- *Corresponding Author:
- Xuewen. Kang
Department of Orthopaedics, Lanzhou University Second Hospital, No.82 Cuiyingmen, Chengguan District, Lanzhou, Gansu, 730000, China
E-mail: zhuixian163661@163.com
| This article was originally published in a special issue, “Biomedical Research in Clinical and Preclinical Pharmaceutics” |
| Indian J Pharm Sci 2020:82(3)Spl issue7;31-38 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Previous studies have shown that the decalcified bone matrix was a potential candidate for bone regeneration. However, the decalcified bone matrix has insufficient bone induction ability, so it cannot repair bone defects alone. Osteoblasts were the main cellular component of bone tissue, and the extracellular matrix secreted by the osteoblasts was one of the main components of bone formation microenvironment. Therefore, this study aims to combine decalcified bone matrix and extracellular matrix to construct a new type of bone reconstruction scaffold. In this study, osteoblasts were cultured on the surface of decalcified bone matrix and then osteoblasts extracellular matrix modified decalcified bone matrix were isolated and prepared to compare and evaluate the characteristics of the two materials, including the extracellular matrix and extracellular matrix residues, cytotoxicity, and bone induction ability. Low cell residue and low content of deoxyribonucleic acid were observed in osteoblasts extracellular matrix modified decalcified bone matrix. Compared with decalcified bone matrix, osteoblasts extracellular matrix modified decalcified bone matrix has more organic matrix proteins in bone tissue, such as type I collagen, osteopontin, and osteocalcin. When cultured on two kinds of materials, rat bone marrow mesenchymal stem cells have good viability. Compared with decalcified bone matrix group, the expression of osteogenic gene and protein of rat bone marrow mesenchymal stem cells in the osteoblasts extracellular matrix modified decalcified bone matrix group was significantly up-regulated. Our results suggest that osteoblasts may be the ideal seed cells to improve the performance of engineered bone scaffolds.
Keywords
Bone marrow mesenchymal stem cells, bone regeneration, new biomaterials, bone matrix
The bone defect was a common problem after trauma and tumor operation, which will reduce the quality of life of the patients[1]. Vascularized autograft was the standard treatment for bone defect[2]. However, this method has inevitable defects, which limited the application of vascularized autograft, such as the lack of donor, incidence and infection of the donor site. Bone tissue engineering was considered to be a promising method to reconstruct the damaged bone. As the most famous scaffold in bone tissue engineering, decalcified bone matrix (DBMs) has been widely used as a candidate material for bone defect repair[3,4]. Because of its good biological properties, including biomechanical strength, biocompatibility, and osteogenic induction, DBMs shows its advantages as an excellent scaffold for bone tissue engineering. However, the effect of DBMs on the repair of bone defects was still controversial. Through the previous study on the bone healing of DBMs, it was found that neither DBMs could not reconstruct the bone defects alone[5]. Therefore, more and more studies have found that DBMs combined with other bioactive factors may have a better effect on bone healing. In addition to the growth factors, inorganic compounds and polymers were also used to enhance the osteogenic ability of DBMs. The combination of DBMs, nanostructured hydroxyapatite, and poly (N-isopropylacrylamide) can increase the production of Alp and osteocalcin (Ocn) in human bone marrow mesenchymal stem cells[6]. These combination methods can be regarded as the improvements of DBMs. However, so far, there is no widely accepted improvement plan.
Extracellular matrix (ECM) secreted by the cells was considered to be an important part of the tissue microenvironment, which can affect the fate of the stem cells in the microenvironment. ECM secreted by the smooth muscle cells has been shown to have the potential to induce the human mesenchymal stromal cells/stem cells to differentiate into smooth muscle cells[7]. Chondrocytes inoculated on the ECM produced by human bone marrow stromal cells (BMSC) showed an increased proliferation rate and a better phenotype than those inoculated on tissue culture plastics[8]. ECM secreted by the human umbilical cord BMSC has been used as an improver to enhance the ability of DBM to repair bone defects[9]. A large number of studies have shown that ECM secreted by the somatic cells or stem cells has the potential to enhance the properties of tissue engineering scaffolds.
