- *Corresponding Author:
- Sneha Kushwaha
Adarsh Vijendra Institute of Pharmaceutical Sciences, Shobhit University, Gangoh, Saharanpur, Uttar Pradesh 247341, India
E-mail: sneha.kush09@gmail.com
| Date of Received | 16 June 2021 |
| Date of Revision | 26 September 2022 |
| Date of Acceptance | 23 November 2022 |
| Indian J Pharm Sci 2022;84(5):1161-1170 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The anti-arthritic activity of cathepsin S enzyme has been acknowledged and regarded as an emerging target for the development of novel therapeutic agents for the treatment of various autoimmune disorders and other inflammatory diseases. Two dimensional-quantitative structure-activity relationships have been performed on a series of tetrahydropyrido-pyrazole nucleus using TSAR 3.3. Attempts have been made to derive and comprehend a correlation between biological activity (dependent variable) and descriptors (independent variables). The study was performed using 268 compounds (data set) by division into training and test set by the random selection method. 179 compounds generated a final quantitative structure-activity relationship model with the leave-out one row method of cross-validation to evaluate the predictive ability of the model. The most significant model with n=179, regression coefficient (0.851), correlation coefficient (0.725), cross-validated correlation coefficient (0.709), standard error (0.349), Fischer statistic value (114.706) was developed using multiple linear regression analysis. For partial least squares, a statistical significance value of 0.988 and a fraction of variance explained 0.723 were observed. A comparable partial least squares model with correlation coefficient (0.723) and neural model with correlation coefficient (0.731) indicated good internal predictability of the model. External test set validation provided correlation coefficient values of 0.708 and 0.706 for multiple linear regressions and partial least squares analysis respectively. Quantitative structure-activity relationship model indicated the importance of hydrophobic (log P (substituent 1)), topological (kier chi 5 (path) index (whole molecule)), electronic (bond dipole moment (substituent 5)) and (dipole moment Z component (substituent 1)) descriptors for the activity of Cathepsin S inhibitors. This study will be effective in designing novel and more potent cathepsin S inhibitors.
Keywords
Anti-arthritic activity, quantitative structure activity relationship, multiple linear regression, partial least square, tetrahydropyrido-pyrazole.
The “Cathepsin” term was extracted from the Greek word ‘kathepsein’ meaning “to digest”[1,2]. It was discovered in the 20th century[3]. A total of 11 human cysteine cathepsins are expressed in the human genome[4]. Out of these 11, cathepsins L, V, S, K and F are endopeptidases, cathepsins X, B, C and H are exopeptidases and cathepsins O and W of unknown category[1,5,6]. Cathepsin S is highly expressed in dendritic cells, macrophages, spleen, lymph nodes, monocytes and/or thymic cortical epithelial cells. The enzyme is involved in antigen processing and presentation. The unique distribution pattern indicates its deep involvement in the immune response[7].
All papain-like cysteine proteases, comprising cathepsin S, are composed of a signal peptide, a propertied and a catalytic domain[8]. Its highly conserved active site consists of Cysteine, Histidine and Asparagine residues[1]. The structure of cathepsin S comprises a single chain monomeric protein with left and right domain[9]. The cleft of the active site lies in between the two domains and contains the residues Cys25 and His159[9]. Cathepsin S plays important role in the development of various inflammation-associated ailments such as cancer[8,10-15], arthritis[9,16], periodontitis[17], psoriasis[9,18], lung diseases[19-25], cardiovascular disease in patients with chronic kidney disease[26-30], bone[31], Sjogren’s Syndrome[32,33] and immune disorders[34]. Inhibitors of cathepsin S, furthermore, act as immunomodulators[35,36].
Consequently, there is a requirement to progress research efforts concentrated on cathepsin S use in diagnostics and as therapeutic targets in diseases[37,38]. Cathepsin S inhibitors of dipeptidyl nitrile are an emerging target for the abolition of tumors[15].
Rheumatoid Arthritis (RA) is a chronic systemic autoimmune inflammatory disease of unknown etiology affecting all joints covered by synovium. The Major Histocompatibility Complex (MHC) molecule, also known as or Human Leukocyte Antigen complex (HLA) molecule and plays a central role in the pathology of RA[39]. The Antigen-Presenting Cells (APCs), usually a macrophage in the synovium, engulf the antigen. Enzymes (peroxides) inside the APCs break down the antigen into fragments[40]. The molecular mechanism starts with the synthesis of MHC II Alpha-Beta (αβ) heterodimers in the endoplasmic reticulum and the association of a protein, the invariant chain (Li) in the peptide-binding cleft. The αβLi complex gets transferred to the lysosome, where a part of the Li is cleaved by cathepsin S leaving a short fragment, Corticotropin Like Intermediate Peptide (CLIP) in the active site preventing any premature binding of antigenic peptides[32,41-43]. Another protein HLA-DM helps in releasing the CLIP from the MHC protein thus providing the binding site for the peptide fragments. After binding to the MHC II molecule, the complex is transferred to the cell surface[41,44,45]. This complex is presented to T-cells (Clusters of Differentiation-4 (CD4) cells i.e. T-helper cell) which the T-Cell Receptor (TCR) recognizes and binds causing APCs to secrete cytokines like Interleukin-1 (IL-1), Interferon-α (IFN-α), IFN-Gamma (γ), Tumor Necrosis Factor (TNF) and other factors that activate lymphocytes and other immune cells to respond to the antigens thus causing inflammation[9,24,39]. More specifically, HLA-DR β1 alleles give a major genetic contribution to RA. HLA-DR β1 loci are dominated by HLA-DR β1*0401, HLA-DR β1*0404, HLA-DR β1*0101, HLA-DR β1*01, HLA-DR β1*04, HLA-DR β1 *0405, HLA-DR β1*0408, HLA-DR β1*1001, HLA-DR β1*1401. These epitope due to the arrangement resemblance in the third hyper variable region of the DR β chain (amino acids 70-74: QKRAA, QRRAA, RRRAA) and therefore expected to present similar antigens to CD4 T cells. Of these, HLA-DR β1*04 alleles pose the strongest genetic susceptibility to RA. This provides a shred of strong evidence for adaptive immunity significant in the pathogenesis of RA via MHC II-dependent T cell activation[46-48].
