- *Corresponding Author:
- Padma Thiagarajan
Department of Biomedical Sciences, School of Biosciences and Technology, Vellore Institute of Technology, Vellore 632014, India
E-mail: padmadk4@gmail.com
| Date of Received | 01 July 2020 |
| Date of Revision | 05 February 2021 |
| Date of Acceptance | 08 June 2021 |
| Indian J Pharm Sci 2021;83(3):556-561 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Favipiravir and remdesivir are investigational drugs for coronavirus disease 2019 that is caused by severe acute respiratory syndrome coronavirus 2. The active forms of these drugs are reported to target and inhibit viral RNA dependent RNA polymerase, which is derived from 3-chymotrypsin like protease, a viral replicase enzyme. The present in silico study explores the comparative efficacy of these drugs to inhibit 3-chymotrypsin like protease and RNA dependent RNA polymerase, to plan therapeutic options for patients based on their disease severity. Active favipiravir and remdesivir molecules bind to 3-chymotrypsin like protease with energies of 6.18 and -6.52 kcal/mol in contrast to -5.62 and -3.91 kcal/mol for RNA dependent RNA polymerase. Further, hydrophobic interactions and salt bridge formations cement drug bindings with 3-chymotrypsin like protease, but not with RNA dependent RNA polymerase. Molecular dynamic simulation experiments, performed under certain experimental constraints reveal that the root mean square flexibilities of active residues in drug complexes with 3-chymotrypsin like protease are lower than in free 3-chymotrypsin like protease making the former more stable than the latter because of their rigidity and stabilities. Both drugs may hence serve as good therapeutic options for early stages of coronavirus disease 2019. However, more severe symptoms may be treated better with favipiravir due to its better binding with RNA dependent RNA polymerase, as compared to remdesivir. The “one drug does not fit all” concept is true for coronavirus disease 2019 as it is being currently realized by clinicians all around the world. Hence precise knowledge about critical interactions of these drugs with the viral enzymes will help medicos make vital therapeutic decisions on interventional options for patients who report to hospitals without over symptoms or with varying degrees of disease severity.
Keywords
Severe acute respiratory syndrome coronavirus 2, coronavirus disease 2019, favipiravir, remdesivir, 3-chymotrypsin like protease, RNA dependent RNA polymerase
Coronavirus disease 2019 (COVID-19) initially flared up at Wuhan in China during December 2019. Since then, it has developed into a pandemic because of the highly contagious nature of its causative organism, viz., severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has crown-like spike proteins on its outer surface[1,2]. The size of the SARS-CoV-2 genome ranges from 29.8-29.9 kilobase pairs. Its 5’ end has the open reading frame 1ab that encodes for the corresponding poly-proteins. Its 3’ end consists of genes encoding for its structural proteins, viz., envelope (E), surface (S), nucleocapsid (N) and membrane (M) proteins. Additionally, ORF3a, ORF6, ORF7a, ORF7b and ORF8 genes, encoding for six accessory proteins, are also present in SARS-CoV-2[3]. The clinical symptoms of COVID-19 are mainly upper and lower respiratory tract infection and at times, severe pneumonia with intense respiratory distress[4].
Extensive studies on the spike protein of SARS-CoV-2 have established that angiotensin-converting enzyme 2 (ACE-2) receptor as its main point of entry into human cells. However, other entry points may also be present for the same[5,6]. The most variable part of the coronavirus genome is the receptor-binding domain that is seen in the spike protein. Its six amino acids L455, S486, Q493, S494, N501 and Y505 are essential for the optimal binding with the ACE-2 receptor[7,8]. A single viral poly-protein, produced upon infection, is cleaved by the main viral protease 3-chymotrypsin like protease (3CLpro) at eleven sites to yield mature functional proteins. Further, non-structural proteins (nsp’s), which are cleavage products of ORF1 viral poly-proteins, facilitate viral replication and transmission[9,10]. Here, RNA dependent RNA polymerase (RdRp), i.e., nsp12, with nsp7 and nsp8 as cofactors, plays a crucial role in the replication cycle of SARS-CoV-2[11].