As the main cellular component of bone, osteoblasts (OB) play an important role in bone development[10,11]. OB stimulated by miR-142-5p (a kind of microRNA) showed enhanced activity, stronger matrix mineralization, and better fracture healing performance, even in the aged mouse models[12]. Therefore, OB was considered to be one of the seed cells in bone tissue engineering. During bone development, OB can secrete a series of organic components, such as type I collagen (Col I), osteopontin (Opn), and osteocalcin (Ocn) [13]. These organic substrates have been proved to be beneficial to osteogenesis and bone healing. Col-1 was used to modify the surface of cultured cells to promote the osteogenic differentiation of dental follicle cells[14]. Opn has been shown to play a role in mineralization, cell adhesion, and migration[15]. It also interacts with collagen and hydroxyapatite minerals to determine the structural characteristics of bone[16]. It seems that ECM secreted by OB may be a candidate for surface modification of DBMs. Therefore, we speculate that DBMs modified by ECM may have a stronger ability of bone induction. In this study, the oxidation-derived DBMs and OBs-ECM modified DBMs (OEDBMs) were prepared. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting (WB) were used to detect the content of Col I, Opn, Ocn in DBMs and OEDBMs. Besides, the cell viability, osteogenic gene and protein expression of rat BMSC were measured by cell counting kit-8, quantitative realtime polymerase chain reaction (qRT-PCR), and WB, respectively.
Materials and Methods
Preparation of DBMs
According to the previous study[16], DBMs from the cow cortical bones were prepared. In short, the cow cortical bones were incubated in the hydrochloric acid (0.6M) for 72 h (room temperature). The DBMs were then washed several times with phosphate buffer solution (PBS) until the pH value was neutral. After desalination and washing, all samples were cut into 200mm slices and freeze-dried, and all samples were disinfected with ethylene oxide.
Preparation of OEDBMs
OBs (BNCC341401, ATCC) (4.0*104 cells/cm2) was inoculated on the surface of DBMs and cultured in DMEM medium (containing 10 % fetal bovine serum). After 10 d of culture, OBs in OBs-DBMs was removed. In short, the OBs-DBMs was washed with PBS for 3 times (10 min each time) and then treated with 0.5 % Triton X-100 (20 mM ammonium hydroxide) for 5 min (37°) to remove OBs. In addition, three times of PBS washing were performed to eliminate the residual reagents.
Qualitative and quantitative evaluation of the secondary decellularization
To qualitatively evaluate the cell residue in OEDBMs, the samples of DBMs, OBs-DBMs, and OEDBMs were fixed in 10 % formaldehyde for 2 h (4º). All the samples were then dehydrated in the graded ethanol and embedded in paraffin to make 5 mm slices. The residue of OBs was observed by hematoxylin-eosin (H&E) staining. The dry weight of all samples was used for quantitative evaluation. Metalloproteinase K was used to digest all samples. The mixture of phenol/ chloroform/isoamyl alcohol was used to extract DNA. The purification process was carried out by sodium acetate and ethanol. Then the DNA contents of DBM (n=5), OBs-DBM (n=5), and OEDBM (n=5) were detected by Quant-iT PicoGreen ds DNA detection kit.
Cytocompatibility of DBM and OEDBM
To evaluate the cytocompatibility of DBM and OEDBM, rBMSC (BNCC340947, ATCC) (1.0* 104 cells/cm2) was inoculated on the two materials. The samples were tested by dead/living cell staining at each time point (1, 3, and 5 d). In short, all samples were washed with PBS and immersed in dead/living cell staining reagents (2mM Calcein acetoxymethyl ester and 4mM propidium iodide) for half an hour, and then washed 3 times with PBS to remove the residual stains. The cytocompatibility of the two materials was observed by a fluorescence inverted microscope. The average integrated optical densities (IOD/mm2) of the living/dead cells in 2 random regions of 5 samples in each group were collected by Image-Pro Plus 6.0.
