprehensive set of biophysical and analytical methods have been used to evaluate the stability and compatibility of these 2 mAbs in a single bag for the duration of about 24 h at 5º or 30º, just indicating no changes of the admixture’s physiochemical and biological properties [12]. However, duration of 24 h at 5º or 30º can cover most but not all the situation for co-infusion of the admixture. Most data shown in this study were qualitative not quantitative, especially for the potency analysis. If the admixture was regarded as a new drug, more stressed stability studies should be conducted according to ICH guideline, like the effect of light exposure, duration at stressed temperature for a longer time, so on. Although the VELVET Cohort 2 study successfully confirm the synergistic effect of co-infusion of pertuzumab and trastuzumab, thorough research of the stability of admixture is needed to be carried out to expand the application of this kind of regimen in any extreme conditions.
An in vitro potency assay system that is capable of evaluating the potency of each component of admixtures was developed, using pertuzumab (named as P) and trastuzumab (named as T) biosimilar products derived admixtures. Firstly, specific enzyme-linked immunosorbent assay (ELISA) based assays were developed and qualified for quantification of P or T, respectively. After that, anti-proliferation of HER2- overexpressed breast cancer cell line MDA-MB-175 VII based potency assay was developed, and qualified to ascertain that this assay could accurately measure the potency of P alone, T alone or their admixtures as well. After the above 2 assays were well established and qualified, the potency of admixtures containing different amount ratios of P and T were measured subsequently. The Half maximal effective concentration (EC50) value of 4-parameter dose-response curves of admixtures of different amount ratios were calculated and analyzed to generate an analysis table. This table could be used to identify the real amount ratio of component P and T in unknown admixtures. In stability study, if the real amount ratio of component P and T changes from the beginning of the study, the assay system could indicate if the loss of potency derives from component P or T.
Trying to qualify our in vitro potency assay system, the forced degraded P and T were prepared by putting normal P and T under high temperature. The forced degraded P and T were then mixed with the normal P and T to prepare unknown degraded admixtures. Using our well established and qualified ELISA based protein quantification assay, cell based biological potency assay and the analysis chart, the property of potency of each component P or T of the unknown admixtures were identified as prepared. This assay system represents a way for potency evaluation of monoclonal antibody admixtures to support their quality control and stability study.
Materials and Methods
Materials
The recombinant mAbs of pertuzumab and trastuzumab biosimilar product (P & T) were produced using Chinese hamster ovary cells followed by purification at Shanghai Pharmaceuticals (Shanghai, China). Although the manufacturing processes are different, their physiochemical and biological properties were comparable with that of the innovator product respectively. The biosimilar and innovator products also share the same formulation constitutes. Protein concentration of P or T is 5mg/ml (nominal) that is measured by UV280nm adsorption method.
Equal amount of P and T were mixed as antibody admixture M (P+T), or M100, where component P and T were mixed with amount ratio of 1:1 and the biological potency is 100 % when treated as a whole. M100 was diluted by mixed formulation buffer to generate M50, M75, M125 and M150, where the amount ratio of component P and T is 1:1, and the theoretical biological potency is 50 %, 75 %, 125 % and 150 % respectively, compared to M100. In the same way, component P or T, also designated as P100 and T100, was diluted by formulation buffer to generate P50, P75, P125, P150 and T50, T75, T125, T150 respectively. Their theoretical biological potencies are 50 %, 75 %, 125 % and 150 % compared to that of P100 or T100. Different ratios of P and T were mixed to generated antibody admixtures M (3P+T), M (2P+T), M (P+2T), M (P+3T), where the ratios of the concentration of P and T are 3:1, 2:1, 1:2, 1:3, respectively. M (3P+T) contains P at 15mg/ ml and T at 5mg/ml. M (2P+T) contains P at 10mg/ml, and T at 5mg/ml. M (P+2T) contains P at 5mg/ml and T at 10mg/ml. M (P+3T) contains P at 5mg/ml and T at 15mg/ml. All the prepared samples are used to qualify the developed potency assay.
For ELISA based protein quantification of P or T, the P specific antigen (extracellular subdomain II of HER2) and T specific antigen (extracellular subdomain IV of HER2) were purchased from QB-Biotech (Shanghai, China). The second antibody Peroxidase-AffiniPure Donkey Anti-Human IgG was purchased from Jackson- Immuno Research (PA, USA).
Cell line and culture medium
The breast cancer cell line MDA-MB-175 VII purchased from ATCC was maintained in vented cap, cell culture flasks (Corning, New York, NY). The growth medium for cell culture is complete DMEM/ F12 medium containing DMEM/F12 (HyClone, Logan, Utah) supplemented with 10 % Fetal bovine serum (FBS) (Gibco, Carlsbad, California). Trypsin (Gibco, Carlsbad, California) was used as the dissociation reagent when passaging these adherent cells. MDAMB- 175 VII cells were sub-cultured once a week with feeding cell density of 5.0×106 cells/ml for 5 d or 4.7×106 cells/ml for 6 d in T-75 flasks. Healthy growing MDA-MB-175 VII cells usually have >95 % viability and doubling time of about 107h. Cells were cryopreserved in two tiered cell banking system, including Master cell bank (MCB) at passage 1-5, and Working cell bank (WCB) at passage >5.
ELISA based protein quantification assay:
For P protein quantification, the P specific antigen (extracellular subdomain II of HER2) was 1:2000 diluted by Citrate-buffered saline (pH 9.5~9.7) and then mixed and immobilized on 96-well ELISA plate (Corning, 9018). The plate was incubated at 4º, overnight (16~18 h). The next day, the plate was washed 5 times with washing buffer (0.01 % Tween 20 in PBS buffer, pH 7.2~7.4) on a plate washer (BioTec). The plate was then blocked with 5 % Bovine serum albumin (BSA) in Phosphate-buffered saline (PBS) at 25º for 2 h. During the blocking periods, P was diluted with sample dilution buffer (0.05 % Tween 20 and 0.5 % BSA in PBS buffer, pH 7.2~7.4) to prepare standard curve samples. Standard curve samples contain 11 points, ranging from 0.2 ng/ml to 25ng/ml. These points distribute equally in logarithmic coordinates. P was also diluted with sample dilution buffer to prepare quality control (QC) samples including higher quality control (HQC) of 15ng/ml, middle quality control (MQC) of 6ng/ml and lower quality control (LQC) of 1.2ng/ml. After blocking, the plate was washed again 5 times with washing buffer followed by the addition of 100μl/well standard curve samples, QC samples, unknown samples and blank. The plate was then incubated at 25º for 2h. After another washing step, the plate was incubated with 100μl/well; 1:30000 diluted Peroxidase-AffiniPure Donkey Anti- Human IgG in sample dilution buffer for an additional 1 h at 25º. Finally the plate was washed again followed by incubation with 100μl/well TMB (Surmodics IVD, MN, USA) for 15 min at 25º in the dark. 2 M H2SO4 was used to stop the enzymatic reaction. Absorbance was measured with a multi-mode microplate reader (M5, Molecular Devices) in a dual wavelength analysis where 450nm was chosen as measurement wavelength and 650 nm as reference wavelength. Absorbance values were calculated by subtracting the measured value at 450 nm with that at 650 nm. The data were then reduced using a 4-P logistic (auto-estimate) regression model in SoftMax Pro. The assay range is defined by the nominal concentration range between 0.4 ng/ml to 20 ng/ml in formulation buffer. The standard curve samples and QC samples must meet the assay acceptance criteria, like the CV % of duplicates should be within 20 % and the recovery for each sample should be with 80 %~120 %. The concentration of P in unknown samples were then calculated using the standard curve.