Quantitative Structure-Activity Relationship (QSAR) technique is employed in biological activity modeling, as well as, calculating Absorption, Distribution, Metabolism and Excretion (ADME)/ toxicity properties[49]. A QSAR model with the help of a mathematical equation correlates the structure/ chemical characteristics of the molecule with their biological activities. This information helps design more potent compounds and the predictions of the biological activities can be done for new entities[50]. A QSAR study has great significance in enzyme inhibition studies and identifying the important active sites in the receptor. Thus, QSAR studies have become important in drug design[51-53]. Presently Two Dimensional (2D) QSAR analysis has been applied as it is simple and less error-prone. As it does not require any conformational search or structural alignment, it is more beneficial than Three-Dimensional (3D) QSAR analysis[54,55]. Moreover, structural descriptors are used in 2D methods encoding all the chemical information[56]. Thus 2D shows superiority over 3D-QSAR[57-58].
In classical 2D-QSAR, the structural similarity i.e., congeneric series of compounds is a strict criterion (chemical diversity must be limited to substitution alone and parent structure must be the same for all compounds). The more number of compounds used in the dataset, the better the results and statistically significant QSAR model. The series used in this study contains 268 congeneric molecules which are quite good in a number. Furthermore, the activity difference (log difference) was found to be 3.658 which are worthy to choose tetrahydropyrido-pyrazole as a scaffold. No 2D-QSAR analysis has been done on this scaffold which created my interest to correlate various physicochemical parameters to the anti-rheumatic activity for the design of some novel tetrahydropyrido-pyrazole derivatives.
Apart from this, highly potent non-covalent inhibitors of human cathepsin S based on tetrahydropyrido-pyrazole heterocycle has been already reported with good in vitro potency against the enzyme, as measured in the enzymatic assay (hCatS Inhibitory Concentration (IC50)) and an invariant chain degradation cellular assay (JY Li IC50)[59-64]. All the above-listed factors were responsible for the selection of tetrahydropyrido-pyrazole scaffolds as cathepsin S inhibitors.
QSAR model can be developed by using Multiple Linear Regression (MLR), Partial Least Squares (PLS) and Artificial Neural Network (ANN). MLR is the most common form of linear regression analysis.
As a predictive analysis, MLR is used to explain the relationship between one continuous dependent variable and two or more independent variables[65]. PLS is multivariate analysis. It is based on principal component analysis. It carries out regression and gives the maximum correlation between the principal components (independent variables) and the dependent variable. The linear equation indicates the relationship between a dependent (activity) variable Y and independent variables X (latent variables or principal components)[65]. Regression analysis and the method of least squares entered the social sciences with the pioneering work by Legendre in 1805 and by Karl Gauss in 1809. Karl Pearson in 1930 has put regression and multiple regressions on a firm mathematical footing[66-68]. The first publication of PLS to regression was given by Harman Wold[69].
Materials and Methods
The project was done at Banasthali Vidyapith University, Jaipur, Rajasthan by Sneha Kushwaha, as a part of the M. Pharm project from July 2013 to June 2014. QSAR model was developed using MLR, PLS and ANN.
Generation of 3D structures:
All the structures of tetrahydropyrido-pyrazole derivatives mentioned in the literature[60-64,70-73] were sketched using Chem Draw Ultra 12.0 software.
The structures were imported on the TSAR worksheet (version 3.3, Accelrys Inc., Oxford, England). The series had five major substituents which were defined by selecting the Whole Molecule|Structures|Define substituents option in the TSAR worksheet toolbar. The molecules now undergo structure optimization. Using CORINA, 2D structures were converted into their 3D forms. It predicts 3D coordinates directly from the molecule’s structural formula. It generates one low-energy conformation for each input structure by default. It is an automatic 3D model building kit[74].
Partial atomic charge calculation of a molecule has been done by using the "charge-2-derive charges" option which is a prerequisite for several structural manipulations. Total molecular energies were calculated using COSMIC by summing all the valence and non-bonded terms for all appropriate sets of atoms. The force-field provided by COSMIC for energy calculations confirms that only the more energetically genuine conformation is considered[65]. Now, the activity data of 268 compounds have been imported into the TSAR worksheet[75]. The inverse logarithmic values log (1/IC50) is used so that higher values are obtained for more effective analogs.
Calculation of the molecular descriptors:
Molecular descriptors were now calculated using TSAR 3.3 software. More than 1250 descriptors were calculated for generating a QSAR model namely molecular attributes molecular indices-topological, connectivity, shape indices, atom counts and Variational Approach for Markov Processes (VAMP).
Data reduction:
Large data set of independent variables may increase the risk of over fitting the data. Thus, there is a significant need to minimize the data pool to eliminate the chance correlation. Descriptors having constant values were deleted initially. Data was reduced by pair-wise correlation. Forward and backward elimination methods were used for the inclusion or rejection of descriptors[76]. A correlation matrix was used to reduce the number of descriptors and to identify the best subset of descriptors with minimum inter-correlation[65].
Next, the data reduction was performed based on t-values by using the backward elimination technique. The stepwise regressions were developed. The descriptors with lower t-values were discarded[65]. Finally, a minimum of three to four descriptors were left which generated good statistical data. Four independent molecular descriptors-Dipole moment Z component (substituent 1), bond dipole moment (substituent 5), log p (substituent 1) and kier chi 5 (path) indexes (whole molecule) were retrieved.
Data set preparation:
The structures of the series were randomly divided into training and test set. The training set consisted of 179 compounds and 85 compounds were included in the test set. The training set was used to generate linear models so that an accurate relationship could be found between structures and biological activity. The molecules of the test set checked the predictive power of the developed model.
Model development and validation:
Model development was done by MLR and validated by PLS and ANN. Various MLR models were generated. The generated models were validated using both internal and external validation techniques. Internal validation was done by applying cross-validation analysis using the Leave One Out (LOO) method. External validation was done using the model developed by the training set through which activities of the test set molecules were predicted[77].