Detection of ECM
The total ECM proteins were extracted by RIPA lysis buffer, and the content of ECM in OEDBMs was quantitatively detected by the enhanced BCA protein detection kit, and DBMs were used as control. After quantification, all samples were mixed with SDS-PAGE sample loading buffer, and then heated at 100° for 5 min to denature all proteins. Subsequently, SDSPAGE protein separation and WB analysis were carried out. In short, all samples were separated with 10 % SDSPAGE, the gel was stained with Coomassie brilliant blue staining solution. For WB, a Bio-Rad wet blotting system (Bio-Rad Laboratories, Hercules, CA) was used to transfer all proteins on the gel to the 0.2 mm PVDF membrane. The PVDF membrane was immersed in 5 % skim milk at room temperature to blocking the antigen sites for 2 h. Then the primary antibody was added and incubated overnight at 4º (mouse anti-glyceraldehyde- 3-phosphate dehydrogenase, mouse anti-Opn, mouse anti-Col I, and mouse anti-Ocn). After washing with TBST for 6 times, remove the excess antibody. The membrane was then incubated with HRP-labeled goat anti-mouse secondary antibody at room temperature for 2 h. The excess secondary antibody was eliminated by TBST washing. After washing, the color was developed with enhanced chemiluminescence (ECL) and observed with a CCD camera gel imaging system.
Osteogenic induction ability of OEDBMs
The bone induction ability of OEDBM was evaluated by quantitative real-time polymerase chain reaction (RT-PCR). rBMSC (1.0±105 cells/cm2) was cultured on OEDBM for 3, 7, and 10 d, and the expression of bone specific genes in rBMSCs was detected at each time point. The rBMSC cultured in 6-well plate was used as control. Total RNA was extracted by TRIzol (Life) and cDNA was synthesized by reverse transcription kit (Tokyo Takara, Japan). The primers for bone specific gene identification (Table 1) were synthesized by iGeneTech Co. Ltd. (Beijing, China). SYBR green Supermix and Roche photocycler 96 systems were used for RT-qPCR. To normalize the expression of the target gene, the GAPDH gene was used as a reference. All the data were
| Genes | 5’-3’ | Primer sequences | Production size (bp) |
|---|---|---|---|
| Alp | Forward | CATCGGACCCTGCCTTAC | 169 |
| Reverse | GGAGACGCCCATACCATC | ||
| Gapdh | Forward | GCAAGTTCAACGGCACAG | 140 |
| Reverse | GCCAGTAGACTCCACGACAT | ||
| On | Forward | GCACCACTCGCTTCTTTG | 103 |
| Reverse | TTGTTGATGTCCTGCTCCT | ||
| Runx2 | Forward | CCCAGTATGAGAGTAGGTGTCC | 149 |
| Reverse | GGGTAAGACTGGTCATAGGACC |
Table 1: The Primers used for the Detection of the Bone Specific Genes
WB analysis of osteogenic proteins
The experiment method was described in the 1.5 section. The antibodies used in this section were anti-GAPDH, anti-Ocn, anti-Runx2, anti-Alp, and HRP labeled goat anti-mouse secondary antibody.
Statistical analysis
All data were expressed as mean±standard deviation (SD) and analyzed with SPSS 22.0. For the comparison between the two groups, 2-tailed t-test was used to detect the significance of the data. For multi-group comparison, one-way analysis of variance was used to analyze the significance of the data. p<0.05 was considered to be statistically significant.
Results
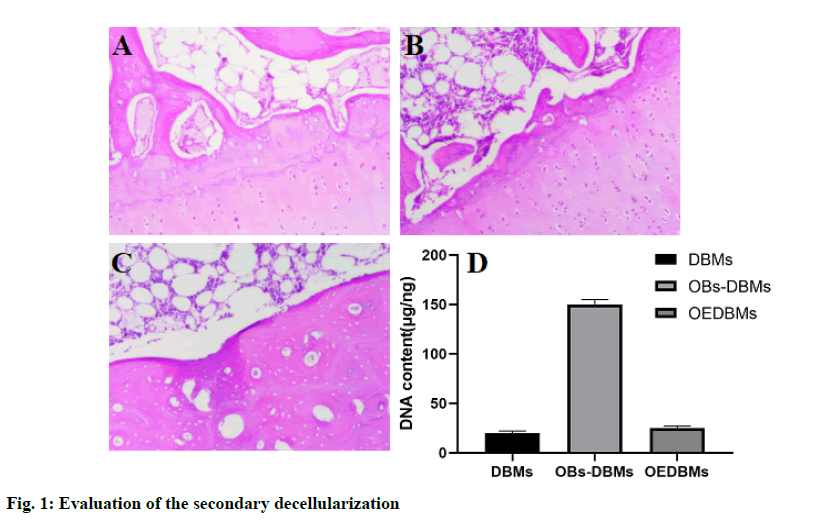
H&E staining was used for qualitative evaluation, and the results showed the role of decellularization in DBM, OBs-DBM, and OEDBM. There were abundant cells on the surface of OBs-DBMs, while low residual cells were observed in DBMs and OEDBMs (fig. 1Afig. 1C). In addition, the DNA content in the three groups was used for the quantitative evaluation of decellularization. The DNA content (mg/ng) of the three groups was 19.70±2.40 (DBMs), 153.77±10.93 (OBs-DBMs), and 23.10±2.12 (OEBMs). Compared with OBs-DBMs, the content of DNA in DBMs (p=0.004) and OEDBMs (p=0.005) groups decreased significantly. There was no significant difference between DBMs and OEDBMs group (p=0.364). The results showed that the decellularization was succeeded in DBMs and OEDBMs groups (fig. 1 D).
