- *Corresponding Author:
- A. P. Malgi and Subarna Roy
Indian Council of Medical Research (ICMR)-National Institute of Epidemiology,
Chennai,
Tamil Nadu 600077,
India
E-mail: drsubarnaroy@gmail.com
| Date of Received | 12 May 2020 |
| Date of Revision | 18 August 2021 |
| Date of Acceptance | 18 March 2022 |
| Indian J Pharm Sci 2022;84(2):328-340 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
There is a constant demand to develop an effective and affordable new drug entity to manage cognitive impairment. Medicinal herbs serve as natural resources for mining of new drug candidates. We used chemoinformatics to identify the bioactive compounds from Amaranthus tricolor L. a common leafy vegetable and often used in traditional medicine to manage cognitive dysfunction. We carried out a series of bioinformatics studies with the phytoconstituents of Amaranthus tricolor to elicit their possible molecular functions in cognitive disorders including Alzheimer’s disease. We first identified the bioactive phytoconstituents from Amaranthus tricolor and predicted their potential protein targets involved in the pathogenesis of cognitive dysfunction using BindingDB (p≥0.7). Gene ontology functional enrichment analysis was performed using search tool for the retrieval of interacting genes/proteins. The pathways that are probably regulated by the identified plant phytoconstituents were analyzed using Kyoto encyclopedia of genes and genomes. Docking studies were carried out with AutoDock4.2v. Molecular dynamics analyses were performed using Schrodinger Desmond 6.1v software for a 50 ns production run. Thirty nine phytoconstituents were identified in Amaranthus tricolor, five of which were predicted to modulate eight potential protein targets involved in cognitive impairment. Gene ontology functional enrichment analysis revealed twenty eight biological processes and 10 molecular functions associated with cognitive impairment. Kyoto encyclopedia of genes and genomes pathway identified eight pathways that are directly related to cognitive impairment. Serotonergic and cholinergic synapse was identified as key pathways. Kaempferol exhibited the highest binding affinity with acetylcholinesterase, monoamino oxidase A and monoamino oxidase B. Molecular dynamics simulation demonstrated stable intermolecular interactions between kaempferol and acetylcholinesterase. Our study identified flavonoids from Amaranthus tricolor as having benefits in managing cognitive impairment and offers a broader scope to mine for potential drug candidates from this natural resource. Our study was limited to computer simulations and calls for wet lab validation of the predicted molecular functions.
Keywords
Amaranthus tricolor, cognitive dysfunction, molecular docking, molecular dynamics, gene ontology, network pharmacology
Cognitive impairment is a complex disorder of old age people characterized by changes in cognitive functions like trouble in remembering, concentrating, learning and decision making. A significant increase in life expectancy in the twentieth century has resulted in conditions like Alzheimer's Disease (AD) becoming the most common neurocognitive disorder with high incidence and intricate pathogenesis. AD is characterized by the presence of extracellular deposits of insoluble amyloidbeta (β) plaques, Neurofibrillary Tangles (NFT) and cholinergic deficits [1]. From a clinical point of view, AD has a strong impact on the lifestyle of patients which is characterized by a prodromal phase with a subsequent progressive loss of memory and decline of cognitive functions, leading to the need for continuous medical care [2]. The primary clinical features of AD are loss of memory and memory impairment at the early stage, subsequent topographical difficulties, loss of attention, confidence and judgment [3]. The five drugs approved by the US Food and Drug Administration (FDA) that are currently used to treat the cognitive manifestations of AD viz. Acetylcholinesterase (ACHE) inhibitors rivastigmine (Exelon), tacrine (Cognex), donepezil (Aricept), galantamine (Razadyne) and N-methyl-Daspartate (NMDA) receptor antagonist memantine (Namenda) [4]. However, these agents lose effectiveness as the disease progresses due to their actions on a specific protein i.e. donepezil acts on ACHE enzyme and memantine on NMDA receptor. These are also associated with numerous side effects i.e. bradycardia, hypotension, increased respiratory secretion, decreased intraocular pressure and vomiting [5,6].
With increasing life expectancy, the number of geriatric people (>60 y of age) is increasing worldwide. In 2015, about 47 million people were living with dementia and it is expected to triple by 2050 [7]. The increase in the number of people with dementia has been exponential in Low Middle-Income Countries (LMICs) like India. Knowledge about the risk factors of developing dementia in LMICs are scanty and very little has been done at ground level for its prevention and treatment. India is going through a significant epidemiological transition with a tremendous increase in noncommunicable diseases. There is a strong correlation between the onset of dementia and age. A study carried out in the United States in 2014 estimated a reduction of dementia cases by 2 million in the USA alone by 2020 if any intervention could delay the onset of dementia only by only 2 y [8]. Therefore dementia cases can be delayed and the number of cases decreased by an effective intervention strategy in an aging population. The economic implication of delaying dementia in a resource-limited country like India is tremendous. Since the pathophysiology of dementia starts years before the actual manifestation of symptoms, there is a window of opportunity to intervene and prevent or delay the clinical manifestations [9]. Therefore attempts are being made to ‘delay’ dementia because there is no ideal drug to prevent it or cure it completely. In the effort to delay dementia/cognitive impairment, several approaches are being tried, some of which rely upon traditional/ alternative/complementary forms of medicine and even their combination [10,11].
Ayurveda is an ancient Indian holistic system of medicine that primarily uses plants and minerals in prescriptions that play an important role in managing various diseases, including cognitive disorders, because of their therapeutic effects, often on multiple targets [12].
The introduction of the concept of systems biology into Ayurveda research has opened up a new vista to gain systematic insights into the holistic understanding of the effects of multiple compounds on multiple targets through traditional medicine’ complex network analysis. Investigation into the role of traditional herbs for the management of complex diseases like dementia and cognitive disorders has enormous potential and can open up opportunities for the discovery of new drugs for these conditions following the concept of multidrug, multi-target and multi-pathway approaches.
‘Gene Ontology (GO) functional enrichment analysis’ and ‘network pharmacology’ are the two emerging areas of pharmacology that deal with the concept of “multicomponent therapeutics and network targets”. These disciplines provide new insights for elucidation of the multi-scale mechanisms of action of herbs in the management of complex diseases like AD [13]. The use of network pharmacology and bioinformatics in research on herbal medicines with classical pharmacognosy and pharmacology has greatly facilitated mechanistic studies on the synergistic/antagonistic actions of herbal phytoconstituents at a molecular level [3,14].
Amaranthus tricolor L. (A. tricolor ) (tambdi bhaji/ lal saag) belongs to the family Amaranthaceae [15]. Amaranth (Amaranthus spp.) is cultivated and consumed as a leafy vegetable [16]. The leaves of this plant contain flavonoids, alkaloids, cardiac glycosides, phenol, amino acids, saponins, tannins, terpenoids, pterocarpans, steroids, quinones, resins and coumarins as major phytoconstituents. A. tricolor also contains amaranthine, kaempferol, ferulic acid, quercetin, apigenin, quercetin 3-o-glucoside, quercetin 3-orutinoside, apigenin 4-o-beta-d-glucopyranoside, feruloylquinic acid, boropinic acid, amarantholidols A, B, C betanin, 4-geranyloxyferulic acid, isoamaranthine, xylofuranosyl uracil, 7-p-coumaroyl, betaxanthin, 7-isopentenyloxycoumarin, etc. The leaves are used as traditional medicine and reported for antioxidant, antiinflammatory and neuroprotective activity [17-21].
