- *Corresponding Author:
- Nisachon Jangpromma
Department of Integrated Science, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
E-mail: nisaja@kku.ac.th
| Date of Received | 19 September 2020 |
| Date of Revision | 14 August 2021 |
| Date of Acceptance | 09 February 2022 |
| Indian J Pharm Sci 2022;84(1):197-206 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Antimicrobial peptides are documented as having a crucial role in innate immune response. Many reports have studied antimicrobial peptides from the crocodile. Thus, the antimicrobial activity of liver protein extract from Crocodylus siamensis was examined. Our results show that P50 and P80, partially purified crocodile liver protein extract at 45 mg/ml, can inhibit gram-positive bacteria with the highest percentages of inhibition at 48.22 % and 37.54 % respectively, while gram-negative bacteria could be inhibited at 47.31 % and 35.32 %, respectively. However, only P80 was selected for further phagocytosis activity investigation since it showed low cytotoxicity in the macrophage cell line. The results show that P80 can kill bacteria by an intracellular killing process in phagocytic cells of macrophages in a dose-dependent manner. Moreover, the internalization of phagocytosis activity of macrophages was assayed using fluorescein isothiocyanate-labeled Escherichia coli and flow cytometry. The results reveal that P80 can degrade bacteria by inducing phagosome formation in a time and dose-dependent manner (200-800 μg/ml). In these findings, the liver protein extract of Crocodylus siamensis had an enhancing effect on macrophage RAW 264.7 cells in the phagocytosis process.
Keywords
Crocodylus siamensis, flow cytometry, liver protein, macrophage, phagocytosis, phagosomey
Innate immunity refers to the first-line host defense that serves to limit infection in the early hours after exposure to a microorganism invasion. The eradication and clearance of invading pathogens requires the complex coordination of multiple innate immune pathways [1]. Basically, the innate immune system includes diverse cells such as Dendritic Cells (DCs), neutrophils, Natural Killer (NK) cells, Innate Lymphoid Cells (ILCs) and macrophages. The macrophages are a type of differentiated tissue cell that originate from blood monocytes. Their primary function is phagocytosis, which depends largely on cell receptors [2]. It offers several functions such as the removal of cell debris, the killing of pathogenic microorganisms and the ability to present antigens to lymphocytes [3]. Therefore, the activation of macrophages is a key event for effective innate and adaptive immunities. Recently, there is a promising attempt towards exploration of novel compounds from natural sources that modulate the immune response, including active compounds from plants and animals, especially reptiles [4,5].
However, it has long been appreciated that blood contains important elements that mediate rapid responses toward infections. Regarding blood systems, blood samples of reptile species have been studied extensively to elucidate immune responses, in order to understand the evolution of the immune system. The reptiles have a broad innate immune response followed by a more moderate adaptive response and as an ectotherm, their immune response is strongly affected by temperature [4]. Studies over the past decade have revealed that the reptiles innate immune responses encompass diverse groups of molecules that include non‐specific leukocytes, lysozymes and Antimicrobial Peptides (AMPs) [6].
The AMPs, or host-defense peptides, are an important component of the innate immune system and play a critical role in the host defense against pathogenic microbes. A previous report showed that the AMP named LL-37 could enhance phagocytosis of human macrophages [7]. Recently, active AMPs were discovered from a wide range of sources such as invertebrates [8], mammals [9] and reptiles, especially in crocodiles [10]. Crocodiles always have injuries from battles for territory, food and breeding. However, any evidence of crocodile disease or death from infection has not been reported. This phenomenon might raise the hypothesis that crocodiles possess an effective immune system. According to previous studies, Crocodylus siamensis (C. siamensis) expresses a variety of small AMPs from crocodile blood, including crocosin [11], leucrocin [12], cathelicidin [13] and hepcidin [14]. In addition, not only antimicrobial activity has been observed, but also other biological properties of C. siamensis blood have been reported, such as antioxidant [15,16], anti- inflammation [15,17], wound healing [18] and anticancer properties [19].
The protein components in the bloodstream are mostly produced from the liver, an organ that is responsible for synthesizing numerous kinds of proteins, including vital proteins for normal function and marker proteins to indicate liver disease. Many proteins are secreted into the bloodstream including albumin, alpha 1 (α1)- globulins, α2-globulins, beta (β)-globulins, blood- clotting factors and transporter proteins, example lipoproteins and albumin [20]. Interestingly, a novel AMP, named Liver-Expressed Antimicrobial Peptide (LEAP) was discovered in vertebrate livers such as fish [21], carp [22], porcine [23] and human [24]. In addition, the liver protein hydrolysates derived from chicken show antimicrobial and antioxidant activity [25]. Previous studies have shown multiple biological properties from crocodilian blood components [10-12]. However, it is not known whether C. siamensis liver is able to generate potential AMPs and mediate phagocytosis. Thus, the primary goal of this study was to evaluate the effects of crocodile liver protein on antimicrobial and phagocytic activity of macrophages. The results from this study will provide some insights into the potential active protein from crocodile liver for finding an active substance for therapeutic use in infectious diseases.
Materials and Methods
Crocodile liver extract:
The crocodile (C. siamensis) liver was kindly provided by Sriracha Moda Farm (Chon Buri, Thailand). The liver was extracted and fractionated following the method previously described by Walker [26]. The liver was cut into small pieces and ground under liquid nitrogen in a mortar using a pestle. The liver powder was then suspended in 10 mM Phosphate Buffer Saline (PBS, pH 7.4) and homogenized on ice using an ultrasonicator. Crude liver protein extract was centrifuged at 12 000´g for 60 min at 4°. The supernatant was collected and kept in lyophilized powder and then stored at -70°.