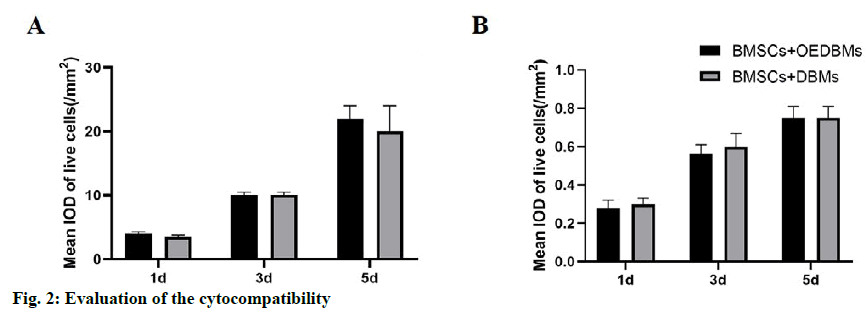
Similar rBMSCs activity was observed at all-time points in DBMs and OEDBMs group. According to the mean IOD analysis, it showed that the cells proliferated well with the extension of culture time (fig. 2A). There was no significant difference between the two groups at each time point (1st d, p=0.346; 3rd d, p=0.902; 5th d, p=0.229). Similarly, there was no significant difference in the red fluorescence mean IOD between the two groups at each time point (fig. 2B) (1st d, p=0.057; 3rd d, p=0.429; 5th d, p=0.452). These results showed that there was no significant difference in the cytocompatibility between DBMs and OEDBMs.
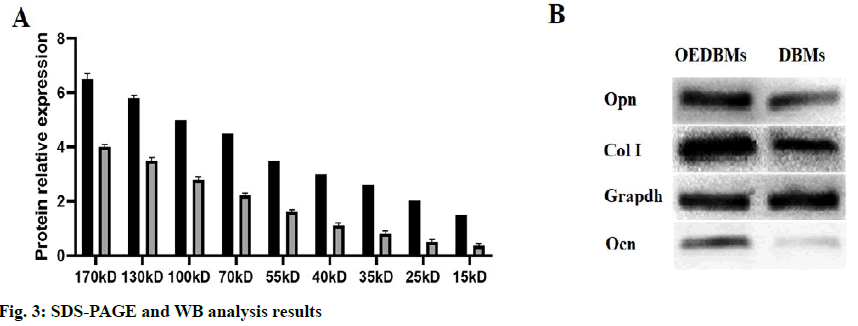
The contents of the total protein and bone-associated protein in DBM group and OEDBM group were detected by SDS-PAGE and WB. As shown by SDS-PAGE (fig. 3A), the expression of the protein in OEDBMs group was significantly higher than that in DBMs group. In the OEDBMs group, a significant increase in protein expression was observed in the range of 15 to 170 kDa. On the contrary, the protein expression in DBMs was significantly low (p<0.05), and the difference between them was significant (p<0.05). Also, compared with DBMs, higher expression levels of Col I, Ocn, and Opn were detected in the OEDBMs group (fig. 3B). This may indicate that OEDBMs has more ECM in the case of low cell residue.