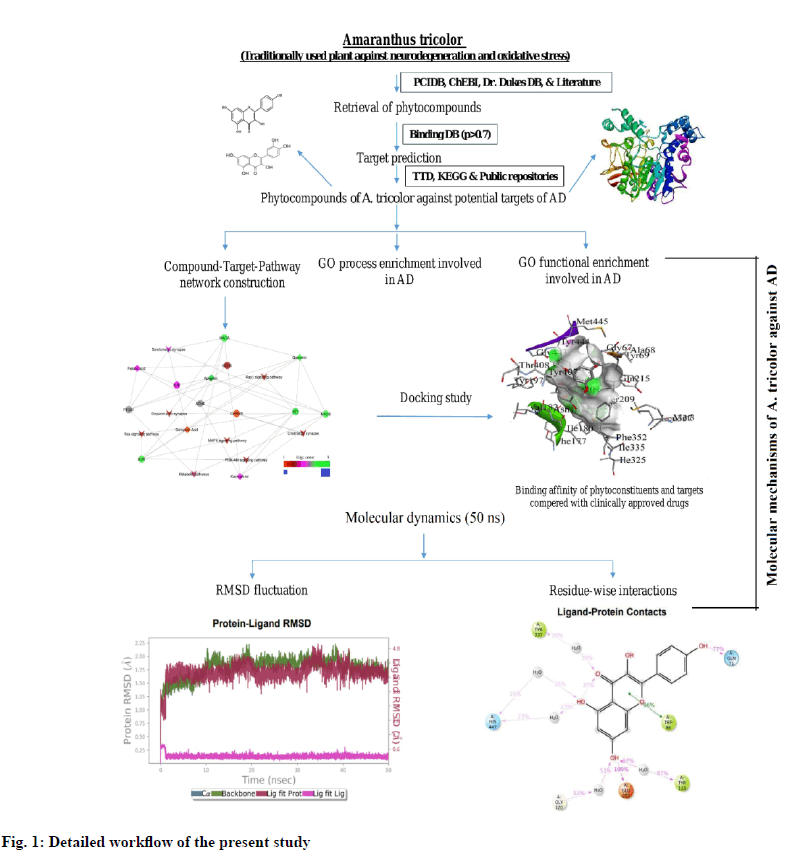
However, there is no credible information on the potential targets regulated by A. tricolor for the management of cognitive impairment. GO functional enrichment analysis, network pharmacology and wet lab experiments have not been reported on this potentially important natural herb. In the current study, we used computational tools and public scientific data to investigate the pharmacological interaction of bioactive phytoconstituents from A. tricolor with potential protein targets and pathways for the management of cognitive impairment. The complete workflow of this study is represented in fig. 1.
Materials and Methods
Identification of bioactive phytoconstituents from A. tricolor and target screening:
Bioactive phytoconstituents of A. tricolor were identified from Dr. Dukes Database (DB), Phytochemical Interaction DB (PCIDB) [22] and scientific journals using the keyword “A. tricolor ”. The Compound Identification number (CID), canonical Simplified Molecular Input Line Entry System (SMILES), Molecular Formula (MF), Molecular Weight (MW), Number of Hydrogen Bond Acceptor (NHBA) and Number of Hydrogen Bond Donors (NHBD) were retrieved from the PubChem chemical database [23]. Canonical SMILES were queried for target prediction in BindingDB [24] at the probability score of ≥0.7 (≥70 %) with respect to the known chemical compounds targeting protein molecules. Gene ID of each protein molecule was retrieved from UniProt [25]. The protein molecules associated with cognitive impairment were separated with reference to the successful and approved targets reported in the Therapeutic Target Database (TTD) [26].
GO functional enrichment analysis and network construction:
Search Tool for the Retrieval of Interacting Genes/ Proteins (STRING) is a universally utilized system for retrieval of protein-protein interaction, known interaction, predicted interaction, process and function of genes, etc. To understand protein interactions systematically, predicted targets were provided as input to STRING 11.0 (Search Tool for the Retrieval of Interacting Genes/Proteins, https://string–db.org) to obtain relevant information on protein interaction. The GO method can efficiently identify the process related to biological phenomena and helps to get more meaningful gene functional information. GO functional enrichment analysis was performed using the functional annotation tool of STRING. The Kyoto Encyclopedia of Genes and Genomes (KEGG), (https://www.genome.jp/kegg/pathway.html) database was used to identify the pathways involved in cognitive impairment. Cytoscape v3.6.1 [27] was used to construct the network between compounds, protein molecules and pathways. The network was analyzed by choosing the command “network analyzer” and the network was treated as direct. Layout algorithm (degree sorted layout) was applied in constructing the network.
Druglikeness and probable side effects of A. tricolor phytoconstituents:
Drug likness property of the phytoconstituents were predicted using Molsoft (http://molsoft.com/mprop/) an online server, which predicts the probable druglike property based on Lipinski’s rule of five, that eliminates compounds having poor absorptivity and bioavailability i.e. if their molecular weight is >500 g/mol, having >5 hydrogen bond donors, >5 log P and >10 hydrogen bond acceptors. ADVERpred (http://www.way2drug.com/adverpred/) an online tool, uses the data of most frequent and severe adverse drug events that have either known or probable relationships to drug consumption and predicts the possible side effects of a compound based on structure-activity relationship. In the current study, we used ADVERpred to predict the probable side effects (Probable activity (Pa) and Probable inactivity (Pi)) of phytoconstituents. The current study utilized the admetSAR2.0 (http:// lmmd.ecust.edu.cn/admetsar2/). This online tool contains 2 10 000 experimental data for 96 000 drug candidates. It contains 27 computational models to predict the ADMET profiles i.e. absorptivity, Blood- Brain Barrier (BBB) permeability, oral bioavailability, cytochrome P450 (CYP450) and isoenzyme inhibitory activity, mutagenicity, plasma protein binding affinity and fish aquatic toxicity.
Ligand-protein docking studies:
The ligands were retrieved from the PubChem chemical database in a Three Dimensional (3D) structure data format (.sdf), minimized using the mmff94 force field and saved in protein data bank (.pdb) file by using MarvinSketch [28]. The protein molecules i.e. ACHE (PDB ID: 4PQE), monoamino oxidase B (PDB ID: 1OJD), monoamino oxidase A (PDB ID: 2Z5X) and Prostaglandin G/H Synthase 1 (PTGS1) (PDB ID: 3N8X) were retrieved from the Research Collaboratory for Structural Bioinformatics PDB (RCSB PDB) (https://www.rcsb.org/). The protein structure was prepared by removing heteroatom and water using Discovery Studio Visualizer v2019 [29]. The docking of ligands with their respective protein molecules was performed using AutoDock4.2 [30]. The ligand protein complex was viewed using Discovery Studio Visualizer v2019.
Molecular dynamics (MD) simulation studies:
MD simulation has become a popular tool in computational biology for analyzing the dynamic behavior of molecular complexes and intermolecular interactions under physiologically realistic circumstances [31]. The stability of intermolecular interactions between kaempferol and ACHE was assessed using MD simulation for 50 ns production run using Schrodinger desmond 6.1v software [32,33]. The complex system was built in a cubic box with 10 Å×10 Å×10 Å dimensions as the periodic boundary, using a preset Simple Point Charge (SPC) water model as the solvent. 5 Sodium (Na+) counter ions (concentration of 6.533 mM) were added to neutralize the system. Water molecule geometry, bond lengths and bond angles of heavy atoms were all restrained using the SHAKE algorithm. To calculate the long range interactions between the molecules, the Particle Mesh Ewald method was used. The Lennard-Jones interactions cut-off was set to 10 Å. For a 100.0 ps production run, the system was minimized. Finally, the isobaric-isothermal ensemble (NPT) was used to maintain a pressure of 1.01325 bar and temperature of 300 K using the Thermostat "Nose- Hoover chain" method with 1.0 ps relaxation time and the Barostat "Martyana-Tobias-Klein" method with 2.0 ps relaxation time. The short range coulomb cut-off radius was adjusted at 9.0 Å. The entire simulation was analyzed for 5000 frames and recorded at an interval of 10.0 ps. The Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF) were used to check the residue-wise interaction fluctuations and the complex compactness was analyzed by the radius of Gyration (rGyr).
