- *Corresponding Author:
- Lizhi Zhao
Department of Cardiology, The Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University, Luzhou, Sichuan 646000, China
E-mail: zhaolizhi@swmu.edu.cn
| This article was originally published in a special issue, “Emerging Therapeutic Interventions of Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(3) Spl Issue “152-162” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Atrial fibrillation is a serious global public health risk. Conventional treatments for atrial fibrillation include surgery, pharmacologic cardioversion, anticoagulation, and ventricular rate control. There have been few studies on the treatment of atrial fibrillation by traditional Chinese medicine, and Yangxin Dingji decoction is one such traditional anti-arrhythmic herbal compound. However, there are few studies on its active ingredients and their possible mechanisms for the treatment of atrial fibrillation. We obtained the active chemical components of Yangxin Dingji decoction from different databases, analysed the bioavailability and pharmacodynamics of each active ingredient of Yangxin Dingji decoction using Swiss absorption, distribution, metabolism and excretion, and predicted its possible targets of action using traditional Chinese medicine systems pharmacology and SwissTargetPrediction. Then, we collected relevant targets of atrial fibrillation through DrugBank, therapeutic targets database, MalaCards, traditional Chinese medicine systems pharmacology, DisGeNet, and online mendelian inheritance in man. Based on this data protein-protein interaction networks were constructed. Finally, Metascape was systematically used to analyse the targets of Yangxin Dingji decoction intervention in atrial fibrillation. 213 active ingredients and 1191 Yangxin Dingji decoction targets were collected, and 362 atrial fibrillation-related targets were predicted, of which 18 intersected with Yangxin Dingji decoction targets. Molecular docking verified that Yangxin Dingji decoction can bind tightly to core targets. Based on the results of network pharmacology and enrichment analysis, we predicted that the main targets of Yangxin Dingji decoction intervention in atrial fibrillation which mainly treat atrial fibrillation through adrenergic signalling pathway, calcium signalling pathway, and voltage-gated potassium channel.
Keywords
Network pharmacology, Yangxin Dingji decoction, atrial fibrillation, molecular docking
Atrial fibrillation is a common heart condition that causes an irregular heartbeat[1]. It can be caused by various factors and appears as rapid oscillation or fibrillation on the Electrocardiogram (ECG) [2]. In recent years, there has been an increase in hypertension and coronary heart disease, which has led to a rise in atrial fibrillation cases[3]. A study has pointed out that patients with atrial fibrillation have a 5-fold increased risk of ischemic stroke[4], which can severely impact their daily lives, reduce their quality of life, and even threaten their life safety. This makes the prevention and treatment of atrial fibrillation more challenging. Western medicine treats atrial fibrillation by controlling the primary disease and risk factors[5]. Specific treatment strategies include medication to reduce and control ventricular rate and prevent thrombosis, as well as surgical treatments such as radiofrequency ablation and surgical atrial maze. However, drug treatment can lead to adverse reactions and unpredictable events, making it difficult to control the medication regimen, while surgical treatments can be expensive and prone to recurrence[6]. Therefore, treating atrial fibrillation poses a significant challenge.
Yangxin Dingji Decoction (YDD) is based on the prescription in clinical use with additions and subtractions, consisting of Codonopsis pilosula, Ophiopogon japonicus (Maitong), Schisandra chinensis, Astragalus membranaceus, Angelica sinesis, Salvia miltiorrhiza, Radix et Rhizoma Polygonatum odoratum, Radix Ziziphus jujuba, Radix et Rhizoma Glycyrrhiza glabra (licorice), and other medicines, among which the Codonopsis pilosula, Maitong and Schisandra chinensis are intended to enrich the qi, nourish the yin, tranquilize the heart and calm the mind. Angelica sinensis, Salvia miltiorrhizae, Radix et Rhizoma Glycyrrhiza glabra act on qi by activating blood circulation, nourishes yin and calms palpitations. In the meta-analysis, it was found that the use of YDD could significantly improve the Chinese medicine symptoms such as palpitation and chest tightness in patients; however, there is little research on the active ingredients of YDD and its potential mechanism for the treatment of atrial fibrillation[7].
Chinese medicine compound treatment has the characteristics of multi-component, multi-pathway and multi-targeting[8]. Chinese medicine treatment is concerned with the overall concept and is difficult to carry out a comprehensive and systematic study from the level of molecules, cells, tissues and other levels of modern medicine. It is this feature of traditional Chinese medicine that emphasizes the need for us to look for a systematic perspective to study the pharmacological system of traditional Chinese medicine, and make full use of all kinds of data to carry out a systematic, based on the overall basis of the compound of traditional Chinese medicine, scientific analysis and exploration, so as to reveal the connotation and essence. Network pharmacology[9] is a research method to analyse and predict the pharmacological mechanism of drugs by using highthroughput screening, network visualization and analysis techniques to reveal the complex biological network relationship among drugs, targets and diseases. In this study, we will analyse the targets of YDD for the treatment of atrial fibrillation through network pharmacology. Primarily, we obtained the bioactive compounds and potential targets of YDD for the treatment of atrial fibrillation. Then, Protein- Protein Interaction (PPI) models of potential targets were constructed. Next, the potential targets and protein complexes were analysed by functional enrichment. Meanwhile, the interactions between bioactive compounds and key targets were visualized by molecular docking.