For T protein quantification, the whole assay procedure is almost the same with that of P protein quantification except that the coating reagent is the T specific antigen (extracellular subdomain IV of HER2). The standard curve samples also constitute 11 points, ranging from 0.2 ng/ml to 15 ng/ml, while QC samples are HQC of 7.5 ng/ml, MQC of 4 ng/ml and LQC of 1.2 ng/ml. The assay range is defined by the nominal concentration range between 0.4 ng/ml to 10 ng/ml in formulation buffer. Just like the P protein quantification assay, only when the standard curve and QC samples meet the acceptance criteria, the T protein quantification results for unknown samples are valid.
Cell based biological potency assay
MDA-MB-175 VII anti-proliferation assay was developed for P, T or their admixture M (P+T). M (P+T) was mixed by equal amount of P and T. The assay procedures for these samples were almost the same. Take the assay procedure for M (P+T) for example, briefly, MDA-MB-175 VII cells in exponential phase were washed with PBS and then harvested through trypsinization and suspended in growth medium containing 10 % FBS (10 % FBS in DMEM/F12). After cell counting by Cedex cell counter (Roche, Basel, Switzerland) to make sure >95% cell viability, the cell density was adjusted to 2×105 cells/ml in assay medium (2 % FBS in DMEM/F12). Aliquots (50 μl/ well) of the prepared cell suspension were then added to each experimental well in a 96-well microtiter plate (Corning, New York, NY). The plate was then incubated for 18-20 h at 37º, 5 % CO2, and 95 % RH. After that, M (P+T) was diluted at 7 various concentrations from 0.002 μg/ml to 60 μg/ml in sample dilution buffer (0.1 % FBS in DMEM/F12). Subsequently, 50 μl M (P+T) diluents of each concentration was added into the plate and mixed with cell suspension. Each experimental well of the plate contained a total of 100 μl volume. The M (P+T) reference standard, test samples were diluted separately, and added in duplicates. The Blank (BL) well contained 50 μl assay medium and 50 μl sample dilution buffer, also in duplicates. The negative control (NC) well contained 50 μl cell suspension and 50 μl assay medium. The plate was incubated again for 95-97 h at 37º, 5 % CO2, and 95 % RH. At the end of the incubation, the plate was added with MTS (Promega, Madison, Wisconsin) at 20 μl/well, followed by an additional incubation of 2.5-3.5 h in the CO2 incubator. The viable cells at each well would interact with MTS and led to a changed color. Finally, the 96-well microtiter plate was put into a multi-mode microplate reader (M5, Molecular Devices) to measure the absorbance at 490 nm and 650 nm simultaneously. The A490nm-450nm of each experimental well was recorded to generate the 4-P dose-response curve, after subtracting the average value of blank wells. The 4-P curve of reference standard, test sample and control were generated separately by the logarithmic plots of absorbance versus concentration to determine EC50 value. The biological activity of test samples was calculated as (EC50 Reference / EC50 Test)×100 %.
The same cell based biological potency assay procedure was used for P potency analysis, where the P protein samples were also diluted into 7 points with the same concentration ranging from 0.002 μg/ml to 60 μg/ml. For T potency analysis, the concentration range of the 7 points was adjusted to be from 0.03 μg/ml to 10 μg/ ml, to avoid redundancy of the 4-P curve, while the rest of procedure was the same as that for P or M (P+T).
Method qualification
Method qualification was carried out for both ELISA based quantification assay and cell based biological potency assay following International Council for Harmonisation and Food and Drug Administration guidelines about analytical method validation [16]. For ELISA based quantification assay, the focus is to identify if the presence of P or T will affect the assay accuracy for T or P protein concentration measurement from antibody admixtures M (3P+T), M (P+3T) and M (P+T). A total of 6 runs were conducted by 2 analysts in 3 d for P or T protein quantification. In each run, M (3P+T), M (P+3T) and M (P+T) were tested 3 times. The results were then compared with the nominal concentrations for inter- or intra- accuracy and precision analysis.
For cell based potency assay, the focus is method accuracy. Take M (P+T) for example, M50, M75, M125 and M150 were subjected to potency analysis by 2 analysts in 3 d. Each sample was repeated once. The measured potency of replicates of each sample was compared with the expected one to evaluate the assay’s accuracy.
Forced degraded sample analysis
Heat degraded samples were prepared and mixed to mimic stability antibody admixtures. P (5 mg/ml) or T (5 mg/ml) were treated at 80º for 30 min to be P* and T*. P and T were then mixed in amount ratio of 1:1 to be S (P+T), where P concentration is 2.5 mg/ml (nominal) and T concentration is 2.5 mg/ml (nominal). S (P*+T), S (P+T*), S (P*+T*) were then prepared in the same way. The protein concentrations of the 4 samples as a whole were determined by UV280nm measurement on a spectrophotometer (MAPADA, Shanghai, China). Extinction coefficient was theoretically calculated based on the amino acid compositions of P and T [17]. The 4 samples were then regarded as unknown samples for potency and concentration measurements using our developed assay system.