Precipitation of crocodile liver protein extract:
The crocodile liver extract was partially purified using the ammonium sulfate precipitation method [27]. Briefly, the lyophilized liver protein was dissolved in 10 mM PBS with a pH of 7.4. The solution was stirred on ice and then ammonium sulfate was gently added to the final concentration to 50 % saturation. After stirring for 2 h, the precipitate was centrifuged at 10 000×g for 20 min at 4°. The pellet was collected and re-suspended in the appropriate buffer. The suspension was further precipitated with 80 % saturated ammonium sulfate and continually stirred for 2 h at 4°. After centrifugation, the pellet was collected and re-suspended in the appropriate buffer. Subsequently, proteins that precipitated at 50 % and 80 % ammonium sulfate saturation were named as the P50 and P80, respectively. Dialysis against double- distilled water at 4° overnight was performed and the protein pattern was analyzed by 15 % Sodium Dodecyl- Sulfate Polyacrylamide Gel Electrophoresis (SDS- PAGE).
Quantitative determination of proteins:
The liver protein concentration was determined by the method of Bradford [28], using bovine serum albumin as the protein standard. After 10 min of incubation at room temperature, the absorbance of the reaction mixture (protein-dye complex) was measured at 595 nm. The protein concentration was then calculated using the standard calibration curve.
Determination of protein pattern:
The SDS-PAGE was performed according to the Laemmli method [29]. The pattern of protein was estimated in 15 % acrylamide gel. Protein samples were prepared by mixing them with 2´ solubilizing buffer (0.5 M Tris (hydroxymethyl) aminomethane Hydrochloride (Tris-HCl) (pH 6.8), 0.5 % (w/v) bromophenol blue, 10 % (v/v) glycerol, 2 % (w/v) Sodium Dodecyl Sulfate (SDS) and 10 % (v/v) β-mercaptoethanol) at a ratio of 1:1 (v/v). The mixtures were then boiled for 5 min. The liver protein extract was separated by SDS-PAGE gel electrophoresis and then protein patterns were visualized using Coomassie Blue R-250 staining. Standard proteins were obtained from a kit (Amersham Bioscience, Sweden).
Refolding gel electrophoresis:
The polyacrylamide gel electrophoresis was performed using a 4 % (w/v) stacking gel and 15 % (w/v) separating gel containing 2 % (w/v) of Micrococcus lysodeikticus (Sigma-Aldrich, United States of America (USA)).20 micrograms of liver protein extract were mixed with 2´ solubilization buffer (62.5 mM Tris-HCl buffer, pH 6.8, 0.006 % (w/v) bromophenol blue as tracking dye, 20 % (v/v) glycerol and 2 % (w/v) SDS), then boiled for 5 min and loaded into each well. The protein sample was run alongside lysozyme as a positive control, using Mini-PROTEAN® Precast Gels (BIO-RAD, USA) at a constant voltage of 150 V until the dye front appeared at the lowest part of the gel. The gel was washed with double-distilled water for 30 min to eliminate residual SDS, submerged in 0.05 M sodium phosphate buffer, pH 7.0 containing 1 % (v/v) Triton X-100 and was shaken until the clear zones appeared.
Antibacterial activity assay:
The antimicrobial activities of the P50 and P80 were investigated against gram-positive (Bacillus cereus (B. cereus) American Type Culture Collection (ATCC) 11778) and gram-negative (Vibrio cholera (V. cholera)) bacteria by the broth-dilution assay using the broadspectrum ampicillin (1 mg/ml) as the positive control [19]. Briefly, 10 μl of the P50 and P80 (1.4-45 μg/ml) were added individually into 100 l of a suspension of bacteria at a starting Optical Density (OD600) nm=0.001 (1×106 colony forming unit (cfu)/ml) in Nutrient Broth (NB) and then incubated at 37 for 16 h. Bacterial growth was measured at OD600 nm using a microplate reader (BIO-RAD, USA). The percentages of bacterial growth inhibition were calculated as follows: Bacterial growth inhibition (%)=100- [(S/N)×100], where S is the OD600 nm of bacteria treated with the liver protein extract samples or positive control and N is the density of 600 nm of untreated bacteria (negative control).
Cell culture and cytotoxicity assay:
Murine macrophage cell lines (RAW 264.7) from the ATCC, USA, were kindly provided by Assistant Professor Dr. Pramote Mahakunakorn, Faculty of Pharmaceutical Sciences, Khon Kaen University, Khon Kaen, Thailand. The cells were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium, supplemented with 10 % heat-inactivated Fetal Bovine Serum (FBS) and 1 % antibiotic:antimycotic (Gibco, USA). Vero cells (normal cells) were cultured in Dulbecco’s Modification of Eagle Medium (DMEM), supplemented with 10 % heat-inactivated FBS and 1 % antibiotic:antimycotic (Gibco, USA). The RAW 264.7 and Vero cells were incubated at 37° in a 5 % Carbon Dioxide (CO2)-humidified atmosphere. The RAW 264.7 and Vero cells (1×105 cells/ml) were seeded on a 96-well plate and allowed to attach at 37° in 5 % CO2 overnight. These cells were incubated with P50 and P80 at different concentrations of 4-1024 g/ml for 24 h. The cell viability was measured after 24 h exposure to P50 and P80 by the 3-(4,5-Dimethylthiazol-2-yl)- 2,5-Diphenyl Tetrazolium Bromide (MTT) assay [30]. The experiments were performed in triplicate. The cell viability of treated cells was evaluated by comparing with the control cells.
Bacterial preparation and Fluorescein Isothiocyanate (FITC) labeling:
Escherichia coli (E. coli) ATCC25922 were grown on a nutrient agar plate for 16-20 h. A single colony was inoculated into the NB and cultured at 37° for 6 h to obtain an exponentially growing culture. The bacteria cells were labeled with FITC as in the previous explanation [31]. Bacteria were washed 3 times in PBS buffer and resuspended in 500 μl of FITC solution (1 mg/ml) with a ratio of 1:1. The solution was incubated at ambient temperature for 60 min in the dark. After that, the bacteria cells were washed with PBS buffer. The FITC-labeled bacteria were diluted to a density of 1×108 cfu/ml in the same buffer prior to further experiments.