Materials and Methods
Acquisition of targets:
Specific information of the Chinese herbal medicines used in this study such as Codonopsis pilosula, Maitong, Nardostachys jatamansi, Schisandra chinensis, Astragalus membranaceus, Radix Angelica sinesis, Salvia miltiorrhiza, Radix Ziziphus jujuba, Pinellia ternata, Rhizoma Polygonatum odoratum and licorice were analyzed according to the Traditional Chinese Medicine Systems Pharmacology (TCMSP) [10] database and the Encyclopedia of Traditional Chinese Medicine (ETCM)[11]. The composition of each herb was analyzed and summarized through these two databases, supplemented by a review of pharmacological literature related to the drug, and then molecular structure diagrams of each important ingredient was obtained from PubChem[12].
Atrial fibrillation target acquisition:
Using "atrial fibrillation" as the keyword, we investigated therapeutic targets for atrial fibrillation from online datasets, such as DrugBank[13], Therapeutic Targets Database (TTD)[14], MalaCards[15], TCMSP[10], DisGeNet[16], and Online Mendelian Inheritance in Man (OMIM)[17]. All these databases are limited to "humans" or "Homo sapiens". In the results, the format of the therapeutic targets in DrugBank was converted to gene symbols using Universal Protein Resource (UniProt)[18] targets, with definitive evidence. Using MalaCards for further targets and analysis, the top 10 % of scores were used in DisGeNet. Once duplicates are removed, all targets are normalized using UniProt.
Component-target-disease-pathway network construction:
Based on the collected data, we constructed a Herb- Component-Target-Disease (H-C-T-D) network using Cytoscape (version 3.8.0)[19] to demonstrate the correlation between bioactive compounds and potential targets and pathways. Targets are used for further PPI exploration and functional enrichment analysis.
PPI analysis:
The relevant targets of atrial fibrillation and YDD were intersected, and the targets of YDD were defined as the potential targets of YDD for the intervention of atrial fibrillation. PPI plays an important role in life processes which are inseparable from PPI, such as Deoxyribonucleic Acid (DNA) synthesis, gene transcription activation, protein translation, cell cycle regulation, signal transduction, etc.,[20,21]. Search Tool for the Retrieval of Interacting Genes (STRING) which is a web-based online tool[22] provides online analysis of PPIs. In order to build PPI network, potential intervention targets were uploaded to STRING. The results were then imported into Cytoscape and the PPI network was evaluated using the CytoNCA[23] plug-in analysis, and the potential functional modules in the PPI network were explored using the Molecular Complex Detection (MCODE)[24]. In addition, to identify the core part of the potential targets more accurately, we imported the potential targets of YDD into the VarElect[25] dataset to analyze the correlation between the genes of the potential targets and atrial fibrillation.
Enrichment analysis of potential intervention targets and potential functional modules:
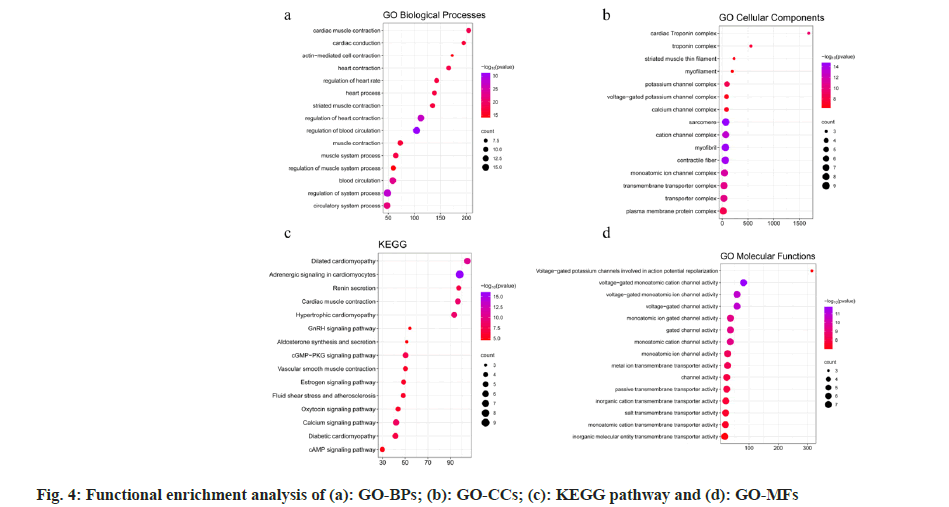
Gene Ontology (GO)[26] and Kyoto Encyclopedia of Genes and Genomes (KEGG)[27] were used to enrich and clarify the potential targets, potential functional modules and their gene functions. Metascape[28] is a web-based platform which is updated monthly, keeping the results of the analysis up to date; it provides gene annotation, feature-rich, and interactive genomic analysis services. The potential targets were uploaded to Metascape for GO-Biological Process (BPs), Cell Components (CC), Molecular Function (MF) and KEGG pathway enrichment analysis, where p<0.01 was considered as significantly enriched, and the top 15 GO enrichment analysis results along with their KEGG pathways were selected.