Results and Discussion
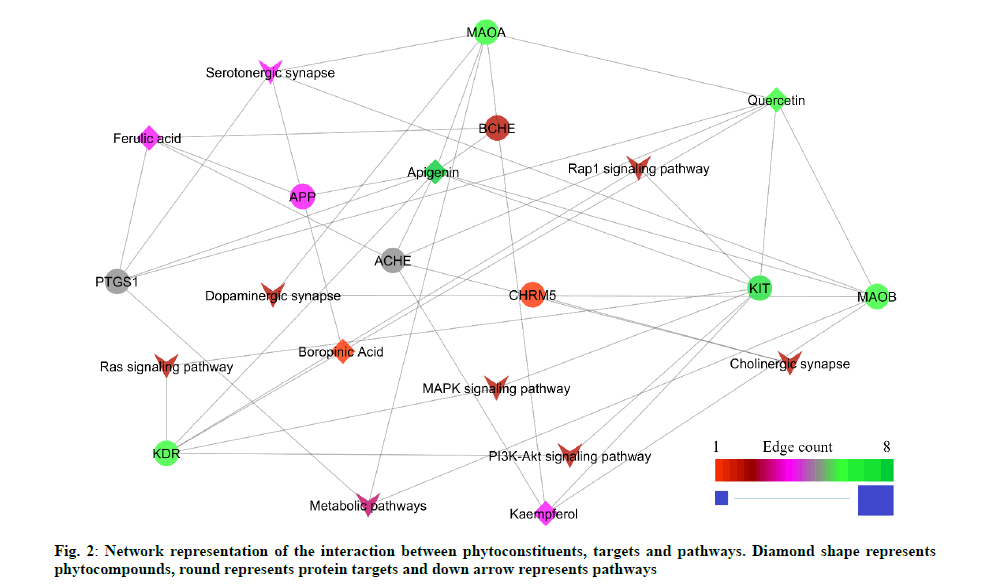
Thirty nine phytoconstituents were identified in A. tricolor from different databases and other open source records. Among them, five compounds were predicted to modulate eight protein molecules involved in cognitive impairment, showing synergistic effect. Apigenin was found to modulate ACHE, Amyloid Precursor Protein (APP), Butyrylcholinesterase (BCHE), KDR, KIT, Monoamine Oxidase A (MAOA), Monoamine Oxidase B (MAOB) and PTGS1; boropinic acid modulated APP; ferulic acid modulated ACHE, APP, BCHE and PTGS1; kaempferol was found to modulate ACHE, KIT, MAOA and MAOB; while quercetin was found modulating ACHE, KDR, KIT, MAOA, MAOB and PTGS1. These phytoconstituents were identified as flavonoids, organoborane and phenolic compounds (Table 1).
| Phytoconstituents | Compound type | PubChem ID | Targets modulated by phytoconstituents |
|---|---|---|---|
| Ferulic acid | Phenolic | 445858 | APP, PTGS1 |
| Quercetin | Flavonoid | 5280343 | ACHE, ADORA2A, MAOA, MAOB, PTGS1, KIT, KDR |
| Apigenin | Flavonoid | 5280443 | APP, ACHE, BCHE, ADORA2A, MAOA, MAOB, PTGS1, KIT, KDR |
| Kaempferol | Flavonoid | 5280863 | ACHE, ADORA2A, MAOA, MAOB, KIT |
| Boropinic Acid | Organoboranes | 10682896 | APP |
Note: AD: Alzheimer's Disease
Table 1: Type of Phytoconstituents with their Probable Targets Involved in AD
The GO functional enrichment analysis result showed 121 biological processes and 18 molecular functions. The peer interpretation revealed 28 biological processes and 10 molecular functions were associated with cognitive impairment (Table 2 and Table 3). The result analyzed from the STRING database showed 19 possible pathways based on the protein-protein interaction. The peer interpretation of the protein-protein interaction using the KEGG pathway identified eight pathways that are directly associated with cognitive impairment. Among 8 pathways, the serotonergic synapse pathway scored the lowest false discovery rate with the highest edge count within the network (Table 4). The constructed network contains 42 edges. Among them, 23 are compound-protein interactions and 19 proteinpathway interactions (fig. 2).
| GO ID | Genes modulated by compounds | Gene count | Biological function | FDR |
|---|---|---|---|---|
| GO:0001505 | ACHE, ADORA2A, APP, BCHE, MAOA, MAOB | 6 | Regulation of neurotransmitter levels | 3.33E-06 |
| GO:0042133 | ACHE, BCHE, MAOA, MAOB | 4 | Neurotransmitter metabolic process | 5.96E-05 |
| GO:0042135 | ACHE, MAOA, MAOB | 3 | Neurotransmitter catabolic process | 8.55E-05 |
| GO:0007271 | ACHE, ADORA2A, CHRM5 | 3 | Synaptic transmission, cholinergic | 0.00013 |
| GO:0065008 | ACHE, ADORA2A, APP, BCHE, KDR, KIT, MAOA, MAOB, PTGS1 | 9 | Regulation of biological quality | 0.00031 |
| GO:0032222 | ACHE, ADORA2A | 2 | Regulation of synaptic transmission, cholinergic | 0.0011 |
| GO:0007612 | APP, BCHE, KIT | 3 | Learning | 0.0029 |
| GO:0006954 | ADORA2A, APP, KIT, PTGS1 | 4 | Inflammatory response | 0.0039 |
| GO:0048167 | ADORA2A, APP, KIT | 3 | Regulation of synaptic plasticity | 0.0039 |
| GO:0048169 | APP, KIT | 2 | Regulation of long-term neuronal synaptic plasticity | 0.0039 |
| GO:0007610 | ADORA2A, APP, BCHE, KIT | 4 | Behavior | 0.0043 |
| GO:0042417 | MAOA, MAOB | 2 | Dopamine metabolic process | 0.0043 |
| GO:0050877 | ADORA2A, APP, BCHE, CHRM5, KIT | 5 | Nervous system process | 0.0069 |
| GO:0008542 | APP, KIT | 2 | Visual learning | 0.0072 |
| GO:0061515 | APP, KIT | 2 | Myeloid cell development | 0.0085 |
| GO:0040012 | ADORA2A, APP, KDR, KIT | 4 | Regulation of locomotion | 0.0137 |
| GO:0007631 | ADORA2A, APP | 2 | Feeding behavior | 0.0156 |