Killing assay:
RAW 264.7 cells (1×106 cells/well) were seeded into a 96-well plate and incubated at 37° in a 5 % CO2 incubator overnight to allow cell attachment. The P80 at a concentration of 50-800 μg/ml was treated with RAW 264.7 cells and incubated at 37° in a 5 % CO2 incubator for 8 h. Then 25 μl of 2×107 cfu/ml E. coli cells were added and incubated at 37° in a 5 % CO2 incubator for 2 h. The phagocytosis was stopped by adding ice-cold PBS buffer and macrophage cells were harvested. The collected macrophages were carefully lysed in ice-cold sterile double-distilled water using a pipette and the lysed cells were prepared with tenfold serial dilutions in a range of 10-10-10 dilutions with the appropriate medium. Then 5 μl of each dilution were spotted on an agar plate and incubated at 37° for 14 h. The colonies of each spot were counted as colonyforming units (cfu/ml).
Phagocytosis activity of C. siamensis liver extract on macrophage cells:
Measurement of phagocytosis of FITC-labeled E. coli was carried out as previously described [32]. Briefly, RAW 264.7 cells (1×106 cells) were seeded into a 12- well plate and allowed to attach overnight. The cells were pre-incubated with P80 (200, 400 and 800 μg/ ml) before incubating them with labeled-E. coli cells at a ratio of 1:10. After the incubation, the internalized reaction with labeled-E. coli cells was then stopped with ice-cold PBS buffer (pH 7.4) and the macrophage cells were harvested. After that, the phagocytosis activity was measured in comparison with untreated cells using the cell-sorting flow cytometry. The phagocytosis activity was evaluated using the Side Scatter (SSC) parameter and displayed as a histogram of FITC intensity of phagocytosis cells.
Cell imaging by IN cell analyzer:
RAW 264.7 cells (1×106 cells/well) were seeded into clear 96-well black plates and incubated at 37° in 5 % CO2 overnight. P80 at a concentration of 200, 400 and 800 μg/ml was added and then incubated at 37° in 5 % CO2 for 8 h. The phagocytosis was investigated by adding the pHrodo green E. coli Bioparticle conjugates into each well and incubating for 60 min. After the incubation, the phagocytosis activity was observed immediately and monitored further every 15 min (75, 90, 105 and 120 min) using a GE/IN Cell analyzer 2000 (GE Healthcare, Sweden).
Statistics analysis:
The collected data were presented as the mean±Standard Deviation (SD) for comparison between control and testing groups of triplicate samples. The differences between the mean values of all data were statistically analyzed by the analysis of variance. Statistical significance was accepted at p<0.05.
Results and Discussion
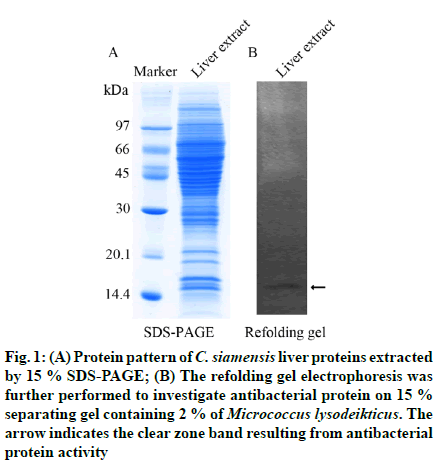
The molecular weight of proteins contained in crude liver protein extract was determined by SDS-PAGE. The protein pattern demonstrated different sizes of proteins ranging over 10-100 kDa (fig. 1A). Since the liver protein extract exhibited an abundance of proteins, the favorable biochemical functions might also be various. However, the antibacterial activity was a key point of this study, thereby the refolding gel was used to investigate the antibacterial activity of liver protein extract against the gram-positive bacteria, Micrococcus lysodeikticus. The clear zone was observed at the protein band of approximately 14 kDa, which indicated antibacterial activity (fig. 1B). However, some antibacterial proteins might be enervated by losing their refolding ability or partially misfolding on the gel. Furthermore, the small active antibacterial peptides (AMPs) might be not detected with 15 % gel, resulting in no sign of antibacterial activity on the refolding gel.