Results and Discussion
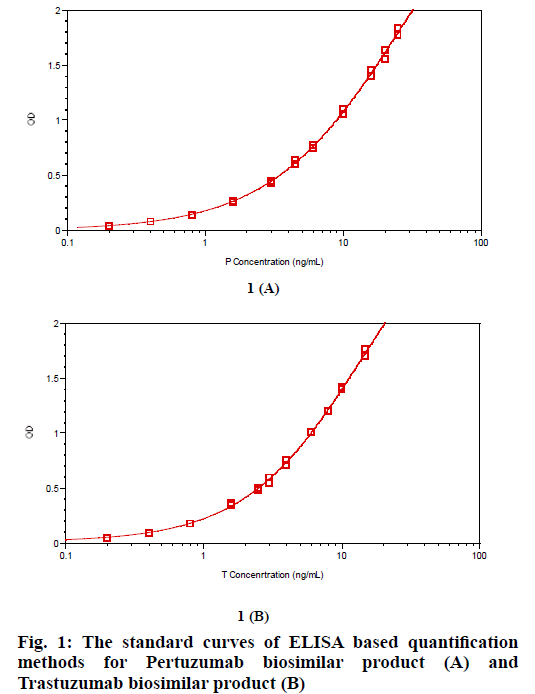
ELISA based P or T specific protein quantification assays were developed using P or T specific target antigens. The detection limit of each method for P or T protein quantification was 0.4 ng/ml, good enough for P or T detection. The standard curves for P or T protein quantification were shown in fig. 1. For each assay, CV % of duplicates of standard curve samples and QC samples are within 10 %. Recovery of standard curve samples and QC samples are within 90 % -110 %, indicating that both methods can accurately measure protein concentration of P or T (Table 1). We repeated the standard curves for P or T quantification several times, and no clear difference of the curves among these repeats was found (data not shown).
| (A) | ||||||
|---|---|---|---|---|---|---|
| Standard Curve Samples | Nominal Conc. (ng/mL) | OD1 | OD2 | CV | Measured Conc.(ng/mL) | Recovery |
| STD1 | 25.0 | 1.827 | 1.772 | 2.2 % | 25.2 | 100.9 % |
| STD2 | 20.0 | 1.636 | 1.551 | 3.8 % | 19.7 | 98.5 % |
| STD3 | 16.0 | 1.455 | 1.400 | 2.7 % | 16.1 | 100.4 % |
| STD4 | 10.0 | 1.097 | 1.048 | 3.2 % | 10.0 | 100.1 % |
| STD5 | 6.0 | 0.769 | 0.739 | 2.8 % | 6.0 | 100.0 % |
| STD6 | 4.5 | 0.633 | 0.600 | 3.8 % | 4.6 | 101.8 % |
| STD7 | 3.0 | 0.448 | 0.425 | 3.7 % | 3.0 | 98.4 % |
| STD8 | 1.6 | 0.263 | 0.256 | 1.9 % | 1.6 | 98.9 % |
| STD9 | 0.8 | 0.141 | 0.141 | N/A | 0.8 | 98.6 % |
| STD10 | 0.4 | 0.077 | 0.076 | 0.9 % | 0.4 | 100.9 % |
| STD11 | 0.2 | 0.041 | 0.042 | 1.7 % | 0.2 | 105.9 % |
| HQC1 | 15.0 | 1.330 | 1.329 | 0.1 % | 14.2 | 94.6 % |
| HQC2 | 15.0 | 1.319 | 1.316 | 0.2 % | 14.0 | 93.1 % |
| HQC3 | 15.0 | 1.347 | 1.331 | 0.8 % | 14.4 | 95.7 % |
| HQC4 | 15.0 | 1.302 | 1.254 | 2.7 % | 13.3 | 88.4 % |
| MQC1 | 6.0 | 0.724 | 0.734 | 1.0 % | 5.7 | 95.5 % |
| MQC2 | 6.0 | 0.717 | 0.728 | 1.1 % | 5.7 | 94.4 % |
| MQC3 | 6.0 | 0.726 | 0.731 | 0.5 % | 5.7 | 95.4 % |
| MQC4 | 6.0 | 0.735 | 0.683 | 5.2 % | 5.5 | 92.0 % |
| LQC1 | 1.2 | 0.191 | 0.195 | 1.5 % | 1.1 | 93.7 % |
| LQC2 | 1.2 | 0.187 | 0.186 | 0.4 % | 1.1 | 90.1 % |
| LQC3 | 1.2 | 0.191 | 0.186 | 1.9 % | 1.1 | 91.2 % |
| LQC4 | 1.2 | 0.189 | 0.176 | 5.0 % | 1.1 | 87.9 % |
| (B) | ||||||
| Standard Curve Samples | Nominal Conc. (ng/mL) | OD1 | OD2 | CV | Measured Conc.(ng/mL) | Recovery |
| STD1 | 15.0 | 1.757 | 1.696 | 2.5 % | 14.9 | 99.5 % |
| STD2 | 10.0 | 1.411 | 1.401 | 0.5 % | 10.2 | 101.9 % |
| STD3 | 8.0 | 1.194 | 1.200 | 0.4 % | 7.9 | 98.2 % |
| STD4 | 6.0 | 1.009 | 1.001 | 0.6 % | 6.1 | 101.3 % |
| STD5 | 4.0 | 0.753 | 0.712 | 4.0 % | 4.0 | 99.7 % |
| STD6 | 3.0 | 0.593 | 0.550 | 5.3 % | 2.9 | 98.0 % |
| STD7 | 2.5 | 0.503 | 0.477 | 3.8 % | 2.5 | 98.0 % |
| STD8 | 1.6 | 0.364 | 0.346 | 3.6 % | 1.7 | 106.1 % |
| STD9 | 0.8 | 0.180 | 0.178 | 0.8 % | 0.8 | 100.0 % |
| STD10 | 0.4 | 0.096 | 0.093 | 2.2 % | 0.4 | 99.3 % |
| STD11 | 0.2 | 0.050 | 0.048 | 2.9 % | 0.2 | 92.5 % |
| HQC1 | 7.5 | 1.035 | 1.116 | 5.3 % | 6.7 | 89.3 % |
| HQC2 | 7.5 | 1.075 | 1.045 | 2.0 % | 6.6 | 87.4 % |
| HQC3 | 7.5 | 1.058 | 1.036 | 1.5 % | 6.4 | 85.9 % |
| HQC4 | 7.5 | 1.044 | 1.051 | 0.5 % | 6.4 | 85.9 % |
| MQC1 | 4.0 | 0.705 | 0.682 | 2.3 % | 3.7 | 93.1 % |
| MQC2 | 4.0 | 0.673 | 0.657 | 1.7 % | 3.5 | 88.3 % |
| MQC3 | 4.0 | 0.676 | 0.658 | 1.9 % | 3.5 | 88.7 % |
| MQC4 | 4.0 | 0.670 | 0.670 | N/A | 3.6 | 89.2 % |
| LQC1 | 1.2 | 0.251 | 0.245 | 1.7 % | 1.1 | 95.1 % |
| LQC2 | 1.2 | 0.243 | 0.237 | 1.8 % | 1.1 | 91.8 % |
| LQC3 | 1.2 | 0.234 | 0.239 | 1.5 % | 1.1 | 90.3 % |
| LQC4 | 1.2 | 0.237 | 0.245 | 2.3 % | 1.1 | 92.2 % |
Table 1: Typical Recovery Of Standard Curve Samples and Qc Samples for Elisa Based Quantification Methods for Pertuzumab Biosimilar Product (A) and Trastuzumab Biosimilar Product (B).