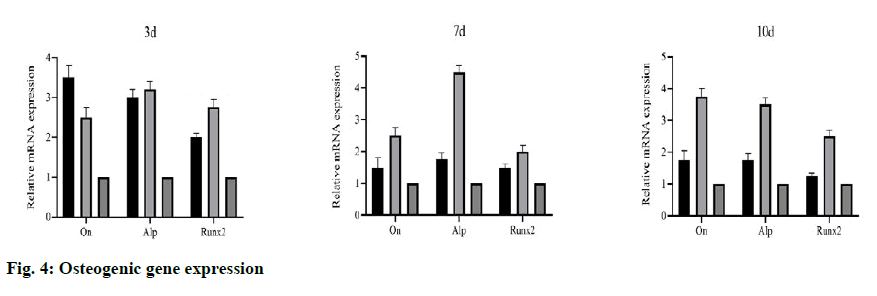
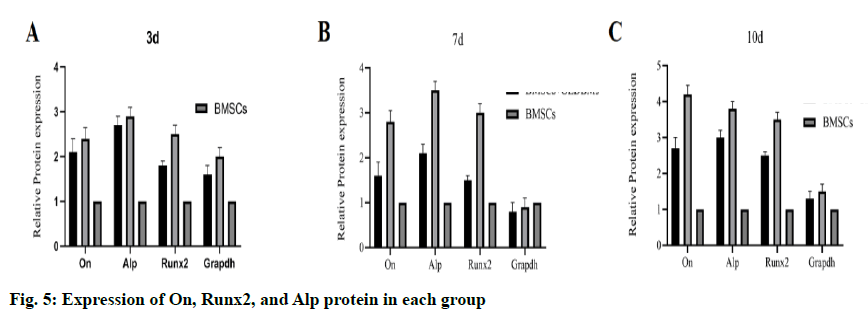
To evaluate the bone induction ability of OEDBMs, the expression levels of On, Runx2, and Alp genes in rBMSCs were analyzed. Compared with the DBMs group, the level of on in the OEDBMs group increased significantly at all-time points (fig. 4A- fig. 4C). A similar trend was observed in the expression level of Alp (fig 4C). The expression level of Runx2 in the OEDBMs group was significantly higher than that in the DBMs group at the 7th and 10th d (fig. 4B). At alltime points, the expression level of all genes in the two experimental groups was higher than that in the control group (fig 4A- fig. 4C).
The result of WB was similar to that of gene expression. At each time point, the protein expression level in the OEDBMs group was the highest (fig. 5A- fig. 4C). The results showed that OEDBMs had stronger osteogenic induction ability than DBMs and petri dish.
Discussion and Results
In this study, two kinds of materials, DBMs and OEDBMs were prepared. The two materials were compared and analyzed to reflect the differences in cell residue, cytocompatibility, ECM, and bone induction. H&E staining and DNA content detection showed that OEDBMs had low cell residue similar to DBMs, and OEDBMs had the same cytocompatibility as DBMs. SDS-PAGE and WB analysis showed that compared with DBMs, Col I, Opn, and Ocn protein was highly expressed in OEDBMs. The results of osteogenic differentiation of rBMSCs in vitro showed that OEDBMs had a stronger ability of bone induction.
Low immunogenicity was the basic requirement of good scaffolds, which was very important for implantation. For decellularization scaffolds, low immunogenicity means low cell residue, which depends on the effective decellularization[17,18]. Detergent (SDS, Triton X-100, sodium deoxycholate) and nucleases (deoxyribonuclease and ribonuclease) were usually used to remove the cells because of their excellent decellularization effect. In this study, detergents and nucleases were combined to prepare DBMs. Our DBMs showed similar results of H&E staining and DNA content to those of previous studies[19,20]. After the first decellularization, DBMs was recellularized with OBs to harvest the secreted ECM, and partially simulate the microenvironment of bone development. After 10 d of culture, Triton X-100 and ammonium hydroxide were used for the second decellularization to reduce the immunogenicity of OEDBMs. In fact, this scheme has been widely used to collect the cell-derived ECM[21,22]. H&E staining and DNA content were also showed that the second decellularization was effective.