Fig. 1: (A) Protein pattern of C. siamensis liver prote ins extracted by 15 % SDS-PAGE; (B) The refolding gel electrophoresis was further performed to investigate antibacterial protein on 15 % separating gel containing 2 % of Micrococcus lysodeikticus. The arrow indicates the clear zone band resulting from antibacterial protein activity
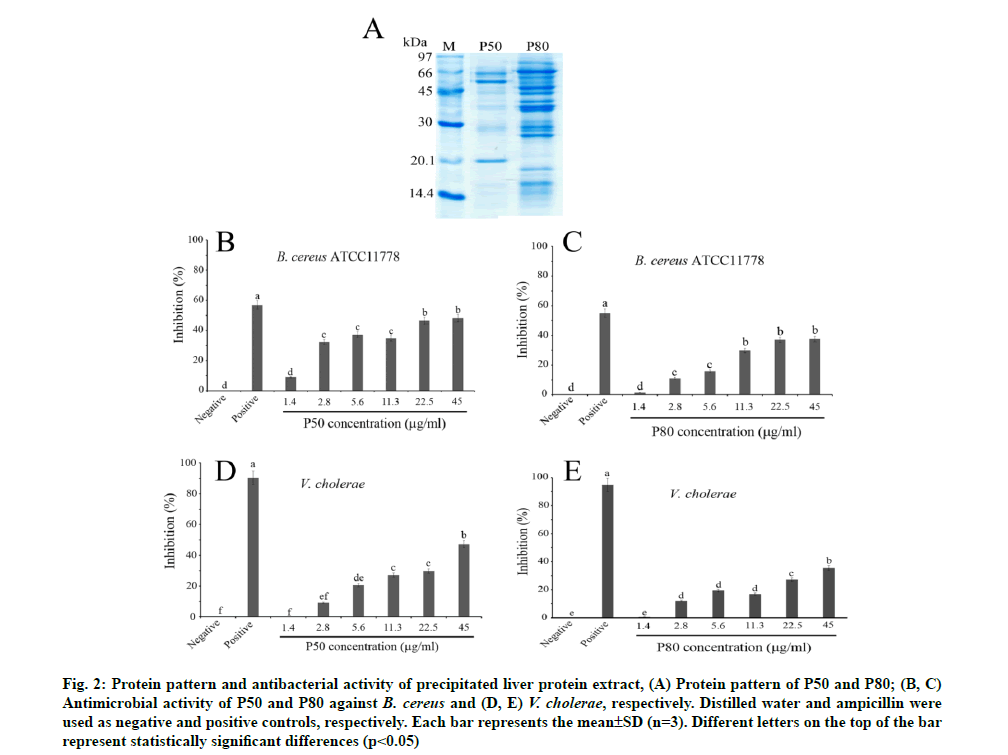
In order to investigate rather active antibacterial proteins and peptides, the crude liver protein extract was precipitated under 50 % (P50) and 80 % (P80) of ammonium sulphate. The results revealed that both P50 and P80 demonstrated higher purified protein patterns than that of crude liver extract. Interestingly, the maintained major proteins appeared in the size ranges of 20.1-66 kDa and 14-80 kDa for P50 and P80, respectively (fig. 2A). The broth dilution assay revealed that at the highest concentration test (45 mg/ml) the percentage of inhibition of P50 and P80 against gram-positive (B. cereus ATCC11778) were at 48.22 % and 37.54 % and against gram-negative (V. cholerae) were at 47.31 % and 35.32 %, respectively (fig. 2B-fig. 2E). The antibacterial activities of both P50 and P80 were also expressed in a dose-dependent manner. Our findings demonstrate that P50 exhibited an excellent antibacterial inhibition more than P80 against B. cereus and V. cholerae (fig. 2). It is important to note that the precipitation with 50 % ammonium sulphate yielded greater total activity of active AMPs than further precipitation with 80 % ammonium sulphate. The higher precipitation level caused excessive precipitates and aggregates, which might maintain an arduous renaturation and misfolding [33], leading to lower activity. In addition, the P80 contained a variety of mixed peptides in a wide range size around 14-80 kDa, thus it might be simply co-aggregation with other proteins, resulting in demonstrating lower activity [34].
Fig. 2: Protein pattern and antibacterial activity of precipitated liver protein extract, (A) Protein pattern of P50 and P80; (B, C) Antimicrobial activity of P50 and P80 against B. cereus and (D, E) V. cholerae, respectively. Distilled water and ampicillin were used as negative and positive controls, respectively. Each bar represents the mean±SD (n=3). Different letters on the top of the bar represent statistically significant differences (p<0.05)
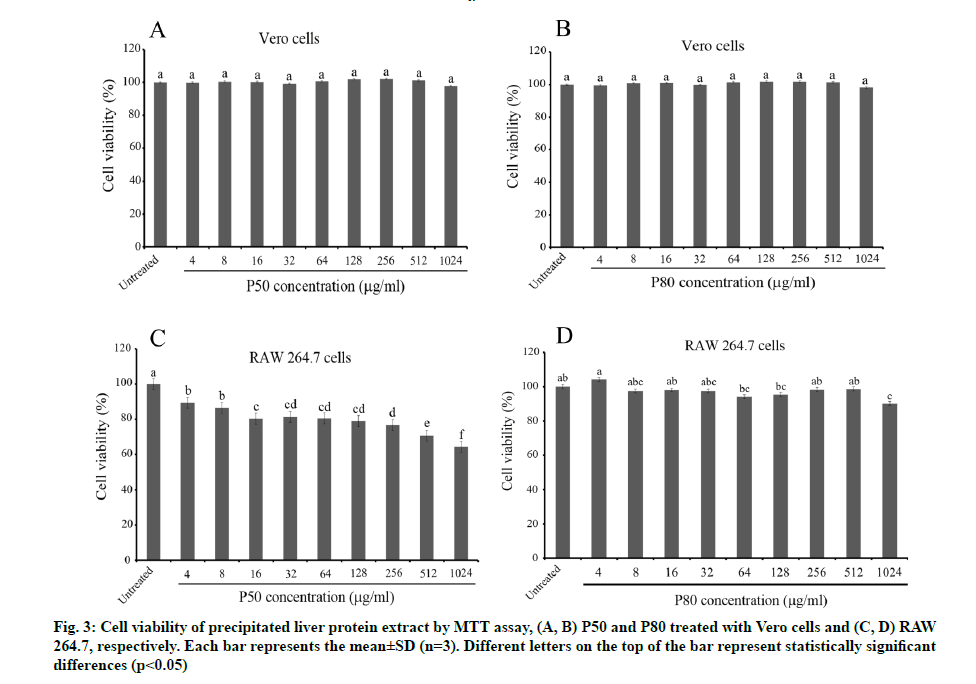
Before the desired effects on the phagocytosis mechanism of macrophages can be investigated, the safety concerns toward macrophages (RAW 264.7) and normal somatic cells (Vero cells) should be determined. The cytotoxicity tests of P50 and P80 (4-1024 µg/ ml) revealed less toxicity on Vero cells without any significant difference compared to the untreated control (fig. 3A and fig. 3B). In contrast, a conspicuous toxicity towards macrophages was presented by P50 in a dose- dependent manner, which expressed lower than 80 % cell viability for the three highest doses (fig. 3C). Interestingly, P80 showed negligible toxicity towards macrophages with higher than 80 % cell viability for all doses (fig. 3D). This finding reveals that some toxic proteins might be misfolded or eliminated at a high concentration of ammonium sulphate, accompanied, nevertheless, with a lower antibacterial activity but less toxicity towards both tested cells, especially macrophages. Due to the lower toxicity of P80 on macrophages, it was chosen to perform all further experiments.