After the method development stage, the ELISA based protein quantification method for P or T was qualified subsequently, especially for accuracy and precision. Because the purpose of these methods was to quantify P or T from antibody admixtures containing P and T and the presence of P or T might affect accuracy of these methods, antibody admixtures M (3P+T), M (P+3T) and M (P+T) were used for method qualification. M (3P+T) was diluted 3 times by formulation buffer to be M (P+1/3T) where the P concentration is 5 mg/ml (nominal), and T concentration is 1.67 mg/ ml (nominal). M (P+3T) was also diluted 3 times by formulation buffer to be M (1/3P+T) where the P concentration is 1.67 mg/ml, and T concentration is 5 mg/ml (nominal). M (P+1/3T), M (P+T) and M (P+3T) were quantified in 6 plates by 2 analysts in 3 d for P quantification. M (1/3P+T), M (P+T) and M (3P+T) were quantified for T quantification in the same way. As we can see in Table 2, the RSD of all 18 replicates of measured P concentration from M (P+1/3T), M (P+T) and M (P+3T) are within 10 %, the recovery is within 90 %-110 % for P quantification assay. The RSD of all 18 replicates of measured T concentration from M (1/3P+T), M (P+T) and M (3P+T) are also within 10 %, the recovery is also within 90 %-110 % for T quantification assay. The results indicate that the ELISA based P or T protein quantification assay shows good accuracy and precision. The presence of more or less P in antibody admixtures is not an issue for T protein quantification, neither is the presence of more or less T in antibody admixtures for P protein quantification.
| (A) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | P Conc.(mg/mL) | Average(mg/mL) | RSD | Recovery | |||||
| P+T | 5.2 | 5.3 | 5.2 | 5.0 | 6.7 % | 100 % | |||
| 5.0 | 5.0 | 4.8 | |||||||
| 4.8 | 4.9 | 4.6 | |||||||
| 4.8 | 4.4 | 5.1 | |||||||
| 5.4 | 5.4 | 5.5 | |||||||
| 4.5 | 4.8 | 5.5 | |||||||
| P+3T | 4.8 | 4.9 | 4.9 | 4.9 | 6.4 % | 98 % | |||
| 5.3 | 5.1 | 4.9 | |||||||
| 4.9 | 4.7 | 4.7 | |||||||
| 5.1 | 4.5 | 5.3 | |||||||
| 5.2 | 5.0 | 4.4 | |||||||
| 4.5 | 5.2 | 5.6 | |||||||
| P+1/3T | 5.0 | 4.7 | 4.9 | 5.0 | 4.9 % | 100 % | |||
| 5.3 | 5.1 | 5.0 | |||||||
| 5.0 | 4.8 | 4.9 | |||||||
| 5.2 | 4.7 | 5.2 | |||||||
| 5.1 | 5.4 | 5.1 | |||||||
| 4.6 | 5.5 | 4.9 | |||||||
| (B) | |||||||||
| Sample | T Conc.(mg/mL) | Average(mg/mL) | RSD | Recovery | |||||
| P+T | 4.5 | 4.7 | 4.8 | 4.8 | 5.4 % | 96 % | |||
| 4.8 | 5.1 | 5.0 | |||||||
| 4.6 | 4.8 | 4.7 | |||||||
| 5.5 | 4.7 | 5.1 | |||||||
| 4.6 | 4.8 | 4.5 | |||||||
| 4.6 | 4.5 | 4.7 | |||||||
| 3P+T | 4.9 | 4.8 | 4.7 | 5.1 | 4.5 % | 102 % | |||
| 5.0 | 5.1 | 5.5 | |||||||
| 5.0 | 4.9 | 5.0 | |||||||
| 5.4 | 4.6 | 5.1 | |||||||
| 5.2 | 5.2 | 5.0 | |||||||
| 5.3 | 5.1 | 5.1 | |||||||
| 1/3P+T | 5.1 | 5.1 | 4.5 | 5.1 | 5.2 % | 102 % | |||
| 5.1 | 5.4 | 5.6 | |||||||
| 5.0 | 4.9 | 5.4 | |||||||
| 5.3 | 4.7 | 5.3 | |||||||
| 5.2 | 5.2 | 5.0 | |||||||
| 5.1 | 5.1 | 4.8 | |||||||
Table 2: Method Qualification of Accuracy and Precision for Pertuzumab Biosimilar Product Quantification (A) and Trastuzumab Biosimilar Product Quantification (B).