Considering the possible cytotoxicity of detergents and nucleases, the cytocompatibility of OEDBMs was tested in this study, with DBMs as the control group[23,24]. According to the results of staining and fluorescence intensity of dead/living cells, DBMs and OEDBMs showed that the green fluorescence intensity of living cells and living cells increased gradually with the extension of culture time. Although the difference was not significant (p>0.05), the green fluorescence intensity of OEDBMs was always higher than that of DBMs at all-time points, which may be related to the expression of Opn, Ocn, and Col I. Therefore, it is speculated that OEDBMs has good cytocompatibility similar to DBMs.
Simulating the microenvironment of bone development is another goal of this study. ECM secreted by OBs and OB plays an important role in bone development and remodeling[25,26]. Therefore, the matrix proteins secreted by OBs were considered to be important components in simulating the microenvironment of bone development. In this study, OBs proliferated well on the surface of DBMs, and they secreted rich ECM. The difference of the total protein between DBMs and OEDBMs was found by SDS-PAGE. DBMs only expressed high molecular weight proteins, while OEDBMs contains a large number of proteins with different molecular weights. Because a large amount of ECM was secreted by the OBs, the organic matrix proteins (Ocn, Opn, and Col I) in OEDBMs were higher than that in DBMs.
There were two main organic phases in bone tissue, collagen and non-collagen protein. Ocn and Opn account for 20 % and 10 % of non-collagen protein, respectively[27]. As the most abundant non-collagen protein in bone, Ocn plays an important role in the bone development[28], especially mineralization[29]. As a recognized marker of osteogenesis, Opn also enhances the osteogenic differentiation and bone formation of stem cells[30]. In addition, Ocn and Opn have synergistic enhancement effect on osteogenic differentiation of human bone marrow mesenchymal stem cells/stromal cells[31]. Both Ocn and Opn have the potential to induce angiogenesis, which was essential for bone development and regeneration[32-34]. Col 1 was also very important for mineralization in the process of bone formation[18]. Previous studies have shown that Col I can be used as a substrate to promote osteogenic differentiation of stem cells[35]. Therefore, the difference of protein expression in Ocn, Opn, and Col I indicates that the bone induction ability of DBMs and OEDBMs may be different.
BMSCs were considered to be an ideal seed cell for bone tissue engineering. Therefore, BMSCs was selected for osteogenic differentiation in this study. As markers of osteogenesis, On, Runx2, and Alp have been used to detect the osteogenic differentiation of stem cells[23,35]. In addition, the inducing ability of DBMs to promote osteogenic differentiation of BMSCs has been widely reported[36]. Similarly, in this study, stronger osteogenic differentiation of DBMs was observed when inoculated with rBMSCs. In terms of gene expression and protein expression, the expression levels of On, Runx2, and Alp were the highest in OEDBMs group. These results were consistent with the previous studies, which have demonstrated the enhanced effect of Opn, Ocn, and Col I on the osteogenic differentiation of stem cells[19,32]. Therefore, OEDBMs had more advantages than DBMs in simulating the microenvironment of bone development, and has a stronger ability to promote the osteogenic differentiation of rBMSCs.
To sum up, OEDBMs preserves OBs-ECM and low cell residues. Compared with DBMs and petri dishes, rBMSCs inoculated on OEDBMs showed stronger osteogenic differentiation potential. OBs-ECM can be used as a promising tool to modify the surface of scaffolds to create a potentially better biological scaffold for bone remodeling.
Authors contributions
Xuewen Kang conceived and designed the experiments; Yong Yang, Yonggang Wang, Xuchang Hu and Bing Ma performed the experiments; Shuai Xing analyzed the data and wrote the paper.
Acknowledgements
This work was supported by Lanzhou University Second Hospital.
Conflict of interests
The authors declared no conflict of interest.
References
- Roseti L, Parisi V, Petretta M, Cavallo C, Desando G, Bartolotti I, et al. Scaffolds for bone tissue engineering: state of the art and new perspectives. Mater Sci Eng C Mater Biol Appl 2017;78:1246-62.
- Mishra R, Bishop T, Valerio IL, Fisher JP, Dean D. The potential impact of bone tissue engineering in the clinic. Regener Med 2016;11:571-87.