Results and Discussion
After data reduction, four independent molecular descriptors viz., dipole moment Z component (substituent 1), bond dipole moment (substituent 5), log P (substituent 1) and kier chi 5 (path) indexes (whole molecule) were left with high correlation with the dependent variable i.e. the biological activity. The statistical values of the regression analysis of this model showed poor predictive ability. The improved model was obtained after data reduction and by removing four compounds-D33, H20, H49, I15 as outliers as shown in Table 1.
| S.no. | Statistical tests | Values before data reduction | Values after MLR |
|---|---|---|---|
| 1 | s value | 0.46 | 0.349 |
| 2 | f value | 58.67 | 114.706 |
| 3 | F probability | 1.7e-035 | 0 |
| 4 | Regression coefficient, r | 0.686 | 0.85 |
| 5 | r2 | 0.471 | 0.725 |
| 6 | Cross validation, r2(cv) | 0.451 | 0.709 |
| 7 | Residual sum of squares | 56.0932 | 21.2565 |
| 8 | Predictive sum of squares | 58.1726 | 22.4783 |
Table 1: Statistical Values Obtained before Data Reduction and after performing MLR Analysis.
The value of correlation coefficient (r2)=0.725 means that the MLR equation accounts for 72.5 % variance in the biological activity which depicts a quite reasonable fit. The cross-validation regression coefficient (r) is greater than 0.6 and the difference between r2 (0.725) and cross-validated correlation coefficient (r2cv) (0.709) is quite small which indicates the good internal predictive ability of the model. Fischer statistic (f) is the measure of the probability of no chance correlation. Fischer statistic value (114.706) was also found significant. It is the ratio of the variance explained by the model and the variance due to the regression error. The high value of F reflects the statistical significance of the model. The value of standard error, (0.34 s) is significantly low for the regression to be significant. It measures the quality of the fit of the model.
The correlation between parameters used and the biological activity is given in Table 2. The statistical significance of the descriptors used in the final QSAR model is given in Table 3. The parameters with t-values greater than 2 indicate their significance in the model. The four highly correlated descriptors were used to generate the regression equation as shown below and analyzed for their relative impacts on the activity of the compounds.
| Log (1/IC50 ) | X1: Dipole moment Z component (Subst. 1) | X2: Bond dipole moment (Subst.5) | X3: log P (Subst.1) | X4: Kier Chi5 (path) index (Whole molecule) | |
|---|---|---|---|---|---|
| Log (1/IC50 ) | 1 | -0.44998 | -0.28614 | 0.56862 | 0.62961 |
| X1: Dipole moment Z component (substituent 1) | -0.44998 | 1 | -0.13913 | -0.25175 | -0.23871 |
| X2: Bond dipole moment (substituent 5) | -0.28614 | -0.13913 | 1 | 0.13363 | -0.11836 |
| X3: log P (substituent 1) | 0.56862 | -0.25175 | 0.13363 | 1 | 0.27559 |
| X4: Kier Chi5 (path) index (Whole molecule) | 0.62961 | -0.23871 | -0.11836 | 0.27559 | 1 |
Table 2: Correlation Matrix showing Correlation between the Biological Activity and the Molecular Descriptors left after Data Reduction.
| Molecular Descriptors | Abbreviation | Jacknife SE | Covariance SE | t-value |
|---|---|---|---|---|
| Dipole moment Z component (substituent 1) | X1 | 0.019759 | 0.01952 | -6.9393 |
| Bond dipole moment (substituent 5) | X2 | 0.024592 | 0.02227 | -8.1963 |
| Log P (substituent 1) | X3 | 0.032695 | 0.030523 | 10.064 |
| Kier Chi5 (path) index (Whole molecule) | X4 | 0.025731 | 0.026444 | 9.3793 |
| Constant | C | 0.25912 |
Table 3: Statistical Significance of Parameters in the best QSAR Model obtained by MLR.
Original equation (by MLR method)
Y=-0.135×X1-0.183×X2+0.307×X3+0.248×X4-2.581
Standardized equation (by MLR method)
Y=-0.193×S1-0.221×S2+0.282×S3+0.264×S4+0.750
Where X1 is dipole moment Z component, X2 is bond dipole moment, X3 is log P, X4 is kier chi 5 (path) indexes and Y is the biological activity.
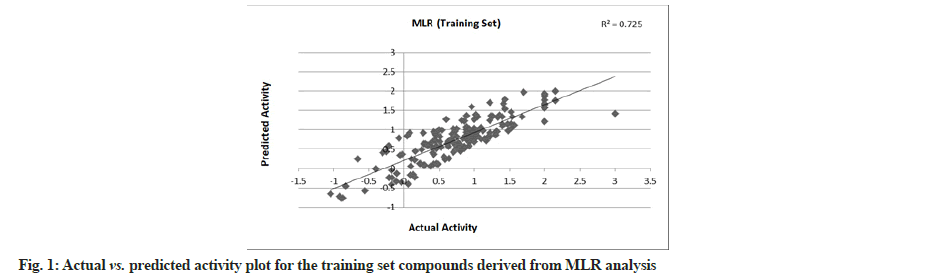
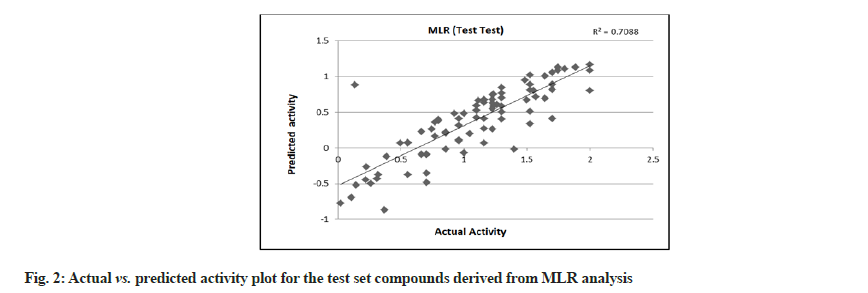
MLR was performed with 179 compounds in the training set and 85 compounds in the test set. The MLR analysis gave satisfactory results with r2=0.725 (training set) and 0.708 (test set) suggesting good external validation shown in fig. 1 and fig. 2. To confirm the liability of the generated model, PLS analysis was performed using the same data set. Both MLR and PLS should have comparable results[78,79]. Table 4 shows the results of PLS. This signifies a 72.387 % variance (greater than 0.6) in the biological activity. A small difference between r2 and r2 cv predicts the good internal predictive ability of the developed model.
| Statistical significance | Fraction of Variance explained, r2 | r2cv | Residual sum of squares | Predictive sum of squares |
|---|---|---|---|---|
| 0.98799 | 0.72387 | 0.71265 | 49.152 | 51.149 |
Table 4: Statistical test Set Values of the Model developed by PLS Analysis.