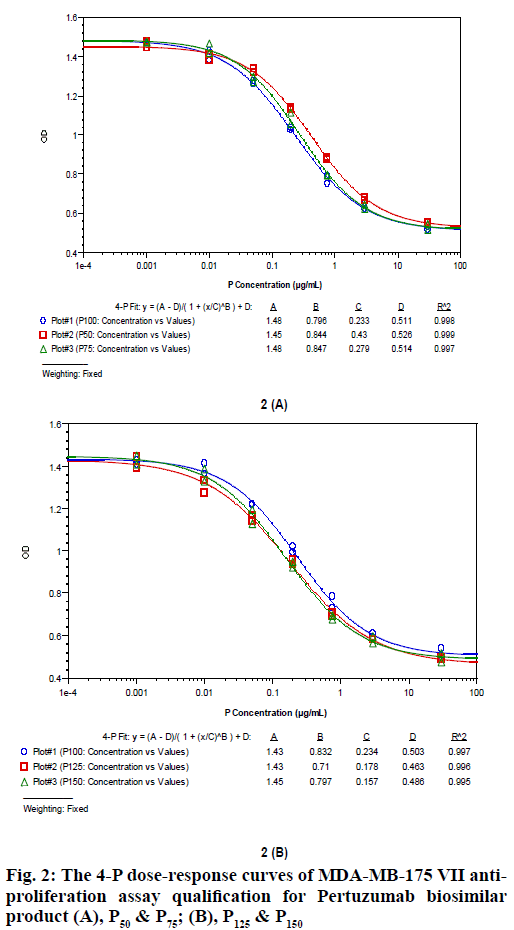
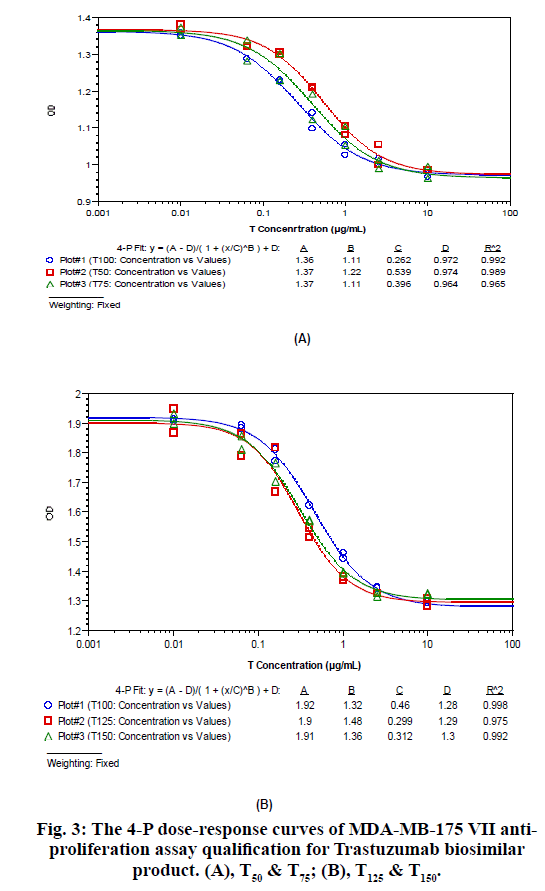
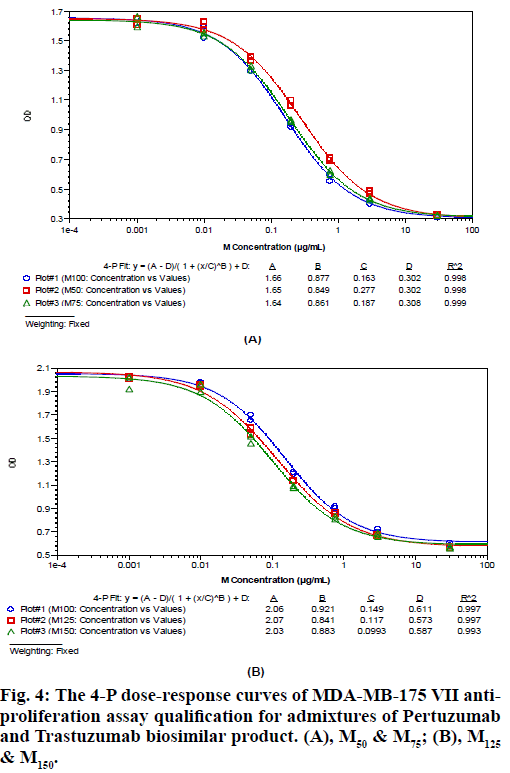
The MDA-MB-175 VII anti-proliferation assay was developed to be suitable for P, T and M (P+T) biological potency evaluation. Key parameters like seeded cell density, FBS concentration, drug concentration, incubation time and cell staining way were optimized. The cell based biological potency assay was then qualified for method accuracy and precision analysis of P, T and M (P+T). Take the biological potency of P100, T100 and M100 as 100%, the relative potency of P50, P75, P125, P150 to P100, T50, T75, T125, T150 to T100, and M50, M75, M125 and M150 to M100 were measured using the same assay. The 4-P dose-response curves for these samples of P, T and M (P+T) are shown in fig. 2, fig. 3, and fig. 4. The measured potency value and recovery of these samples are shown in Table 3, Table 4 and Table 5. It is found that the recovery for all these samples is within 75 %-125 %, indicating that the developed MDA-MD-175 VII anti-proliferation assay is capable of accurately measuring P, T and M (P+T)’s biological potency simultaneously.
| Sample | Nominal Potency | Measured Potency | Recovery |
|---|---|---|---|
| P50 | 50 % | 54 % | 108 % |
| P75 | 75 % | 84 % | 112 % |
| P125 | 125 % | 131 % | 105 % |
| P150 | 150 % | 149 % | 99 % |
Table 3: The MDA-MB-175 Vii Anti-Proliferation Assay Qualification Data For Pertuzumab Biosimilar Product. (A), P50 & P75; (B), P125 & P150
| Sample | Nominal Potency | Measured Potency | Recovery |
|---|---|---|---|
| T50 | 50 % | 49 % | 98 % |
| T75 | 75 % | 66 % | 88 % |
| T125 | 125 % | 154 % | 123 % |
| T150 | 150 % | 147 % | 98 % |
Table 4: The MDA-MB-175 Vii Anti-Proliferation Assay Qualification Data for TRASTUZUMAB Biosimilar Product. (A), T50 & T75; (B), T125 & T150
| Sample | Nominal Potency | Measured Potency | Recovery |
|---|---|---|---|
| M50 | 50 % | 59 % | 118 % |
| M75 | 75 % | 87 % | 116 % |
| M125 | 125 % | 127 % | 102 % |
| M150 | 150 % | 150 % | 100 % |
Table 5: The MDA-MB-175 Vii Anti-Proliferation Assay Qualification Data for Admixtures of Trastuzumab and Pertuzumab Biosimilar Products. (A), M50 & M75; (B), M125 & M150
After confirming that the developed cell based assay could be used for P, T and M (P+T) biological potency detection simultaneously, we tried to find ways to figure out if P’s or T’s potency changes when M (P+T)’s potency is out of the acceptance criteria. Different amount ratio of component P and T were mixed to generate M (P+2T), M (P+3T), M (2P+T), M (3P+T), and subjected to potency analysis by twice.