| GO:0042391 | ADORA2A, APP, KDR | 3 | Regulation of membrane potential | 0.0156 |
| GO:0008360 | KDR, KIT | 2 | Regulation of cell shape | 0.0268 |
| GO:0000187 | APP, KIT | 2 | Activation of MAPK activity | 0.0278 |
| GO:0007166 | ADORA2A, APP, KDR, KIT, MAOA | 5 | Cell surface receptor signaling pathway | 0.0289 |
| GO:0010646 | ACHE, ADORA2A, APP, BCHE, KDR, KIT | 6 | Regulation of cell communication | 0.0289 |
| GO:0007399 | ACHE, ADORA2A, APP, BCHE, KIT | 5 | Nervous system development | 0.0291 |
| GO:0007626 | ADORA2A, APP | 2 | Locomotory behavior | 0.0347 |
| GO:0044237 | ACHE, ADORA2A, APP, BCHE, KDR, KIT, MAOA, MAOB, PTGS1 | 9 | Cellular metabolic process | 0.0365 |
| GO:0007154 | ACHE, ADORA2A, APP, CHRM5, KDR, KIT, MAOA | 7 | Cell communication | 0.0398 |
| GO:0035556 | ADORA2A, APP, KDR, KIT | 4 | Intracellular signal transduction | 0.0401 |
| GO:0065007 | ACHE, ADORA2A, APP, BCHE, CHRM5, KDR, KIT, MAOA, MAOB, PTGS1 | 10 | Biological regulation | 0.0447 |
Note: GO: Gene Ontology and FDR: False Discovery Rate
Table 2: Go Enrichment Analysis of Protein Targets for their Biological Processes Involved in Ad
| GO ID | Genes modulated by compounds | Gene count | Gene function | FDR |
|---|---|---|---|---|
| GO:0003990 | ACHE, BCHE | 2 | ACHE activity | 0.00021 |
| GO:0008131 | MAOA, MAOB | 2 | Primary amine oxidase activity | 0.00025 |
| GO:0042277 | ACHE, APP, BCHE | 3 | Peptide binding | 0.0054 |
| GO:0046983 | ACHE, ADORA2A, APP, KIT, MAOB | 5 | Protein dimerization activity | 0.0054 |
| GO:0001540 | ACHE, BCHE | 2 | Amyloid-beta binding | 0.006 |
| GO:0004714 | KDR, KIT | 2 | Transmembrane receptor protein tyrosine kinase activity | 0.0062 |
| GO:0003824 | ACHE, BCHE, CHRM5, KDR, KIT, MAOA, MAOB, PTGS1 | 8 | Catalytic activity | 0.01 |
| GO:0005178 | APP, KDR | 2 | Integrin binding | 0.0145 |
| GO:0004888 | ADORA2A, CHRM5, KDR, KIT | 4 | Transmembrane signaling receptor activity | 0.0181 |
| GO:0016491 | MAOA, MAOB, PTGS1 | 3 | Oxidoreductase activity | 0.0302 |
Note: GO: Gene Ontology and FDR: False Discovery Rate
Table 3: Go Enrichment Analysis of Protein Targets for their Molecular Functions Involved in AD
| Pathway ID | Pathway description | No. of gene involved in the pathway | FDR | Protein involved in pathways associate with AD |
|---|---|---|---|---|
| hsa04726 | Serotonergic synapse | 4 | 9.33E-06 | APP, MAOA, MAOB, PTGS1 |
| hsa04725 | Cholinergic synapse | 2 | 0.0051 | ACHE, CHRM5 |
| hsa04728 | Dopaminergic synapse | 2 | 0.0062 | MAOA, MAOB |
| hsa04015 | Rap1 signaling pathway | 2 | 0.0129 | KDR, KIT |
| hsa04014 | Ras signaling pathway | 2 | 0.0151 | KDR, KIT |
| hsa04010 | MAPK signaling pathway | 2 | 0.0216 | KDR, KIT |
| hsa04151 | PI3K-Akt signaling pathway | 2 | 0.0283 | KDR, KIT |
| hsa01100 | Metabolic pathways | 3 | 0.0459 | MAOA, MAOB, PTGS1 |
Note: FDR: False Discovery Rate and AD: Alzheimer's Disease
Table 4: AD Pathways Modulated by the Phytoconstituents
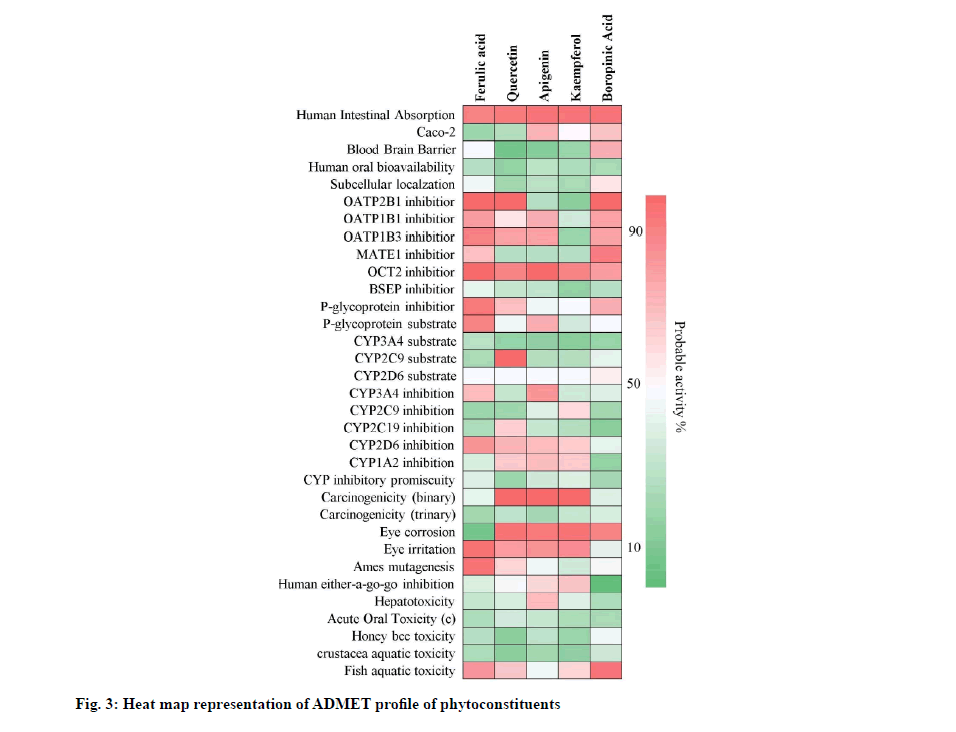
Quercetin, apigenin and kaempferol showed positive drug-likeness scores i.e. 0.93, 0.77 and 0.77 respectively. Ferulic acid and boropinic acid showed negative drug-likeness property i.e. -0.44 and -0.41. Further, quercetin, apigenin and kaempferol were predicted to show hepatotoxicity. Ferulic acid showed myocardial infarction and boropinic acid showed cardiac failure and myocardial infarction. The drug-likeness character and toxicity profiles obtained are shown in Table 5. The predicted probability score for absorptivity, blood-brain barrier permeability, bioavailability, carcinogenicity, isoenzyme inhibition, plasma protein binding affinity and fish aquatic toxicity, etc., of the phytoconstituents of A. tricolor is shown in fig. 3.