The following equation represents the PLS equation (2)
Y=-0.131×X1-0.178×X2+0.289×X3+0.267×X4-2.774
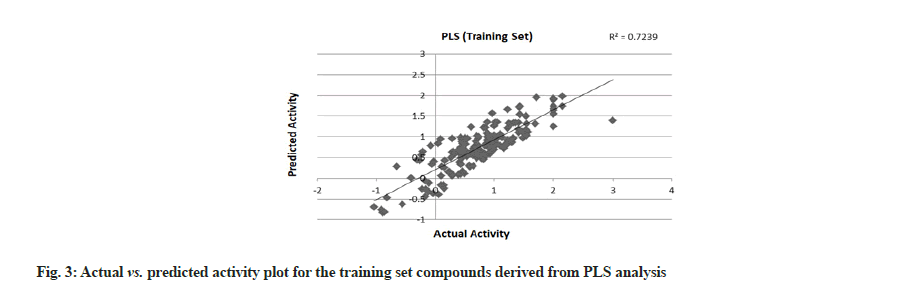
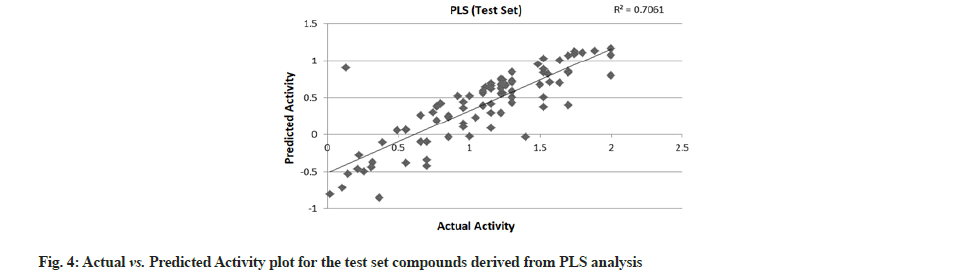
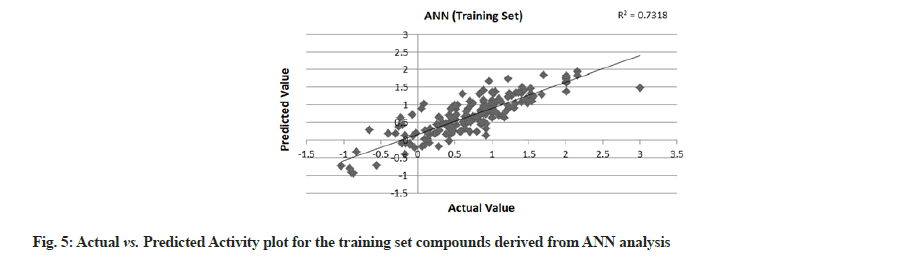
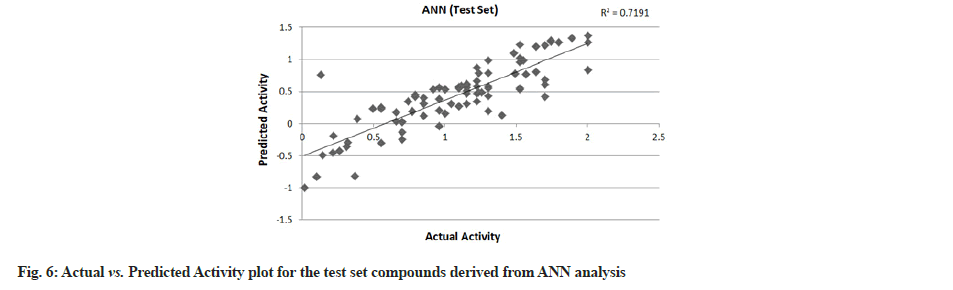
PLS showed perfect results with r2=0.723 (training set) and 0.706 (test set) which further suggested the good external prediction shown in fig. 3 and fig. 4. Further validation was done by performing ANN. The ANN results are shown in fig. 5 and fig. 6. The best Root Mean Square (RMS) fit was found to be 0.0737 at 2440 cycles. Net configuration was 4-15-1 and test RMS fit was 0.0804.
Dipole moment Z component (substituent 1), bond dipole moment (substituent 5), log P (substituent 1) and kier chi 5 (path) indexes (whole molecule) were the inputs and negative log IC50 values were the output for ANN model. As the model suggests, cathepsin S inhibitor activity increases with the increase in log P (substituent 1) and kier chi 5 (path) index (whole molecule) and decreases with an increase in dipole moment Z component (substituent 1) and bond dipole moment (substituent 5). The first descriptor dipole moment Z component (substituent 1) explains the charge distribution and orientation behavior of the molecule. The second descriptor bond dipole moment (substituent 5) uses the concept of electric dipole moment and measures the polarity of a chemical bond within a molecule. Both are negatively correlated with the biological activity as depicted by the plot dependency graphs.
This indicates that adding such groups in a molecule or a lead compound will lead to the increased polarity of the molecule and thus decrease the biological activity. This clearly shows that the active site of cathepsin S enzyme will show some hydrophobic pockets to have hydrophobic interactions. It also provides the fact that the active site of cathepsin S enzyme is lipophilic in nature. The third descriptor log P (substituent 1) explains the lipophilic character of the molecule. The descriptor is positively correlated with the biological activity which is revealed by the plot dependency graph. The less polar groups when introduced will tend to increase the biological activity. It clearly explains that bulky hydrophobic groups at the R1 position are an essential requirement for the cathepsin S inhibitor activity.