- Hinsenkamp M, Collard JF. Growth factors in orthopaedic surgery: demineralized bone matrix versus recombinant bone morphogenetic proteins. Int Orthop 2015;39:137-47.
- Yu SH, Chan HL, Chong LY, Jheng YH, Chang PC. Evaluation of the osteogenic potential of growth factor-rich demineralized bone matrix in vivo. J Periodontol 2015;86:36-43.
- Alidadi S, Oryan A, Bigham-Sadegh A, Moshiri A. Comparative study on the healing potential of chitosan, polymethylmethacrylate, and demineralized bone matrix in radial bone defects of rat. Carbohydr Polym 2017;166:236-48.
- Rodriguez RU, Kemper N, Breathwaite E, Dutta SM, Huber A, Murchison A. Demineralized bone matrix fibers formable as general and custom 3D printed mold-based implants for promoting bone regeneration. Bio fabrication 2016;8:035007.
- Fassbender M, Minkwitz S, Thiele M, Wildemann B. Efficacy of two different demineralized bone matrix grafts to promote bone healing in a critical-size-defect: a radiological, histological and histomorphometric study inrat femurs. Int Orthop 2014;38:1963-9.
- Zhu W, Qiu Y, Sheng F, Yuan X, Xu L, Bao H, et al. An effective delivery vehicle of demineralized bone matrix incorporated with engineered collagen-binding human bone morphogenetic protein-2 to accelerate spinal fusion at low dose. J Mater Sci Mater Med 2017;29:2.
- Saito W, Uchida K, Matsushita O, Inoue G, Sekiguchi H, Aikawa J, et al. Acceleration of call us formation during fracture healing using basic fibroblast growth factor-kidney disease domain-collagen-binding domain fusion protein combined with allogenic demineralized bone powder. J Orthop Surg Res 2015;10:59.
- Nicoletti A, Torricelli P, Bigi A, Fornasari P, Fini M, Moroni L. Incorporation of nano structure dhydroxyapatiteandpoly (N-isopropylacrylamide) in demineralized bone matrix enhances osteoblast and human mesenchymal stem cell activity. Biointerphases 2015;10:041001.
- Rao Pattabhi S, Martinez JS, Keller TC. 3rd Decellularized ECM effects on human mesenchymal stem cell stemness and differentiation. Differentiation 2014;88:131-43.
- Yang Y, Lin H, Shen H, Wang B, Lei G, Tuan RS. Mesenchymal stem cell-derived extracellular matrix enhances chondrogenic phenotype of and cartilage formation by encapsulated chondrocytes in vitro and in vivo. ActaBiomate 2018;69:71-82.
- Deng M, Luo K, Hou T, Luo F, Xie Z, Zhang Z, et al. IGFBP3 deposited in the human umbilical cord mesenchymal stem cell-secreted extracellular matrix promotes bone formation. J Cell Physiol 2018;233:5792-804.
- Souza AT, Freitas GP, Lopes HB, Ferraz EP, Oliveira FS, Beloti MM, et al. Effect of cell therapy with allogeneic osteoblasts on bone repair of rat calvaria defects. Cytotherapy 2018;20:1267-77.
- Wischmann J, Lenze F, Thiel A, Bookbinder S, Querido W, Schmidt O, et al. Matrix mineralization controls gene expression in osteoblastic cells. Exp Cell Res 2018;372:25-34.
- Tu M, Tang J, He H, Cheng P, Chen C. MiR-142-5p promotes bone repair by maintaining osteoblast activity. J Bone Miner Metab 2017;35:255-64.
- Yoon JK, Kim HN, Bhang SH, Shin JY, Han J, La WG, et al. Enhanced bone repair by guided osteoblast recruitment using topographically defined implant. Tissue Eng Part A 2016;22:654-64.
- Fujisawa R, Tamura M. Acidic bone matrix proteins and their roles in calcification. Front Biosci 2012;17:1891-903.
- Yang JL, Yao X, Qing Q, Zhang Y, Jiang YL, Ning LJ, et al. An engineered tendon / ligament bio scaffold derived from decellularized and demineralized cortical bone matrix. J Biomed Mater Res A 2018;106:468-78.