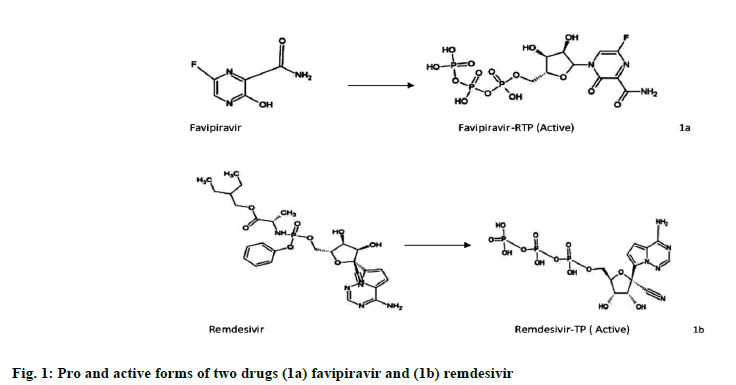
The potential therapeutic options for combating SARS-CoV-2 are now focusing on the human host’s immune system or targeting viral proteins that facilitate its replication[12]. Two investigational drug molecules, viz., favipiravir and remdesivir are currently undergoing clinical trials to treat the viral infection. Favipiravir is an antiviral drug used to treat influenza virus may be effective in COVID-19. Favipiravir is a purine analog and a derivative of pyrazinecarboxamide (6-fluoro-3-hydroxy-2-pirazinecarboxamide). It undergoes ribosylation inside the cells and phosphorylates to a metabolically active favipiravir- 4-ribofuranosyl-5’triphosphate (Favipiravir-RTP) (fig. 1a). It then competes with the purine nucleoside to incorporate into the viral RNA, thus interfering with viral replication and inhibiting RdRp[13]. Remdesivir is an antiviral drug used to treat Ebola but shown effective result in COVID-19. It is also a purine analog that converts into its metabolically active form, Remdesivirtriphosphate (fig. 1b) before competing with adenosine triphosphate (ATP), to incorporate itself into the viral RNA, thus resulting in its premature termination[14,15].
Current reports have mainly focused on the inhibition of RdRp with favipiravir and remdesivir[16,17]. In the present study, mechanistic possibility of these drugs blocking and inhibiting the critical 3CLpro viral enzyme of SARS-CoV-2 has been explored. This valuable information can support clinician decisions to plan therapeutic interventional strategies based on the severity of symptoms in patients.
Materials and Methods
Databases used:
Information about the protein targets was from the UniProt database. The research collaboratory for structural bioinformatics protein data bank (RCSB PDB) provided the structures of target proteins with PDB IDs 6LU7 and 6M71. Information on remdesivir and favipiravir were from PubChem and their active forms viz., favipiravir-4-ribofuranosyl-5-triphosphate and remdesivir triphosphate, which were not present in PubChem, were sketched in ACD/ChemSketch[18]. Open Babel was used to convert the spatial data file (SDF) format of drug compounds, downloaded from PubChem, to program database (PDB) file format[19].
Molecular docking studies and molecular dynamic simulation:
The interactions between 3CLpro and the active drug molecules were studied using AutoDock version 4.2[20]. 3CLpro was first prepared by adding polar hydrogen and then Kollman and Gasteiger charges were added. Active drug molecules as ligands were made ready by adopting the necessary steps. After fixing the active sites with specific residues, a grid box of 75×75×75 Å was used to select all active sites present in the molecule. Lamarckian genetic algorithm was employed for auto docking ligands with proteins. Analysis of hydrogen bonds, binding energies and other interactions like hydrophobicity, salt bridge interactions, etc., were carried out, using AutoDock 4.2, PyMOL 2.3.2 and Protein-Ligand interaction profiler[21]. Molecular dynamics simulation (MDS) was performed in Groningen Machine for Chemical Simulations (GROMACS) 2018[22] for both the free target proteins and their ligand complexes. The topology of the target proteins was generated, with GROMACS96 53a6 force field[23] and the ligand topology file was created, with PRODRG server[24]. A simple point charge water molecule facilitated the solvation process. Counter ions like Na+ and Cl− were used for neutralization of free target proteins and their ligand complexes. After necessary optimizations, the results were obtained, by carrying out MDS, till 2000 picosecond (ps). Root mean square fluctuation (RMSF) and Root mean square deviation (RMSD) were obtained from utility files of GROMACS[25,26].