In the current study, the groups attached at the R1 position are morpholine, piperidine, or piperazine or substitutions on these rings. Morpholine ring has the presence of a weak basic nitrogen atom and an oxygen atom at the opposite position provides a peculiar pKa value and a flexible conformation to the ring. This allows it to take part in several lipophilic-hydrophilic interactions and to improve blood solubility and permeability of the overall structure, thus enhancing biological activity[80-83]. Piperazine ring has two primary nitrogen atoms which improve the pharmacokinetic features of drug candidates due to their appropriate pKa. These nitrogen sites lead to the essential increase in water solubility of the drug-like molecules, thus enhancing the bioavailability. Piperazine side chains serve as a handle to modulate lipophilicity. It maintains a balance between pharmacodynamics and pharmacokinetic profiles of drug-like molecules and high affinity for their targets, thus increasing biological activity[84,85]. Piperidine ring from piperidine based analogs may enhance the important pharmacokinetic properties such as lipophilicity and metabolic stability when attached to molecular scaffolds. Thus, the nature of all the three descriptors clearly explains the hydrophobic nature of the active site of the target-cathepsin S enzyme. In addition to this, cathepsin S enzyme contains a hydrophobic core which will facilitate the binding of the lipophilic groups of the molecule[86].
The fourth descriptor is the kier chi 5 (path) indexes (whole molecule) is a well-known topological indices. It explains the atom’s identity, bonding environment and the number of hydrogen bonds. It explains the molecular connectivity of a molecule. As it is positively correlated, the presence of such groups is beneficial for the increase the biological activity as shown by the plot dependency graph. A 2D-QSAR study has been done on a series of tetrahydropyrido-pyrazole based cathepsin S inhibitors. A statistically significant QSAR model was generated. The model has r, r2 and r2cv as 0.85, 0.72 and 0.71 respectively. Dipole moment Z component (substituent 1), bond dipole moment (substituent 5), log P (substituent 1) and kier chi 5 (path) indexes (whole molecule) contributed to the inhibitory activity. Thus, this study will facilitate to design of new cathepsin S inhibitors with increased potency.
Acknowledgement:
The author thanks the vice chancellor for fulfilling all the requirements and Banasthali Vidyapith for providing all the computational resources.
Conflict of interest:
The authors declare no conflict of interest in this study.
References
- Bromme D, Wilson, S: Parks WC, Mecham RP editors. Extracellular matrix degradation. Canada: Springer Link; 2011. p. 23-51.
- Reiser J, Adair B, Reinheckel T. Specialized roles for cysteine cathepsins in health and disease. J Clin Invest 2010;120(10):3421-31.
[Crossref] [Google Scholar] [PubMed]
- Guha S, Padh H. Cathepsins: Fundamental effectors of endolysosomal proteolysis. Indian J Biochem Biophys 2008;45(2):75-90.
[Google Scholar] [PubMed]
- Mohamed MM, Sloane BF. Cysteine cathepsins: Multifunctional enzymes in cancer. Nat Rev Cancer 2006;6(10):764-75.
[Crossref] [Google Scholar] [PubMed]
- Li YY, Fang J, Ao GZ. Cathepsin B and L inhibitors: A patent review (2010-present). Exp Opin Ther Pat 2017;27(6):643-56.
[Crossref] [Google Scholar] [PubMed]
- Sena BF, Figueiredo JL, Aikawa E. Cathepsin S as an inhibitor of cardiovascular inflammation and calcification in chronic kidney disease. Front Cardiovasc Med 2018;4:88.
[Crossref] [Google Scholar] [PubMed]
- Van Dalen FJ, Bakkum T, Van Leeuwen T, Groenewold M, Deu E, Koster AJ, et al. Application of a highly selective cathepsin S two-step activity-based probe in multicolor bio-orthogonal correlative light-electron microscopy. Front Chem 2021;8:628433.
- Huang CC, Lee CC, Lin HH, Chang JY. Cathepsin S attenuates endosomal EGFR signalling: A mechanical rationale for the combination of cathepsin S and EGFR tyrosine kinase inhibitors. Sci Rep 2016;6(1):29256.
[Crossref] [Google Scholar] [PubMed]
- Löser R, Pietzsch J. Cysteine cathepsins: Their role in tumor progression and recent trends in the development of imaging probes. Front Chem 2015;3:37.
[Crossref] [Google Scholar] [PubMed]
- Huang CC, Lee CC, Lin HH, Chen MC, Lin CC, Chang JY. Autophagy-regulated ROS from xanthine oxidase acts as an early effector for triggering late mitochondria-dependent apoptosis in cathepsin S-targeted tumor cells. PloS One 2015;10(6):e0128045.
[Crossref] [Google Scholar] [PubMed]
- Hsieh MJ, Lin CW, Chen MK, Chien SY, Lo YS, Chuang YC, et al. Inhibition of cathepsin S confers sensitivity to methyl protodioscin in oral cancer cells via activation of p38 MAPK/JNK signaling pathways. Sci Rep 2017;7(1):45039.
[Crossref] [Google Scholar] [PubMed]
- Zhang L, Wang H, Xu J. Cathepsin S as a cancer target. Neoplasma 2015;62(1):16-26.
[Crossref] [Google Scholar] [PubMed]
- da Costa AC, Santa-Cruz F, Mattos LA, Aquino MA, Martins CR, Ferraz ÁA, et al. Cathepsin S as a target in gastric cancer. Mol Clin Oncol 2020;12(2):99-103.
[Crossref] [Google Scholar] [PubMed]
- Liu WL, Liu D, Cheng K, Liu YJ, Xing S, Chi PD, et al. Evaluating the diagnostic and prognostic value of circulating cathepsin S in gastric cancer. Oncotarget 2016;7(19):28124.
[Crossref] [Google Scholar] [PubMed]
- Wilkinson RD, Young A, Burden RE, Williams R, Scott CJ. A bioavailable cathepsin S nitrile inhibitor abrogates tumor development. Mol Cancer 2016;15(1):1-1.