The EC50 values of these samples and their relative potency compared to M (P+T) were recorded, as shown in Table 6. 2 tables were generated in different ways. In Table 6(A), the component P is regarded as active pharmaceutical ingredient, the component T is regarded as excipient, and thus the EC50 value and biological potency are calculated accordingly based on the P protein concentration. It is found that as the amount of component T increases, the average EC50 values of admixtures of P and T decreases gradually, and thus the average relative potency compared to M (P+T) increases from 79 % to 188 %. The result indicates that component T could provide synergistic effect to enhance the biological potency of admixtures. In Table 6(B), the component T is regarded as active pharmaceutical ingredient, the component P is regarded as excipient, and thus the EC50 value and biological potency are calculated accordingly based on the T protein concentration. It is found that as the amount of component P increases, the average EC50 values of admixtures of P and T decreases gradually, and thus the average relative potency compared to M (P+T) increases from 63 % to 239 %. The results indicate that component P could also provide synergistic effect to enhance the biological potency of admixtures. So from Table 6, firstly we find that P and T both have synergistic effect with each other in the cell based biological potency. Secondly, we also find that the amount percent of component P or component T in admixture of P and T was proportional to the admixture’s measured relative potency compared to M (P+T), calculated based on either P protein concentration (Table 6(A)) or T protein concentration (Table 6(B)). The 2 findings represent the basis for further analysis of real samples of admixtures that could help to clarify if some unknown changes happen to the potency of P or T.
| (A) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Amount ratio of P and T | Amount percentage of P | EC50 (μg/mL) | Potency compared to M (P+T) | ||||||
| P | T | Repeat 1 | Repeat 2 | Average | |||||
| 3 | 1 | 75 % | 0.18 | 0.18 | 0.18 | 79 % | |||
| 2 | 1 | 66.7 % | 0.16 | 0.175 | 0.168 | 85 % | |||
| 1 | 1 | 50 % | 0.138 | 0.147 | 0.143 | 100 % | |||
| 1 | 2 | 33.3 % | 0.0855 | 0.0911 | 0.088 | 163 % | |||
| 1 | 3 | 25 % | 0.0687 | 0.0824 | 0.076 | 188 % | |||
| (B) | |||||||||
| Amount ratio of P and T | Amount percentage of T | EC50 (μg/mL) | Potency compared to M (P+T) | ||||||
| P | T | Repeat 1 | Repeat 2 | Average | |||||
| 1 | 3 | 75 % | 0.205 | 0.247 | 0.226 | 63 % | |||
| 1 | 2 | 66.7 % | 0.171 | 0.182 | 0.177 | 81 % | |||
| 1 | 1 | 50 % | 0.139 | 0.147 | 0.143 | 100 % | |||
| 2 | 1 | 33.3 % | 0.0801 | 0.0675 | 0.0738 | 194 % | |||
| 3 | 1 | 25 % | 0.0598 | 0.0599 | 0.0599 | 239 % | |||
Table 6: The EC50 Values of Antibody Admixtures on MDA-MB-175 VII Anti-Proliferation Assay Analysis and their Relative Potency Compared to M (p+t). (a), Calculation Based On The Concentration of Pertuzumab Biosimilar Product; (b), Calculation Based on the Concentration Of Trastuzumab Biosimilar Product.
In order to test the utility of our in vitro potency assay system for real sample analysis, 4 forced degraded admixtures of P and T by heat were prepared. They are S (P+T), S (P*+T), S (P+T*), S (P*+T*). Among them, S (P+T) is normal sample, in which equal amount of P and T were mixed together, the concentration of P or T is 2.5 mg/ml (nominal), while the whole P+T protein concentration is 5mg/mL (nominal). The other 3 samples containing heat treated P, or heat treated T or both are regarded as unknown samples, they are also could be regarded as stability study samples derived from S (P+T). Before the biological potency measurement started, the potency of all the above 4 samples are unknown.
First, all the 4 samples are regarded as a whole, their protein concentrations were measured by UV280 method, the concentration was calculated as the ratio of A280 and extinction co-efficient of M (P+T). The results were shown in Table 7. As predicted, the protein concentrations of all the 4 samples are all almost 5 mg/ml, just the same as that of S (P+T) (nominal). The common used UV280 protein quantification method could not monitor the structural changes of protein itself, the A280 value was determined by the presence of Trp, Tyr and Cys (disulfide bonds), so the protein concentration of heat treated samples and normal samples were measured to be the same with each other, although there might exist some degradation or structural conformational changes in samples that contain heat treated P* or T*.
| Sample | Measured Protein Concentration (mg/mL) |
|---|---|
| S (P+T) | 5.0 |
| S (P*+T) | 4.8 |
| S (P+T*) | 5.1 |
| S (P*+T*) | 5.3 |
Table 7: Protein Concentration of 4 Forced Degraded Admixture Samples Measured By UV280
Based on the protein concentrations measured by UV280, the relative biological potency of these 4 samples were measured subsequently using the MDA-MB-175 VII anti-proliferation assay, with M (P+T) as the reference sample. Because in M (P+T), the amount ratio of P and T is 1:1, so the relative potency of S (P+T) compared to M (P+T) is expected to be 100 %. The results are shown in Table 8. Except S (P+T), whose measured potency is 99 % comparable with M (P+T), the other 3 sample’s measured potency are lower especially for that of S (P*+T*). The results show that the biological potency of the 3 samples containing heat treated P* or T* decreases. However, in these samples, if the component P or component T’s biological potency or both decreases are still not identified.
| Sample | EC50 (μg/mL) (Calculated based on UV280 Concentration) | Relative potency compared to M (P+T) | |
|---|---|---|---|
| Plate 1 | Plate 2 | ||
| M (P+T) | 0.23 | 0.236 | 100 % |
| S (P+T) | 0.233 | N/A* | 99 % |
| S (P*+T) | 0.434 | N/A* | 53 % |
| S (P+T*) | N/A* | 0.313 | 76 % |
| S (P*+T*) | N/A* | 0.584 | 40 % |
*N/A means not run in this plate.
Table 8: Biological Potency of 4 Forced Degraded Admixture Samples CompareD tO M (P+T), Measured By MDA-MB-175 VII Anti-Proliferation Assay.
In order to investigate the decease of potency of these samples more deeply, all the 4 samples were subjected to P or T specific ELISA based protein quantification analysis. For each sample, the protein concentrations of P, T, P*, T* of antibody admixtures were measured, as shown in Table 9. The measured concentrations of P and T of the 4 samples are comparable to 2.5 mg/ml as nominal. The measured concentration of P* is about 1.2 mg/ml in both S (P*+T) and S (P*+T*), while the measured concentration of T* is about 0.8 mg/ml in both S (P+T*) and S (P*+T*). The decrease in P* or T*’s ELISA based concentration is not due to precipitation, because no precipitation was observed in these samples and the UV280 based concentration of all the 4 samples are almost the same as prepared. So the decrease indicates some kind of protein structural changes that will affect the binding of P or T to their target specific antigens. The loss in P* or T*’s protein concentration might reduce the biological potency of antibody admixtures that contains P* or T*. If so, the relationship between ELISA based protein quantification data and the biological potency data of these samples should follow the relationship represented in the analysis table.