- Liu S, Wang Y, Wang J, Qiu P, Wang S, Shi Y, et al. A cancellous bone matrix system with specific mineralization degrees for mesenchymal stem cell differentiation and bone regeneration. Biomater Sci 2019;7:2452-67.
- Farag A, Vaquette C, Theodoropoulos C, Hamlet SM, Hutmacher DW, Ivanovski S. Decellularized periodontal ligament cell sheets with recellularization potential. J Dent Res 2014;93:1313-9.
- Farag A, Hashimi SM, Vaquette C, Volpato FZ, Hutmacher DW, Ivanovski S. Assessment of static and perfusion methods for decellularization of PCL membrane-supported period on tall ligament cell sheet constructs. Arch Oral Biol 2018;88:67-76.
- Dozza B, Lesci IG, Duchi S, Della Bella E, Martini L, Salamanna F. When size matters: differences in demineralized bone matrix particles affect collagen structure, mesenchymal stem cell behavior, and osteogenic potential. J Biomed Mater Res 2017;105:1019-33.
- Leszczak V, Place LW, Franz N, Popat KC, Kipper MJ. Nanostructured biomaterials from electrospun demineralized bone matrix: a survey of processing and crosslinking strategies. ACS Appl Mater Interfaces 2014;6:9328-37.
- Carvalho MS, Cabral JM, da Silva CL, Vashishth D. Synergistic effect of extracellularly supplemented osteopontin and osteocalcin on stem cell proliferation, osteogenic differentiation, and angiogenic properties. J Cell Biochem 2019;120:6555-69.
- Liu X, Xu Q, Liu W, Yao G, Zhao Y, Xu F. Enhanced migration of murine fibroblast-like3 T3-L1 preadipocytes on type I collagen-coated dish is reversed by silibinin treatment. Mol Cell Biochem 2018; 441: 35-62.
- Filipowska J, Tomaszewski KA, Niedźwiedzki Ł, Walocha JA, Niedźwiedzki T. The role of vasculature in bone development, regeneration and proper systemic functioning. Angiogenesis 2017;20:291-302.
- Kusumbe AP, Ramasamy SK, Adams RH. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature 2014;507:323-8.
- Zhang Q, Nakamoto T, Chen S, Kawazoe N, Lin K, Chang J, et al. Collagen/Wollastonite nano wirehybrid scaffolds promoting osteogenic differentiation and angiogenicfactor expression of mesenchymal stem cells. J Nanosci Nanotechnol 2014;14:3221-7.
- Caliari SR, Harley BA. Structural and biochemical modification of acollagen scaffold to selectively enhance MSC tenogenic, chondrogenic,and osteogenic differentiation. Adv Healthc Mater 2014;3:1086-96.
- Qin Y, Wang L, Gao Z, Chen G, Zhang C. Bone marrow stromal/stem cell-derived extracellular vesi cles regulate osteoblast activity and differentiation in vitro and promote bone regeneration in vivo. Sci Rep 2016;6:21961.
- Liu Y, Ming L, Luo H, Liu W, Zhang Y, Liu H. Integration of a calcined bovine bone andBMSC-sheet 3D scaffold and the promotion of bone regeneration inlarge defects. Biomaterials 2013;34:9998-10006.
- Esmailian S, Irani S, Bakhshi H, Zandi M. Biodegradable bead-on-springnanofibers releasing beta-carotene for bone tissue engineering. Mater Sci Eng C Mater Biol Appl 2018;92:800-6.
- Midha S, Kumar S, Sharma A, Kaur K, Shi X, Naruphontjirakul P, et al. Silk fibroin-bioactive glass basedadvanced biomaterials: towards patient-specific bone grafts. BiomedMater 2018;13:055012.
- Kim MO, Jung H, Kim SC, Park JK, Seo YK. Electromagnetic fields and nanoma gnetic particles increase the osteogenic differentiation of human bone marrow-derived mesenchymal stem cells. Int J Mol Med 2015;35:153-60.
- Chen Y, Ye SH, Sato H, Zhu Y, Shanov V, Tiasha T, et al. Hybrid scaffolds of Mg alloy meshreinforced polymer/extracellular matrix composite for critical-sizedcalvarial defect reconstruction. J Tissue Eng Regen Med 2018;12:1374-88.