Results and Discussion
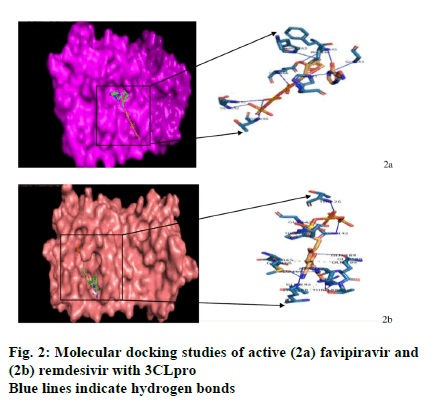
Molecular docking analysis will help to predict the binding energies and hydrogen bonds formed after the binding of free protein and ligand[27]. Active forms of both drugs were docked with the RdRp and 3CLPro using AutoDock4.2 (fig. 2a and fig. 2b). The binding energies for active favipiravir and remdesivir with 3CLpro at -6.18 and -6.52 kcal/mol are lower than those with RdRp, at -5.62 and -3.91 kcal/mol, respectively (Table 1), showing the greater binding efficiency of both drugs with 3CLpro. Also, the nine hydrogen bonds that exist in the active favipiravir-3CLpro complex are all located at the active binding residues (seven at His41, Phe140, Gly143, Cys145, His163, Glu166, Thr190 residues and two at Gln192). In contrast, in its complex with RdRp, none of these nine bonds are located at the active residues. No further interactions are seen here except for one salt-bridge formation in its complex with RdRp. Similarly, for the active Remdesivir-3CLpro complex, nine out of ten hydrogen bonds are formed at the active site residues (five at Tyr54, Asn142, Gly143, Gln189, Gln192, two at Arg188 and two at Thr190), in contrast to one (Cys645) out of five with RdRp. Additionally, four hydrophobic interactions at active sites (Met165, Glu166, Pro168, Gln189) and a salt-bridge (His41) also exist in its 3CLpro complex. Although, two hydrophobic interactions and four saltbridge interactions are present in its RdRp complex, none of them are at the active sites.
| Drug | Protein | Binding energy (Δg) kcal/mol | H bond | Inhibition constant (µm) | Inter- molecular energy |
Van der Waals (VDW)-H bond desolvation energy | Ligand efficiency |
Electro-static energy |
|---|---|---|---|---|---|---|---|---|
| Active favipiravir | 3CLpro | -6.18 | 9 | 29.51 | -8.38 | -8.94 | -0.19 | 0.52 |
| RdRp | -5.43 | 4 | 104.24 | -7.63 | -6.29 | -0.17 | -1.34 | |
| Active remdesivir | 3CLpro | -6.52 | 10 | 16.76 | -8.71 | -8.47 | -0.2 | -0.24 |
| RdRp | -4.58 | 9 | 440.87 | -6.77 | -5.62 | -0.14 | -1.15 |
Table 1: Molecular Docking Analysis of Active Favipiravir and Remdesivir with 3clpro and Rdrp
The main protease of SARS-CoV-2 is 3CLpro (nsp5). It has domains, one (residues 8-101), two (residues 102- 184) and three (residues 201-303), with an active site that is present between the first and second domains and containing a Cys-His catalytic dyad. The polyproteins cleave naturally to release the mature 3CLpro, which then cleaves the downstream nsp’s at eleven sites, releasing nsp4-nsp16[26]. The set includes nsp12, which is the RdRp. RdRp catalyzes the maturation of nsp’s that are essential for the viral life cycle. Targeting the early 3CLpro with these investigational drugs can prevent the cleavage of the downstream nsp’s at these eleven sites, eventually blocking the formation of nsp4- nsp16, including the vital RdRp and their maturation, which is essential for the viral replication.