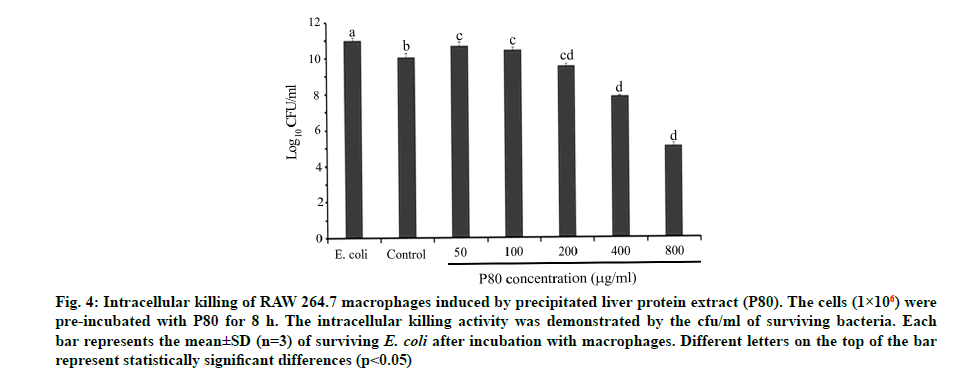
For an investigation of the effect of C. siamensis liver proteins on bacterial phagocytosis, the intracellular killing ability of macrophages was firstly determined. The results demonstrate that the total viable count decreased in a dose-dependent manner for P80 (50-800 µg/ml). Our results show that the dose ranging over 200-800 µg/ml expressed a significantly lower viable count than the control. The intracellular killing assay revealed that the P80 directly affects the intracellular killing of macrophages by dropping the viable counts to a half (at 800 µg/ml) compared with the control (fig. 4). This evidence could be supported by two hypotheses being the P80, a cocktail of potential AMPs from C. siamensis liver extract, expressed strong extracellular and/or intracellular antibacterial activities and the P80 might help to indirectly promote the microbicidal properties through many immunomodulatory compound productions like other AMPs such as cathepsin [34] and cathelicidin LL-37 [35]. However, it is necessary to note that the P80 clearly reinforced the intracellular killing ability of macrophages. In general phagocytic cells such as macrophages express receptors for opsonizing the large particles of antigens (≥0.5 µm) [36,37]. This receptor- mediated process is called phagocytosis and it begins when the phagocytic receptors recognize the bacterial surface. With many signalling cascades, the membrane- bound vacuole termed the phagosome is created and transported to fuse with an endo-lysosome, yielding phagolysosome, which is a degradative organelle with effective microbicidal properties [35]. Therefore, both phagocytosis and intracellular killing comprise the final steps in the resolution of extracellular microbial infections of macrophages.
Fig. 4: Intracellular killing of RAW 264.7 macrophages induced by precipitated liver protein extract (P80). The cells (1×106) were pre-incubated with P80 for 8 h. The intracellular killing activity was demonstrated by the cfu/ml of surviving bacteria. Each bar represents the mean±SD (n=3) of surviving E. coli after incubation with macrophages. Different letters on the top of the bar represent statistically significant differences (p<0.05)
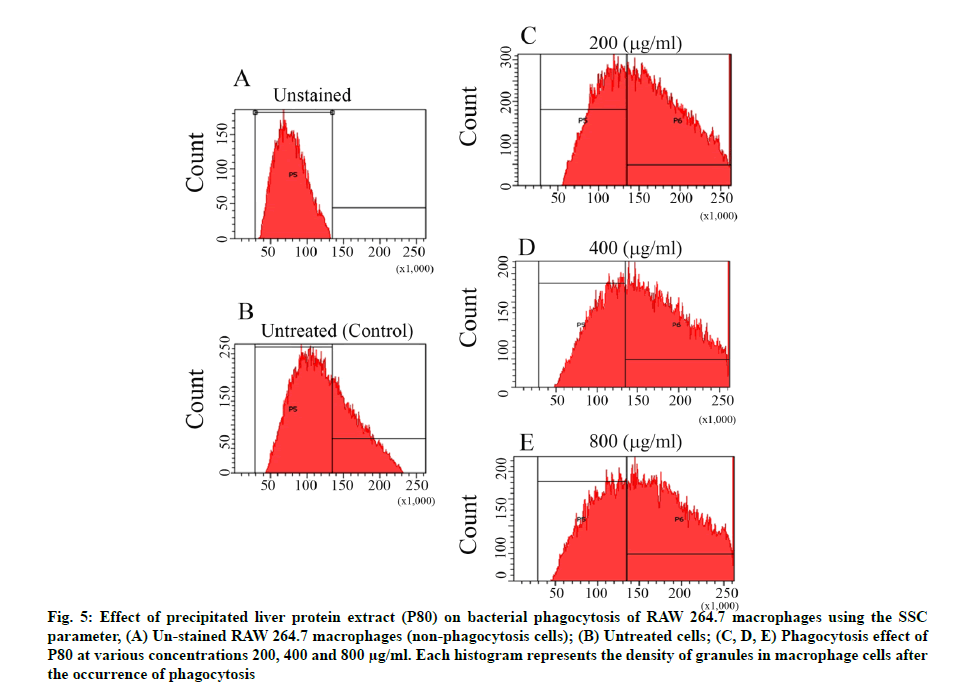
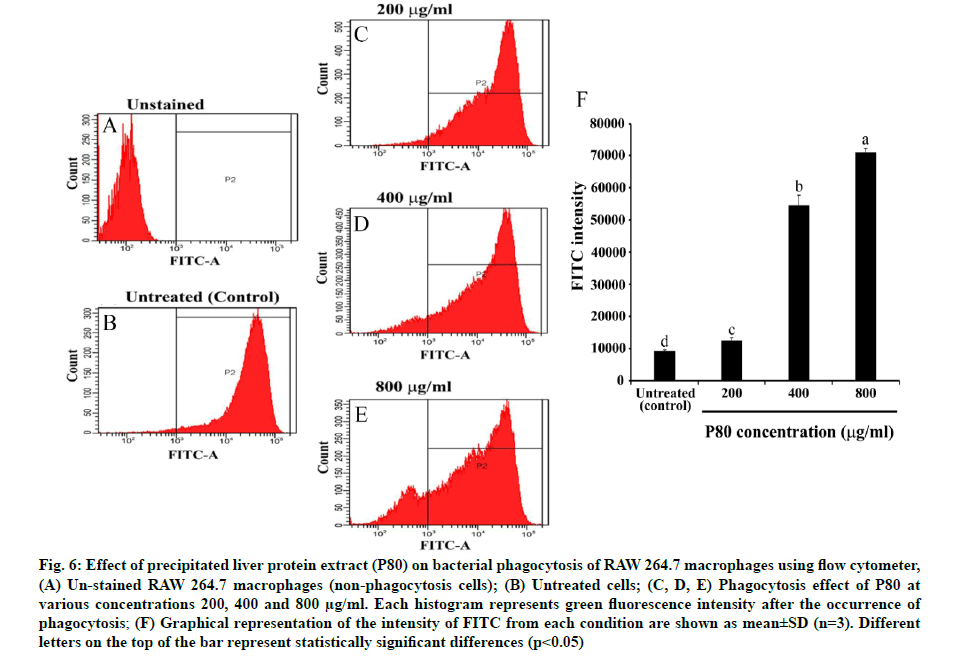
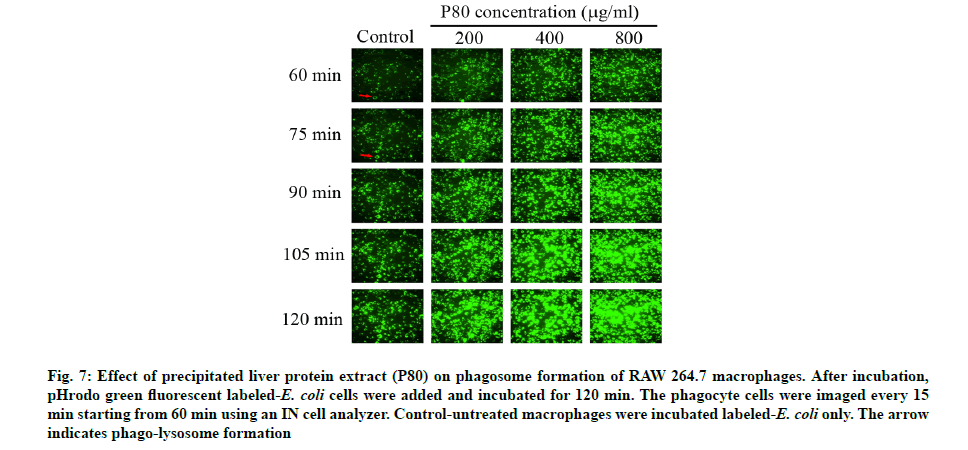
An investigation of P80 on the phagocytic capacity of macrophages was made. The SSC parameter usually aids in determination of the density of macrophage cells or their internal granules, which normally contain the phagocytic bacterial cells resulting from the phagocytosis mechanism. According to fig. 5, the pre-incubation of P80 increased the granule density of macrophages in a dose-dependent manner when compared to the control. Furthermore, the green