| Phytocompounds | PubChem ID | MF | MW (g/mol) | HBA | HBD | Log P | DL score | Probable side effect(s) |
|---|---|---|---|---|---|---|---|---|
| Ferulic acid | 445858 | C10 H10 O4 | 194.06 | 4 | 2 | 2.04 | -0.44 | Myocardial infarction |
| Quercetin | 5280343 | C15 H10 O7 | 302.04 | 7 | 5 | 2.11 | 0.93 | Hepatotoxicity |
| Apigenin | 5280443 | C15 H10 O5 | 270.05 | 5 | 3 | 3.06 | 0.77 | Hepatotoxicity |
| Kaempferol | 5280863 | C15 H10 O6 | 286.05 | 6 | 4 | 2.49 | 0.77 | Hepatotoxicity |
| Boropinic acid | 1.10E+07 | C15 H18 O4 | 262.12 | 4 | 1 | 4.05 | -0.41 | Cardiac failure, myocardial infarction |
Note: MF: Molecular formula; MW: Molecular weight; HBA: Hydrogen Bond Accepter; HBD: Hydrogen Bond Donor; Log P: Partition coefficient and DL score: Druglikeness score
Table 5: Drug Likness and Probable Side Effects Profile of Phytoconstituents from A. Tricolor
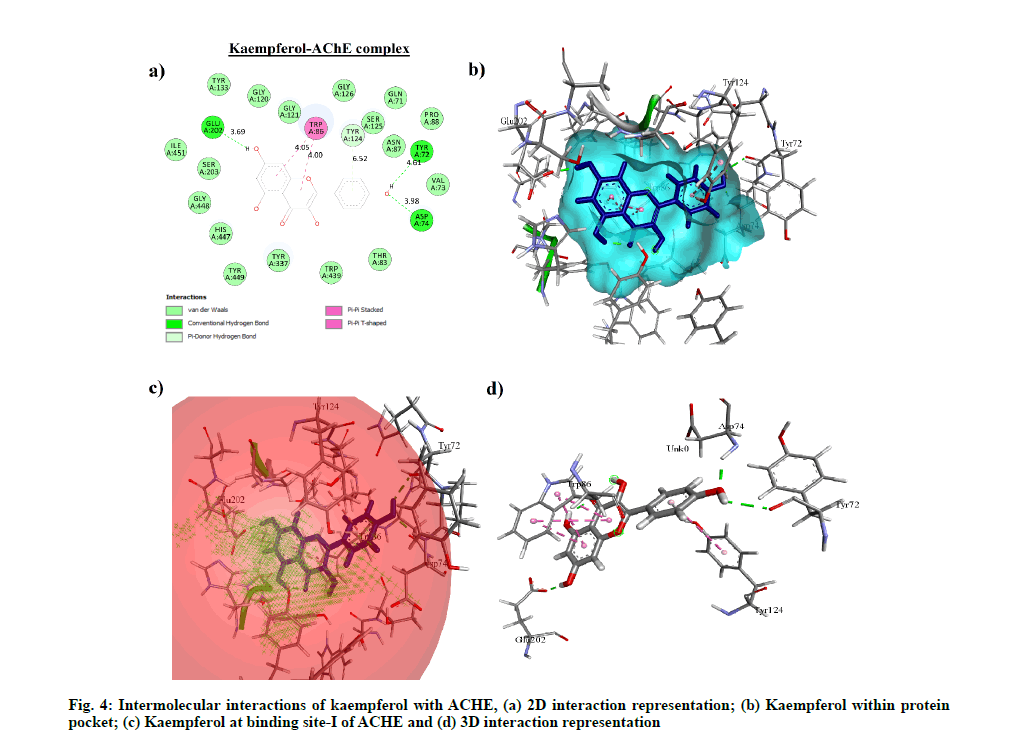
Molecular docking performed on phytocompounds having drug-like property i.e. kaempferol, quercetin, apigenin and clinically approved drug candidates with ACHE, MAOB, MAOA and PTGS1 showed kaempferol having the highest binding affinity with ACHE, MAOA and MAOB i.e. -7.8 kcal/mol (1.92 μM), -8.28 kcal/mol (0.850 μM) and -8.6 kcal/ mol (0.498 μM) respectively. Meloxicam showed the highest binding affinity with PTGS1 i.e. -7.64 kcal/mol (2.52 μM) with four hydrogen bond interactions. However, among apigenin and quercetin, apigenin showed the highest binding affinity with PTGS1 i.e. -4.4 kcal/mol (591.39 μM) with four hydrogen bond interactions. The binding energy, Half-Maximal Inhibitory Concentration (IC50) and hydrogen bond interactions of each compound with their respective protein target are compared with clinically accepted standard drug molecule (Table 6). The interaction of kaempferol with ACHE is shown in fig. 4.
| Targets | PDB ID | Compound | p | BE (kcal/mol) | IC50 (µM) | Amino acid…ligand interactions |
|---|---|---|---|---|---|---|
| ACHE | 4PQE | Kaempferol | 0.7 | -7.8 | 1.92 | Tyr72…OH, Asp74…OH, Glu202…OH |
| Apigenin | 0.7 | -6.76 | 11.1 | Asp74…OH, Phe295…O, Tyr337…=O | ||
| Quercetin | 0.7 | -6.03 | 38.27 | Pro88…OH, Asp131…=O | ||
| Donepezil* | 1 | -5.15 | 168.86 | Thr436…O- | ||
| MAOA receptor | 1OJD | Kaempferol | 0.7 | -8.28 | 0.85 | Asn181…OH |
| Apigenin | 0.86 | -3.97 | 1023 | Asn133…=O, Ile164…OH | ||
| Quercetin | 0.71 | -6.09 | 34.46 | Arg109…OH, Asn125…OH, Thr205…=O, Thr205…OH | ||
| Moclobemide* | 1 | -6.32 | 23.22 | Nil | ||
| MAOB receptor | 2Z5X | Kaempferol | 0.7 | -8.6 | 0.498 | Tyr188…=O, Cys172…OH |
| Apigenin | 0.97 | -4.64 | 393.85 | Lys73…=O, Glu466…OH, AspP471…OH | ||
| Quercetin | 0.7 | -4.31 | 697.85 | Glu366…OH, Lys370…=O | ||
| Safinamide* | 1 | -6.87 | 9.16 | Arg42…O-, Ala263…NH | ||
| PTGS1 | 3N8X | Quercetin | 0.7 | -3.87 | 1044 | Leu92…OH, His95…OH |
| Apigenin | 0.92 | -4.4 | 591.39 | Thr60…OH, Arg61…OH, Ile151…OH, Arg469…OH | ||
| Meloxicam* | 0.78 | -7.64 | 2.52 | Phe210…OH, Thr212…=O, Thr212…S, His386…=O |
Note: ACHE: Acetylcholinesterase; MAOA: Monoamino Oxidase A; MAOB: Monoamino Oxidase B and PTGS1: Prostaglandin G/H Synthase 1, *Standard drug of specific protein for AD; BE: Binding Energy and IC50: Half-Maximal Inhibitory Concentration
Table 6: Probable Score, Binding Affinity, Inhibitory Constant and Hydrogen Bond Interaction of Compounds with Respective Targets
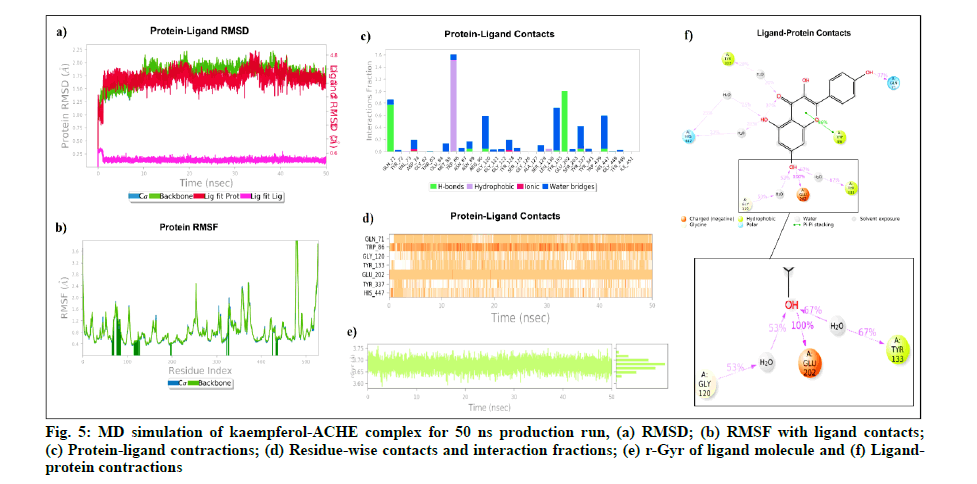
Kaempferol and ACHE complex includes 49 862 atoms with 13 916 water molecules. The system was neutralized by adding 5 Na+ ions (6.533 mM concentration) and was simulated for 50 ns with a 10 ps recording interval. The RMSD of protein alpha Carbon (Cα) (0.738 Å to 2.219 Å) and backbone (0.764 Å to 2.23 Å) was found to be stable throughout the 50 ns simulation. The ligand RMSD values were within the range of 0.114 Å to 0.818 Å from 0 to 50 ns. The average RMSD of ligand with respect to protein was 3.806 Å and ligand with respect to ligand was 0.297 Å. The rGyr deviation of kaempferol with ACHE was found within 3.588 Å to 3.759 Å, which indicates the higher compactness of the kaempferol-ACHE complex.