Interesting and critical inferences arise from the results of the docking studies with these drugs. Both drugs bind efficiently to 3CLpro to prevent viral replication. Inhibition of 3CLpro, as compared to RdRp, will be a better strategy to block the early-stage viral replication that starts soon after infection. Remdesivir, with a lower binding energy (-6.52 kcal/mol), is more effective at this stage, as it binds to 3CLpro with greater efficiency than favipiravir (-6.18kcal/mol). The formation of nine hydrogen bonds at the active site residues for both drugs further supports this binding. Since this binding is additionally cemented by four active site interactions and a salt bridge for the active Remdesivir- 3CLpro complex, treatment of asymptomatic and pre-symtomatic patients with this drug will be a good interventional strategy.
Late stage treatment requires inhibition of both 3CLpro and RdRp, due to the increased viral load and circulation of the late-stage RdRp in the infected cells. In this case, favipiravir, with a binding energy of -5.62 kcal/mol is seen as a better treatment option than remdesivir with -3.91 kcal/mol even if there are no supporting hydrogen bonds in its complex with RdRp, except a salt bridge. No such bonds cement the active remdesivir-RdRp complex at active sites.
Since blocking high concentrations of late-stage RdRp require proportionally high dosages of drugs to combat the severe viral load, it is a matter of grave clinical concern. An option would be, to try combination therapies with early and late-stage enzyme blocker drugs in appropriate dosages, like remdesivir/favipiravir with other potential but less potent ones, including bioactives with good bioavailability, from dietary sources, as currently being envisaged by clinicians. However, adopting such approaches must be done with caution as the synergistic and more importantly, the antagonistic activities of drug molecules in the human host systems may not have been well documented.
MDS[25] is one of the ways of analyzing the feasibility of drug-target interactions under actual physiological conditions that exist in the biological systems. Both RMSF and RMSD serve to interpret the results of this study. Based on the molecular docking analysis, MDS was carried out only for the free 3CLpro and its complexes with the active drug molecules, due to its better binding efficiency as compared to RdRp.
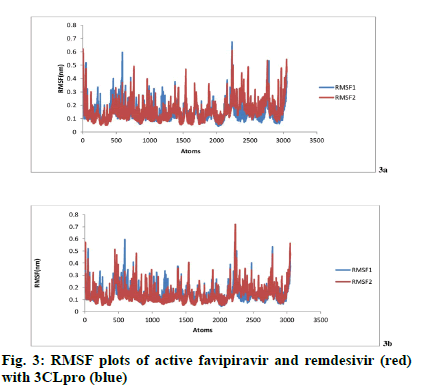
RMSF graph determines the flexibility[25] of residues between active form two drugs (remdisivir and favipiravir) and two target proteins (3CLpro, RdRp). RMSF of free protein i.e., before binding (blue) and after binding with ligand (red) is shown in fig. 3a and fig. 3b. High RMSF value signifies high flexibility and vice versa.
Fig. 3a shows the RMSF values of single amino acid residues of free protein 3CLpro (blue) and 3CLpro-favipiravir complex (red). Favipiravir showed hydrogen bonds with the active site residues of the free protein His41, Phe140, Glu166, Gly143, Cys145, His163,His164, His172, Thr190 and Gln192. RMSF values of free 3CLpro on these active sites are 0.3782 nm, 0.1282 nm, 0.0955 nm, 0.1839 nm, 0.1042 nm, 0.1242 nm, 0.1244 nm, 0.1067 nm, 0.0825 nm and 0.1364 nm respectively. The corresponding residue of 3CLpro-active favipiravir complex shows RMSF values of 0.3431 nm, 1286 nm, 0.0699 nm, 0.1809 nm, 0.086 nm, 0.0816 nm, 0.0897 nm, 0.0924 nm, 0.0738 nm and 0.1101 nm respectively. Except Phe140, all the active site residues of the 3CLproactive favipiravir complex shows lower RMSF values compared to corresponding residues of free protein 3CLpro. It can hence be concluded that the ligand favipiravir binds tightly with the protein 3CLpro. Fig. 3b shows the RMSF values of free protein 3CLpro (blue) and 3CLpro-active remdesivir complex (red). Remdesivir showed hydrogen bonds with the active site residues of the free protein Tyr54, Asn142, Gly143, Arg188, Gln189, Thr190 Gln192. RMSF values of free 3CLpro on these active sites are 0.1247 nm, 0.1723 nm, 0.1839 nm, 0.0788 nm, 0.0823 nm, 0.0825 nm and 0.1364 nm respectively. The corresponding residue of 3CLpro-active remdesivir complex shows RMSF values of 0.1214 nm, 0.2142 nm, 0.0244 nm, 0.0712 nm, 0.0727 nm, 0.0755 nm and 0.1019 nm respectively. Since the active site residues of 3CLproactive remdesivir complex shows lower RMSF values excepting for Asn142 and Gly143 compared to corresponding residues of the free protein 3CLpro, it can be concluded that the ligand i.e., active remdesivir binds tightly with the 3CLpro.