fluorescence measurement of the FITC asserted that the increasing granules certainly contain the phagocytic FITC-labeled E. coli cells. Interestingly, the broader peaks of the histograms were observed to be in a dose-dependent manner for P80 (fig. 6A-fig. 6E). The quantification intensity of FITC at a 200-800 mg/ml also showed a statistically significant increase from untreated cells in a dose-dependent manner (fig. 6F). This indicates that C. siamensis liver proteins promote the phagosome formation of macrophages. The phagosome formation in macrophages induced by C. siamensis liver proteins (P80) was also explored using an IN cell analyzer. The fluorescence measurement of the pHrodo green revealed the increasing density of the phago-lysosome inside macrophages. The results demonstrate that the increase in P80 concentrations (200-800 µg/ml) and incubation times (60-120 min) support the phago-lysosome formation by expressing higher fluorescence density compared to the control at each time point (fig. 7). In detail, the phagosome itself did not provide microbicidal properties, but when it is fused with endo-lysosome to create an effective degradative organelle termed a phagolysosome, the degradation processes were driven [37,38]. Phagocytic bacteria are commonly killed by two different mechanisms. The first mechanism depends on the presence of oxygen, a respiratory burst and the generation of reactive oxygen species. The second mechanism uses granule- containing proteolytic enzymes to kill microbes [39]. These processes naturally cause an acidic condition (low pH) inside the phagolysosome leading to the activation of the pHrodo green dye. Therefore, phagolysosomes could be distinguished and detected as a bright green fluorescence [40]. Altogether, this evidence shows that the P80 did not only provide antibacterial activity which promotes intracellular killing, but also possesses a phagocytic activity contributing to an increase in phagolysosome formation. The understanding of the phagocytosis mechanism induced by AMPs, which usually possess cationic and hydrophobic properties, remains unclear. Interestingly, there have been some reports revealing that cationic and hydrophobic peptides can promote phagocytosis through the enhancement of the binding between phagocytic receptors with the bacterial surface and the induction of cytokine [41,42]. Moreover, the macrophages engulfment could be indirectly promoted by opsonin molecules (e.g., Immunoglobulin G (IgG) and complement component 3 (c3b)). The opsonin receptor on the macrophage surface can bind to opsonin molecules coating the bacterial surface and induce phagocytosis of the bacteria [43]. Therefore, these components might be contained in C. siamensis liver protein extract, which might support the phagocytosis activity of the AMPs.