Further, kaempferol-ACHE contact analysis concluded that Glu202 and Gln71 to form stable hydrogen-bonded interactions for 100 % and 77 % of the duration with the Hydroxy (OH) group of kaempferol, respectively.
Tyr133, Gly120, His447, Tyr337 were found to be involved in hydrophobic and water bridge interactions and showed around 67 %, 53 %, 25 % and 20 % of the time with interaction fraction, respectively.Fig. 5 represents the ligand-protein complex RMSD, residuewise contacts and rGyr of the kaempferol-ACHE complex.
Our study showed flavonoids, organoboranes, flavone glycosides and phenolic phytocompounds from A. tricolor are likely to interact with numerous protein targets involved in the pathogenesis of AD [34] viz., ACHE [35], MAOA [36], MAOB [36], etc. Further, we identified a number of protein molecules that regulate biological processes, molecular functions and pathways as likely targets of bioactive phytoconstituents of A. tricolor . GO functional, process and pathway enrichment analysis was the mainstay of our study. It deals with the interaction and behavior of the biological entities, focus on understanding the relationship between the drug molecules with the biological targets and helps to know the functions of protein complexes, gene regulatory networks and significant pathways associated with disease pathogenesis [37]. The GO analysis identified regulation of neurotransmitter levels, neurotransmitter metabolic process, learning, inflammatory response, behavior, regulation of locomotion, cell communication and many more biological processes associated with cognitive impairment. Results obtained from the GO enrichment analysis in our study revealed various molecular functions viz. ACHE activity, primary amine oxidase activity, peptide binding activity, protein dimerization activity, amyloid-beta binding activity, transmembrane receptor protein tyrosine kinase activity, catalytic activity, integrin binding activity, transmembrane signaling receptor activity and oxidoreductase activity are associated with cognitive impairment and AD. We constructed the network interaction between predicted active phytoconstituents of A. tricolor with their probable targets and enriched pathways thus identified. Ferulic acid, quercetin, apigenin, kaempferol and boropinic acid were identified as the key bioactive phytoconstituents from A. tricolor having modulatory activities on a number of pathways involved in AD e.g. serotonergic synapse (hsa04726) [38], cholinergic synapse (hsa04725) [39], metabolic pathway (hsa01100) [40], dopaminergic pathway (hsa04728) [41], etc. Among five compounds, kaempferol, apigenin and quercetin (flavonoids) showed the highest potential in the pharmacotherapy of AD by targeting ACHE, APP, BCHE, KDR, KIT, MAOA, MAOB and PTGS1 protein molecules within the network. Flavonoids enhance cognitive function at a behavioral stage and attenuate the cognitive decline promoted by brain disorders [42]. The constructed network showed that kaempferol, apigenin and quercetin having the highest edge count (Table 1), which suggests higher probability of these compounds in the modulation of pathogenesis associated with cognitive impairment. Previous literature suggests ACHE as a potential target for AD. It terminates signal transduction at the neuromuscular junction by rapid hydrolysis of the acetylcholine released into the synaptic cleft and plays a role in the neuronal apoptosis and cognitive impairment [43]. The ACHE enzyme degrades the freely available acetylcholine in the cerebral cortex and hippocampus, leading to the impairment of cognitive function in AD patients [35]. PTGS1 converts arachidonate to Prostaglandin H2 (PGH2) in the stomach and platelets, Prostaglandin E2 (PGE2) in gastric epithelial cells and Thromboxane A2 (TXA2) in the plates [44]. Further, these prostaglandin derivatives play a crucial role in cytoprotection, activation, aggregation of platelets, vasoconstriction, proliferation of vascular smooth muscle cells, etc [45]. MAO enzyme catalyzes the oxidative deamination of xenobiotic amines. It has a vital role in the breakdown of neuroactive and vasoactive amines in the peripheral tissues and central nervous system leading to the development of neurodegenerative diseases, anxiety, schizophrenia, mood disorders, depression, migraine and sexual maturation [36].
The affinity of the ligand molecule within the protein pocket is explained by the binding energy and number of hydrogen bond interactions. Docking studies revealed kaempferol to have more affinity towards ACHE, MAOA and MAOB compared to the clinically approved drugs donepezil, moclobemide and safinamide used in the study. Kaempferol showed three hydrogen bonds through the interaction with ACHE i.e. Tyr72… OH, Asp74…OH, Glu202…OH, one with MAOA i.e. Asn181…OH, and two bonds with MAOB i.e. Tyr188…=O, Cys172…OH. Moreover, MD simulation demonstrated the stable interaction of kaempferol with ACHE. Glu202 residue showed 100 % hydrogen bond interaction fraction with the OH group of the kaempferol; which suggests kaempferol as a potential inhibitor of ACHE. Previous studies have demonstrated kaempferol as a potent drug candidate for managing cognitive deficit via the modulation of spatial learning and memory, glutathione level, apoptosis marker cytochrome c inflammatory marker Tumour Necrosis Factor alpha (TNF-α), endogenous antioxidants Superoxide Dismutase (SOD) and lipid peroxidation marker Malondialdehyde (MDA) [46,47]. A previous study by Bahrani et al. isolated and characterized the ACHE inhibitors from Aquilaria subintegra for the treatment of AD and reported kaempferol as a major ACHE inhibitor (85.8 % inhibition) [48]. Further, a study by Hupparage et al. reported that hydroalcoholic extract of A. tricolor L. leaves restore the cholinergic system’s function, inhibit oxidative stress and improve the memory function in the scopolamine treated rats [49]. Although several researchers reported a decrease in the risk of development of AD with intake of dietary flavonoids. Although, several researchers have reported dietary flavonoids decreases the risk of development of AD [50-52] to the best of our knowledge, despite having high content of flavonoids, such molecular mechanism of action for the leafy vegetable A. tricolor L.is not reported so far. An earlier study on A. tricolor L. showed a neuroprotective effect on gene expression of Receptor for Advanced Glycation End-Products (RAGE) during oxidative stress in SH-SY5Y cells [15]. Importantly, Ayurveda formulations often contain traditional herbs and dietary plants that are widely used to treat numerous diseases/disorders due to bioactive phytocompounds [53,54]. The current study identified mechanisms of action of phytocompounds in A. tricolor and their possible roles in the management of AD mostly attributed to the presence of flavonoids (kaempferol, apigenin and quercetin) [17,20].