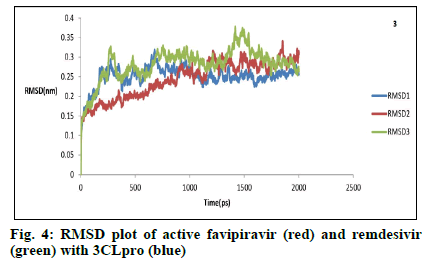
RMSD analysis helps to compare the equilibrium and stability of molecules before and after binding of the free protein with the ligand[25]. Fig. 4 shows the RMSD values for free 3CLpro (blue) and its complexes with the active favipiravir (Red) and remdesivir (green), as a function of time till 2000 ps. Initial structural re-organizations occur, at times with RMSD rising above 0.30, for both 3CLpro and its active favipiravir complex, till 1350 and 1850 ps, respectively. After that, the structures settle down with short-range oscillations around average values at 2000 ps to attain equilibrium. Active remdesivir-3CLpro complex undergoes significantly higher perturbations, as RMSD values rise till 0.37 at 1500 ps, (probably due to the usage of Chem-Sketch figures of molecules). Equilibrium is, however attained at similar values as free 3CLpro and its active favipiravir complex, at 2000 ps. Remdesivir, with a molecular weight of 602.6 g/mol, incorporates one phosphorous, eight oxygen and six nitrogen atoms. Naturally, its complex with 3CLpro will take sufficient time and turbulence to reorient, in contrast to favipiravir, which has a molecular weight of only 157.1 g/mol and contains one fluorine, two oxygen and three nitrogen atoms. The continuity of this state of equilibrium beyond 2000 ps and analysis of RMSD values of drug complexes, relative to the 3CLpro molecules are yet to be experimented.
The results here are strongly supported by a double blind, placebo controlled multicenter trial[28], which found that remdesivir is not associated with statistically significant clinical benefits in patients admitted to hospitals for severe COVID-19. Also patients with symptom durations of 10 d or less (early stage of infection) had a numerically faster time to clinical improvement with remdesivir than those receiving placebo (although larger trials may be required here). These practically observed clinical outcomes are exactly seen in our docking studies that project remdesivir to be more effective in early stage infection due to 3CLpro inhibition, rather than in severe infection wherein inhibition of RdRp is required and may be facilitated by favipiravir.
This in silico study, based on AutoDock and MDS, shows that active forms of investigational drugs favipiravir and remdesivir bind more efficiently to 3CLpro, an early-stage viral protease, as compared to a later stage viral enzyme RdRp, which is in fact derived from 3CLpro. Remdesivir inhibits 3CLpro better than favipiravir, which in turn is better for inhibiting RdRp, a late-stage viral enzyme but the latter hypothesis needs further validation. Clinicians have lately realized in actual hospital settings that different drugs may be required for each stage of COVID-19 based on the degree of severity of symptoms that each stage reflects in infected individuals. Their internal milieu is further complicated by the immune-modulatory conditions and more so in case of co-morbid patients. Hence one drug may not fit all patients at all stages of the disease. Combination therapeutics with these drugs and also with other potential and investigational molecules, including bio-actives from dietary sources, is a future area of immediate research.