Fig. 5: Effect of precipitated liver protein extract (P80) on bacterial phagocytosis of RAW 264.7 m acrophages using the SSC parameter, (A) Un-stained RAW 264.7 macrophages (non-phagocytosis cells); (B) Untreated cells; (C, D, E) Phagocytosis effect of P80 at various concentrations 200, 400 and 800 µg/ml. Each histogram represents the density of granules in macrophage cells after the occurrence of phagocytosis
Fig. 6: Effect of precipitated liver protein extract (P80) on bacterial phagocytosis of RAW 264.7 macrophages using flow cytometer, (A) Un-stained RAW 264.7 macrophages (non-phagocytosis cells); (B) Untreated cells; (C, D, E) Phagocytosis effect of P80 at various concentrations 200, 400 and 800 µg/ml. Each histogram represents green fluorescence intensity after the occurrence of phagocytosis; (F) Graphical representation of the intensity of FITC from each condition are shown as mean±SD (n=3). Different letters on the top of the bar represent statistically significant differences (p<0.05)
Fig. 7: Effect of precipitated liver protein extract (P80) on phagosome formation of RAW 264.7 macrophages. After incubation, pHrodo green fluorescent labeled-E. coli cells were added and incubated for 120 min. The phagocyte cells were imaged every 15 min starting from 60 min using an IN cell analyzer. Control-untreated macrophages were incubated labeled-E. coli only. The arrow indicates phago-lysosome formation
Author’s contributions:
Preeyanan Anwised and Khantheeranut Suttee contributed equally to this work.
Acknowledgements:
This work was financially supported by the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission, Khon Kaen University, Thailand (Grant no. NRU571004). Additionally, we would like to thank the Protein and Proteomics Research Center for Commercial and Industrial Purposes (ProCCI), Department of Biochemistry, Faculty of Science, Khon Kaen University for providing essential laboratory facilities. The authors thank the Sriracha Moda Co. Ltd., Chon Buri, Thailand for providing crocodile livers.
Conflict of interests:
The authors declared no conflict of interest.
References
- Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: A cell biological perspective. Annu Rev Immunol 2015;33:257-90.
[Crossref] [Google Scholar] [PubMed]
- Aderem A, Underhill DM. Mechanisms of phagocytosis in macrophages. Annu Rev Immunol 1999;17(1):593-623.
[Crossref] [Google Scholar] [PubMed]
- Gordon SB, Read RC. Macrophage defences against respiratory tract infections: The immunology of childhood respiratory infections. Br Med Bull 2002;61(1):45-61.
[Crossref] [Google Scholar] [PubMed]
- Rios FM, Zimmerman LM. Immunology of reptiles. In: eLS. John Wiley, Sons editor. Chichester Publishers; 2015. p. 1-7.
- Patil KS, Jalalpure SS, Wadekar RR. Effect of Baliospermum montanum root extract on phagocytosis by human neutrophils. Indian J Pharm Sci 2009;71(1):68-71.
[Crossref] [Google Scholar] [PubMed]
- Zimmerman LM, Vogel LA, Bowden RM. Understanding the vertebrate immune system: Insights from the reptilian perspective. J Exp Biol 2010;213(5):661-71.
[Crossref] [Google Scholar] [PubMed]
- Wan M, van der Does AM, Tang X, Lindbom L, Agerberth B, Haeggström JZ. Antimicrobial peptide LL‐37 promotes bacterial phagocytosis by human macrophages. J Leukoc Biol 2014;95(6):971-81.
[Crossref] [Google Scholar] [PubMed]
- Tang F, Zhang Y, Shaoa Y, Zhua F, Huanga P, Baia X. Isolation and identification of a new Bombyx mori nucleopolyhedrovirus strain isolated from Yunnan, China. ScienceAsia 2017;43(1):26-32.
- Wang G. Human antimicrobial peptides and proteins. Pharmaceuticals 2014;7(5):545-94.
- Merchant ME, Leger N, Jerkins E, Mills K, Pallansch MB, Paulman RL, et al. Broad spectrum antimicrobial activity of leukocyte extracts from the American alligator (Alligator mississippiensis). Vet Immunol Immunopathol 2006;110(3):221-8.
[Crossref] [Google Scholar] [PubMed]
- Preecharram S, Jearranaiprepame P, Daduang S, Temsiripong Y, Somdee T, Fukamizo T, et al. Isolation and characterisation of crocosin, an antibacterial compound from crocodile (Crocodylus siamensis) plasma. Anim Sci J 2010;81(3):393-401.
[Crossref] [Google Scholar] [PubMed]
- Pata S, Yaraksa N, Daduang S, Temsiripong Y, Svasti J, Araki T, et al. Characterization of the novel antibacterial peptide leucrocin from crocodile (Crocodylus siamensis) white blood cell extracts. Dev Comp Immunol 2011;35(5):545-53.
[Crossref] [Google Scholar] [PubMed]
- Tankrathok A, Punpad A, Kongchaiyapoom M, Sosiangdi S, Jangpromma N, Daduang S, et al. Identification of the first Crocodylus siamensis cathelicidin gene and RN15 peptide derived from cathelin domain exhibiting antibacterial activity. Biotechnol Appl Biochem 2019;66(2):142-52.
[Crossref] [Google Scholar] [PubMed]
- Hao J, Li YW, Xie MQ, Li AX. Molecular cloning, recombinant expression and antibacterial activity analysis of hepcidin from Simensis crocodile (Crocodylus siamensis). Comp Biochem Physiol B Biochem Mol Biol 2012;163(3-4):309-15.
[Crossref] [Google Scholar] [PubMed]
- Phosri S, Mahakunakorn P, Lueangsakulthai J, Jangpromma N, Swatsitang P, Daduang S, et al. An investigation of antioxidant and anti-inflammatory activities from blood components of crocodile (Crocodylus siamensis). Protein J 2014;33(5):484-92.
[Crossref] [Google Scholar] [PubMed]
- Theansungnoen T, Yaraksa N, Daduang S, Dhiravisit A, Thammasirirak S. Purification and characterization of antioxidant peptides from leukocyte extract of Crocodylus siamensis. Protein J 2014;33(1):24-31.
[Crossref] [Google Scholar] [PubMed]
- Kommanee J, Phosri S, Daduang S, Temsiripong Y, Dhiravisit A, Thammasirirak S. Comparisons of anti-inflammatory activity of crocodile (Crocodylus siamensis) blood extract. Chiang Mai J Sci 2014;41(3):627-34.