| Sample | P and T protein concentration (mg/mL) | |
|---|---|---|
| P | T | |
| S (P+T) | 2.5 | 2.5 |
| S (P*+T) | 1.1 | 2.5 |
| S (P+T*) | 2.7 | 0.8 |
| S (P*+T*) | 1.2 | 0.8 |
Table 9: Protein Concentration of P and T from 4 Forced Degraded Admixtures, Measured By Elisa Based P or T Quantification Methods
Finally, the ELISA based protein quantification data of the 4 samples was compared with their biological potency data under the base of biological potency analysis table of antibody admixtures (Table 6). Based on Table 6 where the EC50 value and the relative biological potency compared to M (P+T) was calculated based on P protein concentration or T protein concentration, the biological potency data of S (P+T), S (P*+T), S (P+T*) and S (P*+T*) was also adjusted accordingly based on P or T protein concentration. From the relative biological potency data of these 4 samples compared to M (P+T), the amount ratio of P and T in these samples could be predicted based on the analysis table. The predicted amount ratio of P and T in these samples and that generated from measured ELISA based P or T quantification data, were compared and summarized in Table 10. Whether the calculation was based on P protein concentration (Table 10(A))or T protein concentration (Table 10(B)), the predicted amount ratio of P and T is found in accordance with that measured by ELISA based P or T quantification.
| (A) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Sample | P and T con. Measured by ELISA (mg/mL) | Relative potency compared to M (P+T) (Calculated based on P Concentration) |
Amount ratio of P and T (Calculated based on P Concentration) |
||||||
| Predicted | Measured | ||||||||
| P | T | P | T | P | T | ||||
| S (P+T) | 2.5 | 2.5 | 99 % | 1 | 1 | 1 | 1 | ||
| S (P*+T) | 1.1 | 2.5 | 121 % | 1 | 1~2 | 1 | 2.3 | ||
| S (P+T*) | 2.7 | 0.8 | 71 % | 1 | ~1/3 | 1 | 0.3 | ||
| S (P*+T*) | 1.2 | 0.8 | 85 % | 1 | 0.5 | 1 | 0.67 | ||
| (B) | |||||||||
| Sample | P and T con. Measured by ELISA (mg/mL) | Relative potency compared to M (P+T) (Calculated based on T Concentration) |
Amount ratio of P and T (Calculated based on T Concentration) |
||||||
| Predicted | Measured | ||||||||
| P | T | P | T | P | T | ||||
| S (P+T) | 2.5 | 2.5 | 99 % | 1 | 1 | 1 | 1 | ||
| S (P*+T) | 1.1 | 2.5 | 53 % | 1 | 3~4 | 1 | 2.3 | ||
| S (P+T*) | 2.7 | 0.8 | 239 % | 1 | 1/3 | 1 | 0.3 | ||
| S (P*+T*) | 1.2 | 0.8 | 127 % | 1 | 0.5~1 | 1 | 0.67 | ||
Table 10: The Predicted and Measured Amount Ratio of P and T in 4 Forced Degraded Admixture Samples and their Relative Potency Compared to M (P+T). (A), Calculation Based on the Concentration of Pertuzumab Biosimilar Product; (B), Calculation Based on the Concentration of Trastuzumab Biosimilar Product.
Finally, the ELISA based protein quantification data of the 4 samples was compared with their biological potency data under the base of biological potency analysis table of antibody admixtures (Table 6). Based on Table 6 where the EC50 value and the relative biological potency compared to M (P+T) was calculated based on P protein concentration or T protein concentration, the biological potency data of S (P+T), S (P*+T), S (P+T*) and S (P*+T*) was also adjusted accordingly based on P or T protein concentration. From the relative biological potency data of these 4 samples compared to M (P+T), the amount ratio of P and T in these samples could be predicted based on the analysis table. The predicted amount ratio of P and T in these samples and that generated from measured ELISA based P or T quantification data, were compared and summarized in Table 10. Whether the calculation was based on P protein concentration (Table 10(A)) or T protein concentration (Table 10(B)), the predicted amount ratio of P and T is found in accordance with that measured by ELISA based P or T quantification. and T’s concentrations, also called the decrease of both P*’s and T*’s potency is the cause for bad potency of S (P*+T*).
Because P* and T* are heat treated degraded samples, their biological potency are most probably worse than normal. The admixtures containing P* or T* will have a worse potency. In this study, using our in vitro potency assay system, the decrease of potency of 4 unknown degraded samples are measured using ELISA based P and T quantification assay, and cell based biological potency assay. By analyzing the data under the analysis table, the decrease of P* and T*’s potency could be monitored and identified from their admixtures as predicted.
The purpose of developing the cell based biological potency assay system for antibody admixtures is try to investigate in detail the potency of each component of admixtures, especially in stability study. The assay system needs specific and accurate protein quantification first, while the traditional UV280 based protein quantification method cannot specifically measure the concentration of each component of admixtures. In this case, High Performance Liquid Chromatography (HPLC) or Capillary Electrophoresis (CE) based methods like RP-HPLC, Ion-exchange chromatography (IEC)- HPLC or Capillary zone electrophoresis (CZE) that can separate P and T by their different hydrophobicity or charge properties were tried first for specific protein quantification in antibody admixtures of P and T [12]. However, it was shown that not only the main peaks of P and T but also their product related impurities could be separated in the HPLC or CE chromatograms. It is not easy to clarify if or not the total area of the 2 main peaks could definitely represent the content of P or T respectively. Actually, the 2 main peaks of P or T might contain with each other, which make the quantification more difficult. In conclusion, the HPLC and CE based protein quantification methods are not suitable for admixtures of P or T. Apart from HPLC or CE, enzyme linked immunosorbent assay (ELISA) is another choice that has been broadly used in the area of bioanalysis of biologics [18]. The key point is specificity. In this case, the P or T specific antigens (extracellular subdomain II and IV of HER2) are used in assay development. The assay shows good specificity, accuracy and precision to measure P and T protein concentration of their admixtures.
The combinational use of Pertuzumab and Trastuzumab has long been proven having synergistic effect against breast cancer. Pertuzumab and Trastuzumab admixture was referred to as homo-combination. Their Fab fragments are targeting different extra cellular locus of HER2 that both can in vitro inhibit the proliferation of HER2 overexpressing breast tumor cells and induce ADCC. So in this case, it is not difficult to find a cell based assay that is capable of measuring potency of P and T as well as their admixtures. Their synergistic effect against MDA-MB-175 VII proliferation was also confirmed as predicted. The ELISA based protein quantification assay also depends on the binding of the specific antigens of extra cellular locus of HER2 to P or T’s Fab fragments, this is probably why the data generated from protein quantification is in accordance with that generated from the cell based anti-proliferation potency when analyzing the amount ratio of component P and T in their admixtures. On the other hand, this accordance also indicates the accountability of the data generated from both protein quantification and potency analysis in our assay system. To some extent, the use of P or T specific antigens of extra cellular locus of HER2 for ELISA based protein quantification method development might be an essential choice in our assay system.