- Weitoft T, Larsson A, Manivel VA, Lysholm J, Knight A, Rönnelid J. Cathepsin S and cathepsin L in serum and synovial fluid in rheumatoid arthritis with and without autoantibodies. Rheumatology 2015;54(10):1923-8.
[Crossref] [Google Scholar] [PubMed]
- Memmert S, Damanaki A, Nogueira AV, Eick S, Nokhbehsaim M, Papadopoulou AK, et al. Role of cathepsin S in periodontal inflammation and infection. Mediators Inflamm 2017;2017:4786170.
[Crossref] [Google Scholar] [PubMed]
- Ainscough JS, Macleod T, McGonagle D, Brakefield R, Baron JM, Alase A, et al. Cathepsin S is the major activator of the psoriasis-associated proinflammatory cytokine IL-36γ. Proc Natl Acad Sci 2017;114(13):E2748-57.
[Crossref] [Google Scholar] [PubMed]
- Andrault PM, Samsonov SA, Weber G, Coquet L, Nazmi K, Bolscher JG, et al. Antimicrobial peptide LL-37 is both a substrate of cathepsins S and K and a selective inhibitor of cathepsin L. Biochemistry 2015;54(17):2785-98.
[Crossref] [Google Scholar] [PubMed]
- Brown R, Nath S, Lora A, Samaha G, Elgamal Z, Kaiser R, et al. Cathepsin S: Investigating an old player in lung disease pathogenesis, comorbidities, and potential therapeutics. Respir Res 2020;21(1):1-7.
[Crossref] [Google Scholar] [PubMed]
- Wartenberg M, Andrault PM, Saidi A, Bigot P, Nadal-Desbarats L, Lecaille F, et al. Oxidation of cathepsin S by major chemicals of cigarette smoke. Free Radic Biol Med 2020;150:53-65.
[Crossref] [Google Scholar] [PubMed]
- Andrault PM, Schamberger AC, Chazeirat T, Sizaret D, Renault J, Staab-Weijnitz CA, et al. Cigarette smoke induces overexpression of active human cathepsin S in lungs from current smokers with or without COPD. Am J Physiol Lung Cell Mol Physiol 2019;317(5):L625-38.
[Crossref] [Google Scholar] [PubMed]
- Nakajima T, Nakamura H, Owen CA, Yoshida S, Tsuduki K, Chubachi S, et al. Plasma cathepsin S and cathepsin S/cystatin C ratios are potential biomarkers for COPD. Dis Markers 2016;2016:4093870.
[Crossref] [Google Scholar] [PubMed]
- Zhou PP, Zhang WY, Li ZF, Chen YR, Kang XC, Jiang YX. Association between SNPs in the promoter region in cathepsin S and risk of asthma in Chinese Han population. Eur Rev Med Pharmacol Sci 2016;20(10):2070-6.
[Google Scholar] [PubMed]
- Small DM, Brown RR, Doherty DF, Abladey A, Zhou-Suckow Z, Delaney RJ, et al. Targeting of cathepsin S reduces cystic fibrosis-like lung disease. Eur Respir J 2019;53(3):1801523.
[Crossref] [Google Scholar] [PubMed]
- Sena BF, Figueiredo JL, Aikawa E. Cathepsin S as an inhibitor of cardiovascular inflammation and calcification in chronic kidney disease. Front Cardiovasc Med 2018;4:88.
[Crossref] [Google Scholar] [PubMed]
- Figueiredo JL, Aikawa M, Zheng C, Aaron J, Lax L, Libby P, et al. Selective cathepsin S inhibition attenuates atherosclerosis in apolipoprotein E–deficient mice with chronic renal disease. Am J Pathol 2015;185(4):1156-66.
[Crossref] [Google Scholar] [PubMed]
- Ahmad S, Siddiqi MI. Insights from molecular modeling into the selective inhibition of cathepsin S by its inhibitor. J Mol Model 2017;23(3):1-7.
- Figueiredo JL, Aikawa M, Zheng C, Aaron J, Lax L, Libby P, et al. Selective cathepsin S inhibition attenuates atherosclerosis in apolipoprotein E–deficient mice with chronic renal disease. Am J Pathol 2015;185(4):1156-66.
[Crossref] [Google Scholar] [PubMed]
- Wu H, Du Q, Dai Q, Ge J, Cheng X. Cysteine protease cathepsins in atherosclerotic cardiovascular diseases. J Atheroscler Thromb 2018;25(2):111-23.
[Crossref] [Google Scholar] [PubMed]
- Repnik U, Starr AE, Overall CM, Turk B. Cysteine cathepsins activate ELR chemokines and inactivate non-ELR chemokines. J Biol Chem 2015;290(22):13800-11.
[Crossref] [Google Scholar] [PubMed]
- Hargreaves P, Daoudlarian D, Theron M, Kolb FA, Manchester Young M, Reis B, et al. Differential effects of specific cathepsin S inhibition in biocompartments from patients with primary Sjögren syndrome. Arthritis Res Ther 2019;21(1):175.
[Crossref] [Google Scholar] [PubMed]
- Edman MC, Janga SR, Meng Z, Bechtold M, Chen AF, Kim C, et al. Increased Cathepsin S activity associated with decreased protease inhibitory capacity contributes to altered tear proteins in Sjögren’s Syndrome patients. Sci Rep 2018;8(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Vizovišek M, Fonovi? M, Turk B. Cysteine cathepsins in extracellular matrix remodeling: Extracellular matrix degradation and beyond. Matrix Biol 2019;75:141-59.
[Crossref] [Google Scholar] [PubMed]
- Gupta S, Singh RK, Dastidar S, Ray A. Cysteine cathepsin S as an immunomodulatory target: Present and future trends. Expert Opin Ther Targets 2008;12(3):291-9.
[Crossref] [Google Scholar] [PubMed]
- Chen SJ, Chen LH, Yeh YM, Lin CC, Lin PC, Huang HW, et al. Targeting lysosomal cysteine protease cathepsin S reveals immunomodulatory therapeutic strategy for oxaliplatin-induced peripheral neuropathy. Theranostics 2021;11(10):4672-87.