Several attempts have been made to understand the molecular mechanisms of action of herbal constituents in the management of complex diseases like AD. Exploration of herbal medicines through GO functional, process and pathway enrichment, network pharmacology and other computational methods for the management of complex diseases like AD is a well-accepted [55-57]. Although, herbs contain a complex mixture of pharmacologically active phytoconstituents, this complexity may up/down-regulate the various protein molecules and may cause the synergistic or additive effect to reduce the complications associated with AD [58]. Identification of herbal toxicity plays a critical role in the minimization of organ damage and severe Adverse Drug Events (ADEs). The current study utilized these approached and available bioinformatics methodologies to identify the toxicity associated with phytoconstituents based on their structure-activity relationships. However, the predicted toxicities have not been found in literature through molecular biological experiments and/or clinical investigations.
In conclusion, the current study utilized the drugtarget- pathway relationship to elucidate the molecular mechanism of action of A. tricolor L. against AD. Kaempferol, quercetin and apigenin as were identified as major compounds that are likely to interact with the potential protein targets involved in the pathogenesis of AD. Enrichment analysis of genes showed serotonergic synapse, cholinergic synapse, dopaminergic synapse, AD, Ras-proximate-1 (Rap1) signaling pathway, Rat sarcoma (Ras) signaling pathway, Mitogen- Activated Protein Kinases (MAPK) signaling pathway, Phosphatidylinositol 3 Kinase-Protein Kinase B (PI3KAkt) signaling pathway and metabolic pathways as major pathways regulated by these phytoconstituents, thereby exerting their effects. The probable intermolecular interactions among the predicted therapeutic targets of AD with phytocompounds of A. tricolor L. was demonstrated through molecular docking while MD simulation identified stable interactions of the kaempferol-ACHE complex. GO functional enrichment analysis and network pharmacology-based approach offer an easy and reliable strategy for identifying potential therapeutic targets of traditional herbal medicines. The present study should help design wet-lab studies to experimentally validate the findings, which should go a long way in finding one of the multipronged solutions to tackle AD in the future.
Acknowledgements
The authors are thankful to the Principal KLE College of Pharmacy, Belagavi and Biomedical Informatics Centre of ICMR-NITM Belagavi for providing the necessary facilities to conduct the work. Thanks are also due to ICMR for sustaining the activities of the BIC of NITM through extramural projects (2018-3009 and 2019-0045).
Conflict of Interest
The authors declared no conflicts of interest.
References
- Huang HC, Jiang ZF. Accumulated amyloid-β peptide and hyperphosphorylated tau protein: Relationship and links in Alzheimer's disease. J Alzheimers Dis 2009;16(1):15-27.
[Crossref] [Google Scholar] [Pub Med]
- Bäckman L, Small BJ. Cognitive deficits in preclinical Alzheimer's disease and vascular dementia: Patterns of findings from the Kungsholmen project. Physiol Behav 2007;92(1-2):80-6.
- Zeng Q, Li L, Jin Y, Chen Z, Duan L, Cao M, et al. A network pharmacology approach to reveal the underlying mechanisms of Paeonia lactiflora pall. On the treatment of Alzheimer’s disease. Evid Based Complementary Altern Med 2019;2019.
- Shen ZX. Brain cholinesterases: II. The molecular and cellular basis of Alzheimer's disease. Med Hypotheses 2004;63(2):308-21.
[Crossref] [Google Scholar] [Pub Med]
- Ali TB, Schleret TR, Reilly BM, Chen WY, Abagyan R. Adverse effects of cholinesterase inhibitors in dementia, according to the pharmacovigilance databases of the United-States and Canada. PLoS One 2015;10(12):e0144337.
- Terry AV, Buccafusco JJ. The cholinergic hypothesis of age and Alzheimer's disease-related cognitive deficits: Recent challenges and their implications for novel drug development. J Pharmacol Exp Ther 2003;306(3):821-7.
[Crossref] [Google Scholar] [Pub Med]
- Pratchett T. A global assessment of dementia, now and in the future. Lancet 2015;14:691.
[Crossref] [Google Scholar] [Pub Med]
- Lewis F, Karlsberg Schaffer S, Sussex J, O’Neill P, Cockcroft L. The trajectory of dementia in the UK-making a difference. Office of Health Economics Consulting Reports 2014;2013:1-55.
- Livingston G, Sommerlad A, Orgeta V, Costafreda SG, Huntley J, Ames D, et al. Dementia prevention, intervention and care. Lancet 2017;390(10113):2673-734.
[Crossref] [Google Scholar] [Pub Med]
- Leung EL, Cao ZW, Jiang ZH, Zhou H, Liu L. Network-based drug discovery by integrating systems biology and computational technologies. Brief Bioinform 2013;14(4):491-505.
[Crossref] [Google Scholar] [Pub Med]
- Tewari D, Stankiewicz AM, Mocan A, Sah AN, Tzvetkov NT, Huminiecki L, et al. Ethnopharmacological approaches for dementia therapy and significance of natural products and herbal drugs. Front Aging Neurosci 2018;12;10:3.
[Crossref] [Google Scholar] [Pub Med]
- ven Murthy MR, K Ranjekar P, Ramassamy C, Deshpande M. Scientific basis for the use of Indian ayurvedic medicinal plants in the treatment of neurodegenerative disorders: Ashwagandha. Cent Nerv Syst Agents Med Chem 2010;10(3):238-46.
[Crossref] [Google Scholar] [Pub Med]
- Fang J, Wang L, Wu T, Yang C, Gao L, Cai H, et al. Network pharmacology-based study on the mechanism of action for herbal medicines in Alzheimer treatment. J Ethnopharmacol 2017;196:281-92.
[Crossref] [Google Scholar] [Pub Med]
- Zhang H, Yan ZY, Wang YX, Bai M, Wang XB, Huang XX, et al. Network pharmacology-based screening of the active ingredients and potential targets of the genus of Pithecellobium marthae(Britton & Killip) Niezgoda & Nevl for application to Alzheimer’s disease. Nat Prod Res 2019;18;33(16):2368-71.
[Crossref] [Google Scholar] [Pub Med]
- Amornrit W, Santiyanont R. Neuroprotective effect of Amaranthus lividus and Amaranthus tricolor and their effects on gene expression of RAGE during oxidative stress in SH-SY5Y cells. Gen Mol Res 2016;26:15027562.
[Crossref] [Google Scholar] [Pub Med]
- Ramdwar MN, Chadee ST, Stoute VA. Estimating the potential consumption level of amaranth for food security initiatives in Trinidad, West Indies. Cogent Food Agric 2017;3(1):1321475.
- Sable KV, Saswade RR. Preliminary phytochemical analysis of Amaranthus spinosus leaves. Int J Life Sci 2017;5(4):742-5.
- Bihani GV, Bodhankar SL, Kadam PP, Zambare GN. Anti-nociceptive and anti-inflammatory activity of hydroalcoholic extract of leaves of Amaranthus tricolor L. Der Pharm Lett 2013;5(3):48-55.
- Peter K, Gandhi P. Rediscovering the therapeutic potential of Amaranthus species: A review. Egypt J Basic Appl Sci 2017;4(3):196-205.
- Hussain MM. A comprehensive review on the phytoconstituents from six species of the genus Amaranthus. Bangladesh Pharm J 2019;22(1):117-24.
- Kengar S, Thorat D, Jadhav J. Nephroprotective effect of Amaranthus spinosus root extract in carbon tetrachloride induced toxicity in male albino rat. Int J Drug Dev Res 2017;9(2):5-7.