Author’s contributions:
Dipjyoti Dey and Nirban Dey are joint first authors of the manuscript.
Acknowledgements:
The authors are grateful to Vellore Institute of Technology, Vellore for facilitating this study.
Conflicts of interest:
The authors declared no conflict of interest.
References
- Chavez S, Long B, Koyfman A, Liang SY. Coronavirus Disease (COVID-19): A primer for emergency physicians. Am J Emerg Med; 2020.
- Shereen MA, Khan S, Kazmi A, Bashir N, Siddique R. COVID-19 infection: Origin, transmission and characteristics of human coronaviruses. J Adv Res 2020;24:91-8.
- Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep 2020;19.
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020;395:497-506.
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol 2020;5(4):562-9.
- Ortega JT, Serrano ML, Pujol FH, Rangel HR. Role of changes in SARS-CoV-2 spike protein in the interaction with the human ACE2 receptor: An in silico analysis. EXCLI J 2020;19:410-7.
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, et al. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020;581:215-20.
- Hussain M, Jabeen N, Raza F, Shabbir S, Baig AA, Amanullah A, et al. Structural variations in human ACE2 may influence its binding with SARS‐CoV‐2 spike protein. J Med Virol 2020;92(9):1580-6.
- Jin Z, Du X, Xu Y, Deng Y, Liu M, Zhao Y, et al. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature 2020;582:289-93.
- Pillaiyar T, Manickam M, Namasivayam V, Hayashi Y, Jung SH. An overview of severe acute respiratory syndrome–coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem 2016;59(14):6595-628.
- Kirchdoerfer RN, Ward AB. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun 2019;10(1):1-9.
- Liu C, Zhou Q, Li Y, Garner LV, Watkins SP, Carter LJ, et al. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent Sci 2020;6(3):315-31.
- Du YX, Chen XP. Favipiravir: pharmacokinetics and concerns about clinical trials for 2019‐nCoV infection. Clin. Pharmacol Ther 2020;108(2):242-7.
- Furuta Y, Komeno T, Nakamura T. Favipiravir (T-705), a broad spectrum inhibitor of viral RNA polymerase. Proc Jpn Acad Ser B Phys Biol Sci 2017;93(7):449-63.
- Tchesnokov EP, Feng JY, Porter DP, Gotte M. Mechanism of inhibition of Ebola virus RNA-dependent RNA polymerase by remdesivir. Viruses 2019;11(4):326.
- Elfiky AA. Ribavirin, Remdesivir, Sofosbuvir, Galidesivir and Tenofovir against SARS-CoV-2 RNA dependent RNA polymerase (RdRp): A molecular docking study. Life Sci 2020;253:117592.
- Elfiky AA. SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. J Biomol Struct Dyn 2020:1-9.
- Spessard GO. ACD Labs/LogP dB 3.5 and ChemSketch 3.5. J Chem Inf Comput Sci 1998;38(6):1250-3.
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform 2011;3(1):1-4.
- Forli S, Huey R, Pique ME, Sanner MF, Goodsell DS, Olson AJ. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat Protoc 2016;11(5):905-19.
- Salentin S, Schreiber S, Haupt VJ, Adasme MF, Schroeder M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res 2015;43(W1):W443-7.
- Lindahl E, Hess B, Van Der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model 200;7(8):306-17.
- Oostenbrink C, Villa A, Mark AE, Van Gunsteren WF. A biomolecular force field based on the free enthalpy of hydration and solvation: the GROMOS force‐field parameter sets 53A5 and 53A6. J Comput Chem 2004;25(13):1656-76.
- Schuttelkopf AW, Van Aalten DM. PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr 2004;60(8):1355-63.
- Ragunathan A, Malathi K, Ramaiah S, Anbarasu A. FtsA as a cidal target for Staphylococcus aureus: Molecular docking and dynamics studies. J Cell Biochem 2019;120(5):7751-8.
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B 2020;10(5):766-88.
- Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 2011;7(2):146-57.
- Wang Y, Zhang D, Du G, Du R, Zhao J, Jin Y, et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet 2020;395:1569-78.