- Jangpromma N, Preecharram S, Srilert T, Maijaroen S, Mahakunakorn P, Nualkaew N, et al. In vitro and in vivo wound healing properties of plasma and serum from Crocodylus siamensis blood. J Mol Microbiol Biotechnol 2016;26(6):1140-7.
[Crossref] [Google Scholar] [PubMed]
- Theansungnoen T, Maijaroen S, Jangpromma N, Yaraksa N, Daduang S, Temsiripong T, et al. Cationic antimicrobial peptides derived from Crocodylus siamensis leukocyte extract, revealing anticancer activity and apoptotic induction on human cervical cancer cells. Protein J 2016;35(3):202-11.
[Crossref] [Google Scholar] [PubMed]
- Boron W, Boulpaep E. Medical Physiology: A cellular and molecular approach, 2nd ed. Philadelphia, PA: Saunders/Elsevier Health Sciences; 2003.
- Liang T, Ji W, Zhang GR, Wei KJ, Feng K, Wang WM, et al. Molecular cloning and expression analysis of liver-expressed antimicrobial peptide 1 (LEAP-1) and LEAP-2 genes in the blunt snout bream (Megalobrama amblycephala). Fish Shellfish Immunol 2013;35(2):553-63.
[Crossref] [Google Scholar] [PubMed]
- Liu F, Li JL, Yue GH, Fu JJ, Zhou ZF. Molecular cloning and expression analysis of the liver-expressed antimicrobial peptide 2 (LEAP-2) gene in grass carp. Vet Immunol Immunopathol 2010;133(2-4):133-43.
[Crossref] [Google Scholar] [PubMed]
- Sang Y, Ramanathan B, Minton JE, Ross CR, Blecha F. Porcine liver-expressed antimicrobial peptides, hepcidin and LEAP-2: Cloning and induction by bacterial infection. Dev Comp Immunol 2006;30(4):357-66.
[Crossref] [Google Scholar] [PubMed]
- Krause A, Sillard R, Kleemeier B, Klüver E, Maronde E, Conejo‐García JR, et al. Isolation and biochemical characterization of LEAP‐2, a novel blood peptide expressed in the liver. Protein Sci 2003;12(1):143-52.
[Crossref] [Google Scholar] [PubMed]
- Chakka AK, Elias M, Jini R, Sakhare PZ, Bhaskar N. In vitro antioxidant and antibacterial properties of fermentatively and enzymatically prepared chicken liver protein hydrolysates. J Food Sci Technol 2015;52(12):8059-67.
[Crossref] [Google Scholar] [PubMed]
- Walker JM. The Protein Protocols Handbook. New Jersey: Humana Press; 2002.
- Wingfield P. Protein precipitation using ammonium sulfate. Curr Protoc Protein Sci 2001;1-10.
[Crossref] [Google Scholar] [PubMed]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976;72(1-2):248-54.
[Crossref] [Google Scholar] [PubMed]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227(5259):680-5.
[Crossref] [Google Scholar] [PubMed]
- Moro C, Palacios I, Lozano M, D’Arrigo M, Guillamón E, Villares A, et al. Anti-inflammatory activity of methanolic extracts from edible mushrooms in LPS activated RAW 264.7 macrophages. Food Chem 2012;130(2):350-5.
- Campbell PA, Canono BP, Drevets DA. Measurement of bacterial ingestion and killing by macrophages. Curr Protoc Immunol 1994;12(1):14-6.
[Crossref] [Google Scholar] [PubMed]
- Shapiro H. Practical Flow Cytometry. Alan R Liss, editor. 3rd ed. New York; 1995.p. 542.
- Dobson CM. Protein folding and misfolding. Nature 2003;426(6968):884-90.
[Crossref] [Google Scholar] [PubMed]
- Mitsudome T, Moribe M, Obayashi Y, Uchiyama A, Aono M. Influence of low-molecular-weight aggregates on aggregate growth kinetics and physical properties of solid-state proteins during storage. Eur J Pharm Biopharm 2020;146:10-8.
[Crossref] [Google Scholar] [PubMed]
- Flannagan RS, Heit B, Heinrichs DE. Antimicrobial mechanisms of macrophages and the immune evasion strategies of Staphylococcus aureus. Pathogens 2015;4(4):826-68.
[Crossref] [Google Scholar] [PubMed]
- Wan M, Tang X, Rekha RS, Muvva SJ, Brighenti S, Agerberth B, et al. Prostaglandin E2 suppresses hCAP18/LL‐37 expression in human macrophages via EP2/EP4: Implications for treatment of Mycobacterium tuberculosis infection. FASEB J 2018;32(5):2827-40.
[Crossref] [Google Scholar] [PubMed]
- Flannagan RS, Jaumouille V. Grinstein S. The cell biology of phagocytosis. Annu Rev Pathol 2012;7:61-98.
[Crossref] [Google Scholar] [PubMed]
- Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol 1994;124(5):677-88.
[Crossref] [Google Scholar] [PubMed]
- Fairn GD, Grinstein S. How nascent phagosomes mature to become phagolysosomes. Trends Immunol 2012;33(8):397-405.
[Crossref] [Google Scholar] [PubMed]
- Flaherty DK. Phagocytosis and intracellular killing, immunology for pharmacy. Elsevier Publishers; 2012.
- Simons ER. Measurement of phagocytosis and of the phagosomal environment in polymorphonuclear phagocytes by flow cytometry. Curr Protoc Cytom 2010;51(1):9-31.
[Crossref] [Google Scholar] [PubMed]
- MWer S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16(3):1215.
[Crossref] [Google Scholar] [PubMed]
- Easterhoff D, Ontiveros F, Brooks LR, Kim Y, Ross B, Silva JN, et al. Semen-derived enhancer of viral infection (SEVI) binds bacteria, enhances bacterial phagocytosis by macrophages and can protect against vaginal infection by a sexually transmitted bacterial pathogen. Antimicrob Agents Chemother 2013;57(6):2443-50.
[Crossref] [Google Scholar] [PubMed]