In this study, P and T have similar biological functions. They both can inhibit the proliferation of breast cancer cell line MDA-MB-175 VII in a similar mechanism. This is another point for successfully developing such an assay system. Even if the targets of 2 antibodies are totally different with different Fab related functions, a similar assay system could also be developed from Fc related functions like ADCC or complement-dependent cytotoxicity (CDC) that might be similar between the 2 antibodies. In that case, because ADCC or CDC depends on the binding of Fab fragments to their specific target antigens, protein quantification data derived from specific antigen based ELISA can thus be related with the potency of ADCC or CDC. Consequently, a similar biological potency based analysis table could be generated.
The idea might also be applied to bi-specific antibody. Bi-specific antibody contains 2 active binding sites that can simultaneously binds to 2 different types of antigens. The 2 active binding sites of bi-specific antibody usually has biological potency as a whole, while the 2 sites could also be evaluated by 2 specific antigen based ELISA analysis. In a similar way, a biological potency based analysis table could be generated to predict the changes of these 2 sites. The change of certain active binding site of the bi-specific antibody could be monitored.
This study provides a preliminary model for potency analysis of antibody admixtures. More data generated from repeats of experiments performed on certain amount ratio of 2 components will help improve the accuracy and precision of the assay system. Further physiochemical analysis like IEC, CZE, or Reverse phase high-performance liquid chromatography-mass spectrometry (RP-HPLC-MS) are also needed to investigate the structural changes of each component that might cause the change of potency, in order to better understand and confirm the data measured in the assay system.
Acknowledgements
The authors thank Shanghai Pharmaceuticals (Shanghai, China) to provide Pertuzumab and Trastuzumab biosimilar products for this study. This study was supported by Major Science and Technology Project for ‘Significant New Drugs Creation’ 2015ZX09102013 which is also gratefully acknowledged.
References
- Carter PJ, Lazar GA. Next generation antibody drugs: pursuit of the ’high-hanging fruit’. Nat Rev Drug Discov 2018;17(3):197-223.
- Kumler I, Tuxen MK, Nielsen DL. A systematic review of dual targeting in HER2-positive breast cancer. Cancer Treat Rev 2014;40(2):259-270.
- Carvalho S, Levi-Schaffer F, Sela M, Yarden Y. Immnunotherapy of cancer: from monoclonal to oligoclonal cocktails of anti-cancer antibodies: IUPHAR Review 18. Br J Pharmacol 2016;173(9):1407-24.
- Swain SM, Baselga J, Kim SB, Ro J, Semiglazov V, Campone M, et al. Pertuzumab, Trastuzumab, and Docetaxel in HER2-Positive Metastatic Breast Cancer. N Engl J Med 2015;372(8):724-34.
- Andersson M, Lopez-Vega JM, Petit T, Zamagni C, Easton V, Kamber J, et al. Efficacy and Safety of Pertuzumab and Trastuzumab Administered in a Single Infusion Bag, Followed by Vinorelbine: VELVET Cohort 2 Final Results. Oncologist 2017;22(10):1160-8.
- Hurvitz SA, Martin M, Symmans WF, Jung KH, Huang C, Thompson AM, et al. Neoadjuvant Trastuzumab, Pertuzumab, and Chemotherapy Versus Trastuzumab Emtansine Plus Pertuzumab in Patients With HER2-positive Breast Cancer (KRISTINE): A Randomised, Open-Label, Multicentre, Phase 3 Trial. Lancet Oncol 2018;19(1):115-26.
- Junttila TT, Akita RW, Parsons K, Fields C, Lewis Phillips GD, Friedman LS, et al. Ligand-independent HER2/HER3/PI3K complex is disrupted by trastuzumab and is effectively inhibited by the PI3K inhibitor GDC-0941. Cancer Cell 2009;15(5):429-40.
- Franklin MC, Carey KD, Vajdos FF, Leahy DJ, de Vos AM, Sliwkowski MX. Insights into ErbB signaling from the structure of the ErbB2-pertuzumab complex. Cancer Cell 2004;5(4):317-28.
- Kasprzyk PG, Song SU, Di Fiore PP, King CR. Therapy of an animial model of human gastric cancer using a combination of anti-erbB-2 monoclonal antibodies. Cancer Res 1992;52(10):2771-6.
- Spiridon CI, Ghetie MA, Uhr J, Marches R, Li JL, Shen GL, et al. Targeting multiple HER-2 epitopes with monoclonal antibodies results in improved antigrowth activity of a human breast cancer cell line in vitro and in vivo. Clin Cancer Res 2002;8(6):1720-30.
- Scheuer W, Friess T, Burtscher H, Bossenmaier B, Endl J, Hasmann M. Strongly enhanced antitumor activity of trastuzumab and pertuzumab combination treatment on HER2-positive human xenograft tumor models. Cancer Res 2009;69(24):9330-6.
- Glover ZW, Gennaro L, Yadav S, Demeule B, Wong PY, Sreedhara A. Compatibility and stability of pertuzumab and trastuzumab admixtures in i.v. infusion bags for coadministration. J Pharm Sci 2013;102(3):794-812.
- Sreedhara A, Glover ZK, Piros N, Xiao N, Patel A. Stability of IgG1 monoclonal antibodies in intravenous infusion bags under clinical in use conditions. J Pharm Sci 2012;101(1):21-30.
- Wang J, Yiu B, Obermeyer J, Filipe CD, Brennan JD, Pelton R. Effects of temerature and relative humidity on the stability of paper-immobilized antibodies. Biomacromolecules 2012;13(2):559-64.
- Shah DD, Zhang J, Maity H, Mallela KMG. Effect of photo-degradation on the structure, stability, aggregation, and function of an IgG1 monoclonal antibody. Int J Pharm 2018;547(1):438-49.
- Food and Drug Administration, Center for Drug Evaluation and Research (CDER), Center for Biologics Evaluation and Research (CBER). Analytical Procedures and Methods Validation for Drugs and Biologics, Guidance for Industry; 2015.
- Pace CN, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar adsorption coefficient of a protein. Protein Sci 1995;4(11):2411-23.
- Myler HA, Given A, Kolz K, Mora JR, Hristopoulos G. Biotherapeutic bioanalysis: a multi-indication case study review. Bioanalysis 2011;3(6):623-43.