[Crossref] [Google Scholar] [PubMed]
- Vizovišek M, Vidak E, Javoršek U, Mikhaylov G, Bratovš A, Turk B. Cysteine cathepsins as therapeutic targets in inflammatory diseases. Expert Opin Ther Targets 2020;24(6):573-88.
[Crossref] [Google Scholar] [PubMed]
- Wilkinson RD, Williams R, Scott CJ, Burden RE. Cathepsin S: Therapeutic, diagnostic, and prognostic potential. Biol Chem 2015;396(8):867-82.
[Crossref] [Google Scholar] [PubMed]
- Kumar V, Abbas, AK, Fausto N, Aster JC. Pathologic Basis of Disease. 8th ed: Philadelphia: Sauders Elsevier; 2008.
- Villadangos JA, Bryant RA, Deussing J, Driessen C, Lennon?Duménil AM, Riese RJ, et al. Proteases involved in MHC class II antigen presentation. Immunol Rev 1999;172(1):109-20.
[Crossref] [Google Scholar] [PubMed]
- Riese RJ, Mitchell RN, Villadangos JA, Shi GP, Palmer JT, Karp ER, et al. Cathepsin S activity regulates antigen presentation and immunity. J Clin Invest 1998;101(11):2351-63.
[Crossref] [Google Scholar] [PubMed]
- McDowell SH, Gallaher SA, Burden RE, Scott CJ. Leading the invasion: The role of Cathepsin S in the tumour microenvironment. Biochim Biophys Acta Mol Cell Res 2020;1867(10):118781.
[Crossref] [Google Scholar] [PubMed]
- Yadati T, Houben T, Bitorina A, Shiri-Sverdlov R. The ins and outs of Cathepsins: Physiological function and role in disease management. Cells 2020;9(7):1679.
[Crossref] [Google Scholar] [PubMed]
- Kumar P, Mina U. Life Sciences, Fundamentals and Practices. 4th ed: New Delhi: Pathfinder Publication; 2013. p. 460-515.
- Sadegh-Nasseri S, Kim A. MHC class II auto-antigen presentation is unconventional. Front Immunol 2015;6:372.
[Crossref] [Google Scholar] [PubMed]
- Snir O. Specific autoimmunity in rheumatoid arthritis-T cells, antibodies and genetic regulation. Karolinska Institutet (Sweden); 2011.
- Houtman M, Hesselberg E, Rönnblom L, Klareskog L, Malmström V, Padyukov L. Haplotype-specific expression analysis of MHC class II genes in healthy individuals and rheumatoid arthritis patients. Front Immunol 2021;12:3315.
[Crossref] [Google Scholar] [PubMed]
- Trier N, Izarzugaza J, Chailyan A, Marcatili P, Houen G. Human MHC-II with shared epitope motifs are optimal Epstein-Barr virus glycoprotein 42 ligands-relation to rheumatoid arthritis. Int J Mol Sci 2018;19(1):317.
[Crossref] [Google Scholar] [PubMed]
- Hansch C, Fujita T. p-σ-π Analysis, a method for the correlation of biological activity and chemical structure. J Am Chem Soc 1964;86(8):1616-26.
- Jamloki A, Karthikeyan C, Sharma SK. QSAR studies on some GSK-3α inhibitory 6-aryl-pyrazolo-(3, 4-b) pyrimidines. Asian J Biochem 2006;1:236-43.
- Thomas, G. Medicinal Chemistry: An Introduction. 2nd ed: UK: Wiley; 2008. p. 646.
- Gupta SP, Kumaran S. Quantitative structure-activity relationship studies on benzodiazepine hydroxamic acid inhibitors of matrix metalloproteinases and tumour necrosis factor-α converting enzyme. Asian J Biochem 2006;1(1): 47-56.
[Crossref]
- Sahdev AK, Sethi B, Rawat SL, Singh A, Anand N. A review article role of QSAR: Significance and uses in molecular design. J Emerg Technol Innov Res 2018;5(1):426-35.
- Hoffman B, Cho SJ, Zheng W, Wyrick S, Nichols DE, Mailman RB, et al. Quantitative structure−Activity relationship modeling of dopamine d1 antagonists using comparative molecular field analysis, genetic algorithms−Partial least-squares and K nearest neighbor methods. J Med Chem 1999;42(17):3217-26.
[Crossref] [Google Scholar] [PubMed]
- Shen M, LeTiran A, Xiao Y, Golbraikh A, Kohn H, Tropsha A. Quantitative structure activity relationship analysis of functionalized amino acid anticonvulsant agents using k nearest neighbor and simulated annealing PLS methods. J Med Chem 2002;45(13):2811-23.
[Crossref] [Google Scholar] [PubMed]
- Rogers D, Hopfinger AJ. Application of genetic function approximation to quantitative structure-activity relationships and quantitative structure-property relationships. J Chem Inf Comput Sci 1994;34(4):854-66.
- Golbraikh A, Bonchev D, Tropsha A. Novel chirality descriptors derived from molecular topology. J Chem Inf Comput Sci 2001;41(1):147-58.
[Crossref] [Google Scholar] [PubMed]
- Zheng W, Tropsha A. Novel variable selection quantitative structure-property relationship approach based on the k-nearest-neighbor principle. J Chem Inf Comput Sci 2000;40(1):185-94.
- Thurmond RL, Beavers MP, Cai H, Meduna SP, Gustin DJ, Sun S, et al. Nonpeptidic, noncovalent inhibitors of the cysteine protease cathepsin S. J Med Chem 2004;47(20):4799-801.
[Crossref] [Google Scholar] [PubMed]
- Wiener JJ, Wickboldt AT, Nguyen S, Sun S, Rynberg R, Rizzolio M, et al. Pyrazole-based arylalkyne cathepsin S inhibitors. Part III: Modification of P4 region. Bioorg Med Chem Lett 2013;23(4):1070-4.
[Crossref] [Google Scholar] [PubMed]
- Lee-Dutra A, Wiener DK, Arienti KL, Liu J, Mani N, Ameriks MK, et al. Discovery and SAR of novel pyrazole-based thioethers as cathepsin S inhibitors: Part 1. Bioorg Med Chem Lett 2010;20(7):2370-4.