- Duke JA. Database of biologically active phytochemicals and their activity. CRC Press; 2020.
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucleic Acids Res 2015;44(D1):D1202-13.
[Crossref] [Google Scholar] [Pub Med]
- Gilson MK, Liu T, Baitaluk M, Nicola G, Hwang L, Chong J. BindingDB in 2015: A public database for medicinal chemistry, computational chemistry and systems pharmacology. Nucleic Acids Res 2015;44(D1):D1045-53.
[Crossref] [Google Scholar] [Pub Med]
- Boutet E, Lieberherr D, Tognolli M, Schneider M, Bansal P, Bridge AJ, et al. UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: How to use the entry view. Methods Mol Biol 2016;1374:23-54.
[Crossref] [Google Scholar] [Pub Med]
- Li YH, Yu CY, Li XX, Zhang P, Tang J, Yang Q, et al. Therapeutic target database update 2018: Enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res 2018;46(D1):D1121-7.
[Crossref] [Google Scholar] [Pub Med]
- Ilyas M, Salpietro V, Efthymiou S, Bourinaris T, Tariq A, Imdad M, et al. Identification of common genetic markers of paroxysmal neurological disorders using a network analysis approach. Neurol Sci 2020;41(4):851-7.
[Crossref] [Google Scholar] [Pub Med]
- Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization and performance of MMFF94. J Comput Chem 1996;17(5-6):490-519.
- Systèmes D. Biovia, discovery studio modeling environment. Dassault Systèmes Biovia: San Diego, CA, USA; 2016.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 1998;19(14):1639-62.
- Hollingsworth SA, Dror RO. Molecular dynamics simulation for all. Neuron 2018;99(6):1129-43.
[Crossref] [Google Scholar] [Pub Med]
- Bowers KJ, Chow DE, Xu H, Dror RO, Eastwood MP, Gregersen BA, et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. IEEE 2006;43-43.
- Patil VS, Hupparage VB, Malgi AP, Deshpande SH, Patil SA, Mallapur SP. Dual inhibition of COVID-19 spike glycoprotein and main protease 3CLpro by Withanone from Withania somnifera. Chin Herb Med 2021;13(3):359-69.
[Crossref] [Google Scholar] [Pub Med]
- Singhal AK, Naithani V, Bangar OP. Medicinal plants with a potential to treat Alzheimer and associated symptoms. Int J Nutr Pharmacol Neurol Dis 2012;2(2):84.
- H Ferreira-Vieira T, M Guimaraes I, R Silva F, M Ribeiro F. Alzheimer’s disease: Targeting the cholinergic system. Curr Neuropharmacol 2016;14(1):101-15.
[Crossref] [Google Scholar] [Pub Med]
- Cai Z. Monoamine oxidase inhibitors: Promising therapeutic agents for Alzheimer’s disease. Mol Med Rep 2014;9(5):1533-41.
[Crossref] [Google Scholar] [Pub Med]
- Berger SI, Iyengar R. Network analyses in systems pharmacology. Bioinformatics 2009;25(19):2466-72.
[Crossref] [Google Scholar] [Pub Med]
- Kanehisa, M. Post-genome informatics, serotonergic synapse. Oxford University Press; 1995
- Kanehisa, M. Post-genome informatics, cholinergic synapse. Oxford University Press; 1995.
- Kanehisa, M. Post-genome informatics, metabolic pathway. Oxford University Press; 1995.
- Kanehisa, M. Post-genome informatics, dopaminergic pathway. Oxford University Press; 1995.
- Bakoyiannis I, Daskalopoulou A, Pergialiotis V, Perrea D. Phytochemicals and cognitive health: Are flavonoids doing the trick? Biomed Pharmacother 2019;109:1488-97.
[Crossref] [Google Scholar] [Pub Med]
- Downes GB, Granato M. Acetylcholinesterase function is dispensable for sensory neurite growth but is critical for neuromuscular synapse stability. Dev Biol 2004;270(1):232-45.
[Crossref] [Google Scholar] [Pub Med]
- Hoozemans JJ, O'Banion MK. The role of COX-1 and COX-2 in Alzheimer's disease pathology and the therapeutic potentials of non-steroidal anti-inflammatory drugs. Curr Drug Targets CNS Neurol Disord 2005;4(3):307-15.
[Crossref] [Google Scholar] [Pub Med]
- Wallace JL. Prostaglandins, NSAIDs and gastric mucosal protection: Why doesn’t the stomach digest itself? Physiol Rev 2008;88(4):1547-65.
[Crossref] [Google Scholar] [Pub Med]
- Kouhestani S, Jafari A, Babaei P. Kaempferol attenuates cognitive deficit via regulating oxidative stress and neuroinflammation in an ovariectomized rat model of sporadic dementia. Neural Regen Res 2018;13(10):1827.
[Crossref] [Google Scholar] [Pub Med]
- Li S, Pu XP. Neuroprotective effect of kaempferol against a 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine-induced mouse model of Parkinson’s disease. Biol Pharm Bull 2011;34(8):1291-6.
[Crossref] [Google Scholar] [Pub Med]
- Bahrani H, Mohamad J, Paydar MJ, Rothan HA. Isolation and characterisation of acetylcholinesterase inhibitors from Aquilaria subintegra for the treatment of Alzheimer’s disease (AD). Curr Alzheimer Res 2014;11(2):206-14.
[Crossref] [Google Scholar] [Pub Med]
- Hupparage VB, Rasal VP, Patil VS, Patil PP, Mulange SG, Malgi AP, et al. Ameliorative effect of Amaranthus tricolor L. leaves on scopolamine-induced cognitive dysfunction and oxidative stress in rats. J Appl Pharm Sci 2020;10(10):111-20.
- Rendeiro C, Rhodes JS. Dietary flavonoids and brain health in aging: Food for thought. Factors Affecting Neurological Aging 2021:589-601.
- Tahir MS, Almezgagi M, Zhang Y, Bashir A, Abdullah HM, Gamah M, et al. Mechanistic new insights of flavonols on neurodegenerative diseases. Biomed Pharmacother 2021;137:111253.
[Crossref] [Google Scholar] [Pub Med]
- Devi S, Kumar V, Singh SK, Dubey AK, Kim JJ. Flavonoids: Potential candidates for the treatment of neurodegenerative disorders. Biomedicines 2021;9(2):99.
[Crossref] [Google Scholar] [Pub Med]
- Rathor L. Medicinal plants: A rich source of bioactive molecules used in drug development. Evid Based Valid Tradit Med 2021:195-209.
- Patwardhan B, Datta HS. Ayurveda and brain health. Nutraceuticals Brain Health Beyond 2021:41-53.
- Sidders B, Karlsson A, Kitching L, Torella R, Karila P, Phelan A. Network-based drug discovery: Coupling network pharmacology with phenotypic screening for neuronal excitability. J Mol Biol 2018;430(18):3005-15.
[Crossref] [Google Scholar] [Pub Med]
- Biradar P, Patil V, Joshi H, Khanal P, Mallapur S. Experimental validation and network pharmacology evaluation to decipher the mechanism of action of Erythrina variegata L. bark against scopolamine-induced memory impairment in rats. Adv Tradit Med 2020;1-4.
- Patil VS, Deshpande SH, Harish DR, Patil AS, Virge R, Nandy S, et al. Gene set enrichment analysis, network pharmacology and in silico docking approach to understand the molecular mechanism of traditional medicines for the treatment of diabetes mellitus. J Proteins Proteom 2020;11(4):297-310.