[Crossref] [Google Scholar] [PubMed]
- Wiener JJ, Wickboldt Jr AT, Wiener DK, Lee-Dutra A, Edwards JP, Karlsson L, et al. Discovery and SAR of novel pyrazole-based thioethers as cathepsin S inhibitors. Part 2: Modification of P3, P4, and P5 regions. Bioorg Med Chem Lett 2010;20(7):2375-8.
- Wiener DK, Lee-Dutra A, Bembenek S, Nguyen S, Thurmond RL, Sun S, et al. Thioether acetamides as P3 binding elements for tetrahydropyrido-pyrazole cathepsin S inhibitors. Bioorg Med Chem Lett 2010;20(7):2379-82.
[Crossref] [Google Scholar] [PubMed]
- Ameriks MK, Axe FU, Bembenek SD, Edwards JP, Gu Y, Karlsson L, et al. Pyrazole-based cathepsin S inhibitors with arylalkynes as P1 binding elements. Bioorg Med Chem Lett 2009;19(21):6131-4.
[Crossref] [Google Scholar] [PubMed]
- Paliwal SK, Pandey A, Paliwal S. Quantitative structure activity relationship analysis of N-(mercaptoalkanoyl)-and [(acylthio) alkanoyl] glycine derivatives as ACE inhibitors. Am J Drug Discov Dev 2011;1(2):85-104.
- Magnello ME. Karl Pearson and the establishment of mathematical statistics. Int Stat Rev 2009;77(1):3-29.
- Stanton JM. Galton, Pearson, and the peas: A brief history of linear regression for statistics instructors. J Stat Edu 2001;9(3):1-13.
- Panayotova P, Pearson Karl. SAGE Research Methods Foundations; 2020.
- Wold S, Ruhe A, Wold H, Dunn, Iii WJ. The collinearity problem in linear regression. The partial least squares (PLS) approach to generalized inverses. SIAM J Sci Stat Comput 1984;5(3):735-43.
- Ameriks MK, Cai H, Edwards JP, Gebauer D, Gleason E, Gu Y, et al. Pyrazole-based arylalkyne cathepsin S inhibitors. Part II: Optimization of cellular potency. Bioorg Med Chem Lett 2009;19(21):6135-9.
[Crossref] [Google Scholar] [PubMed]
- Gustin DJ, Sehon CA, Wei J, Cai H, Meduna SP, Khatuya H, et al. Discovery and SAR studies of a novel series of noncovalent cathepsin S inhibitors. Bioorg Med Chem Lett 2005;15(6):1687-91.
[Crossref] [Google Scholar] [PubMed]
- Grice CA, Tays K, Khatuya H, Gustin DJ, Butler CR, Wei J, et al. The SAR of 4-substituted (6, 6-bicyclic) piperidine cathepsin S inhibitors. Bioorg Med Chem Lett 2006;16(8):2209-12.
[Crossref] [Google Scholar] [PubMed]
- Wei J, Pio BA, Cai H, Meduna SP, Sun S, Gu Y, et al. Pyrazole-based cathepsin S inhibitors with improved cellular potency. Bioorg Med Chem Lett 2007;17(20):5525-8.
[Crossref] [Google Scholar] [PubMed]
- Sadowski J, Gasteiger J. From atoms and bonds to three-dimensional atomic coordinates: Automatic model builders. Chem Rev 1993;93(7):2567-81.
- Suh JT, Skiles JW, Williams BE, Youssefyeh RD, Jones H, Loev B, et al. Angiotensin-converting enzyme inhibitors. New orally active antihypertensive (mercaptoalkanoyl)-and [(acylthio) alkanoyl] glycine derivatives. J Med Chem 1985;28(1):57-66.
[Crossref] [Google Scholar] [PubMed]
- Patil RB, Sawant SD. 2D QSAR Analysis on B-Ring Trifluromethylated chromenone analogues as anticancer agents. Int J Adv Pharm Biol Chem 2012;1(1):72-80.
- Sharma MC, Smita S. 2D Qsar study of 7-methyljuglone derivatives: An approach to design anti-tubercular agents. J Pharmacol Toxicol 2011;6(5):493-504.
- Paliwal S, Narayan A, Paliwal S. Quantitative structure activity relationship analysis of dicationic diphenylisoxazole as potent anti-trypanosomal agents. QSAR Comb Sci 2009;28(11):1367-75.
- Paliwal SK, Pal M, Siddiqui AA. Quantitative structure activity relationship analysis of angiotensin II AT1 receptor antagonists. Med Chem Res 2010;19(5):475-89.
- Lenci E, Calugi L, Trabocchi A. Occurrence of morpholine in central nervous system drug discovery. ACS Chem Neurosci 2021;12(3):378-90.
- Kumari A, Singh RK. Morpholine as ubiquitous pharmacophore in medicinal chemistry: Deep insight into the structure-activity relationship (SAR). Bioorg Chem 2020;96:103578.
[Crossref] [Google Scholar] [PubMed]
- Kourounakis AP, Xanthopoulos D, Tzara A. Morpholine as a privileged structure: A review on the medicinal chemistry and pharmacological activity of morpholine containing bioactive molecules. Med Res Rev 2020;40(2):709-52.
[Crossref] [Google Scholar] [PubMed]
- Tzara A, Xanthopoulos D, Kourounakis AP. Morpholine as a scaffold in medicinal chemistry: An update on synthetic strategies. ChemMedChem. 2020;15(5):392-403.
[Crossref] [Google Scholar] [PubMed]
- Zhang RH, Guo HY, Deng H, Li J, Quan ZS. Piperazine skeleton in the structural modification of natural products: A review. J Enzyme Inhib Med Chem 2021;36(1):1165-97.
[Crossref] [Google Scholar] [PubMed]
- Wu YJ. Heterocycles and medicine: A survey of the heterocyclic drugs approved by the US FDA from 2000 to present. Progr Heterocyclic Chem 2012;24:1-53.
- Turk V, Stoka V, Vasiljeva O, Renko M, Sun T, Turk B, et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta 2012;1824(1):68-88.
[Crossref] [Google Scholar] [PubMed]