Molecular docking:
The Two-Dimensional (2D) structure files of the core compounds of YDD were imported into Open Babel[29] (version 3.1.1) to convert into MOL2 format, and the structures of related core targets were collected from Protein Data Bank (PDB) [30] and saved in its respective format while for proteins that could not be retrieved from the PDB database, we used AlphaFold[31] for protein structure resolution. The core target protein was removed from water molecules and its ligands using PyMOL[32]. Similarly, AutoDockTools[33] was used for hydrogenation of core target proteins and calculation of Gasteiger charges, saved as PDB, Partial Charge and Atom Type (PDBQT) files, energy minimization of potential core compound ligands, assignment of ligand atomic type, calculation of charge, storage in PDBQT format, and finally, semi-flexible molecular docking using AutoDock Vina[34] to assess the binding energy of ligands to target proteins, where docking score affinity <-4.25 kcal/mol-1 can be considered to have binding activity between the ligand and the target. A score <-5.0 indicates good binding activity, and a score <-7.0 kcal/mol-1 has strong docking activity between the two[35].
Results and Discussion
Target prediction was carried out here, where 623 kinds of ingredients present in YDD were collected from the TCMSP and ETCM retrieval systems and related literature, which included 95 kinds of Salvia, 65 kinds of Codonopsis, 44 kinds of Astragalus membranaceus, 35 kinds of Angelica sinensis, 35 kinds of Ophiopogon japonicus, 25 species of japonicus, 17 species of jatamansi, 49 species of Schisandra chinensis, 14 species of Pinellia ternata kernels, 159 species of boiled licorice, 31 species of sour jujuba kernels, and 54 species of Polygonatum odoratum. After the comparison and deduplication of each component, a total of 213 compounds were verified to have high bioavailability and a total of 1191 targets were obtained by screening the potential targets of these components using SwissTargetPrediction.
Atrial fibrillation target acquisition was also studied. We searched OMIM, GeneCards, and DisGeNET for 25, 230, and 180 atrial fibrillation-related targets, respectively, and merged and de-duplicated them to obtain 362 atrial fibrillation-related genes.
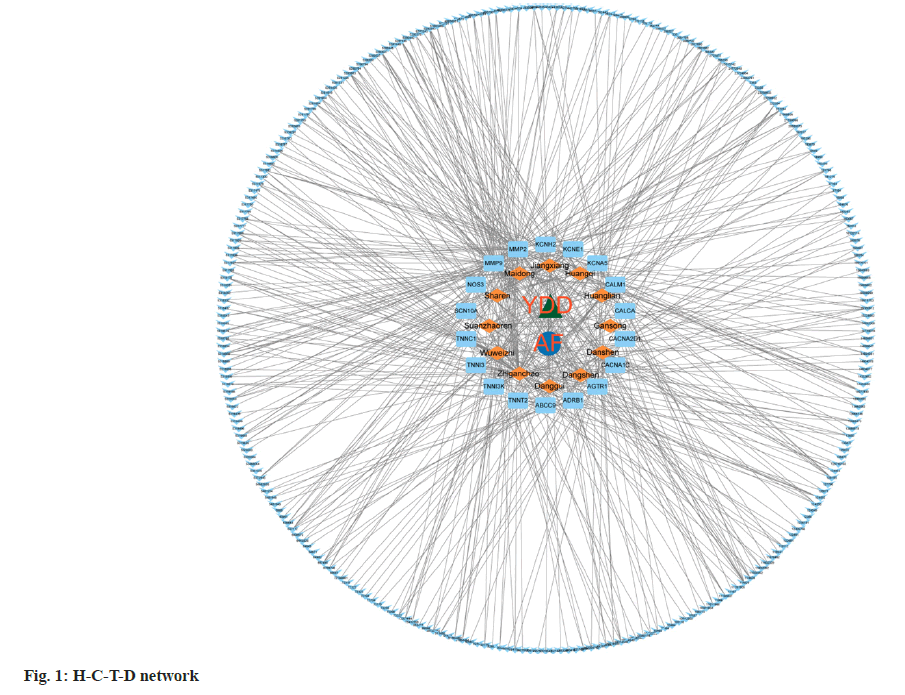
H-C-T-D network was constructed based on the data obtained above and constructed the H-C-T-D network using Cytoscape. The constructed network consisted of 297 nodes, including, 12 herbals, atrial fibrillation, and 18 common targets, with 541 pathways connected between the nodes (fig. 1). This network preliminarily revealed the complex mechanism of a variety of compounds, multiple targets and pathways of YDD decoction in the intervention of atrial fibrillation.
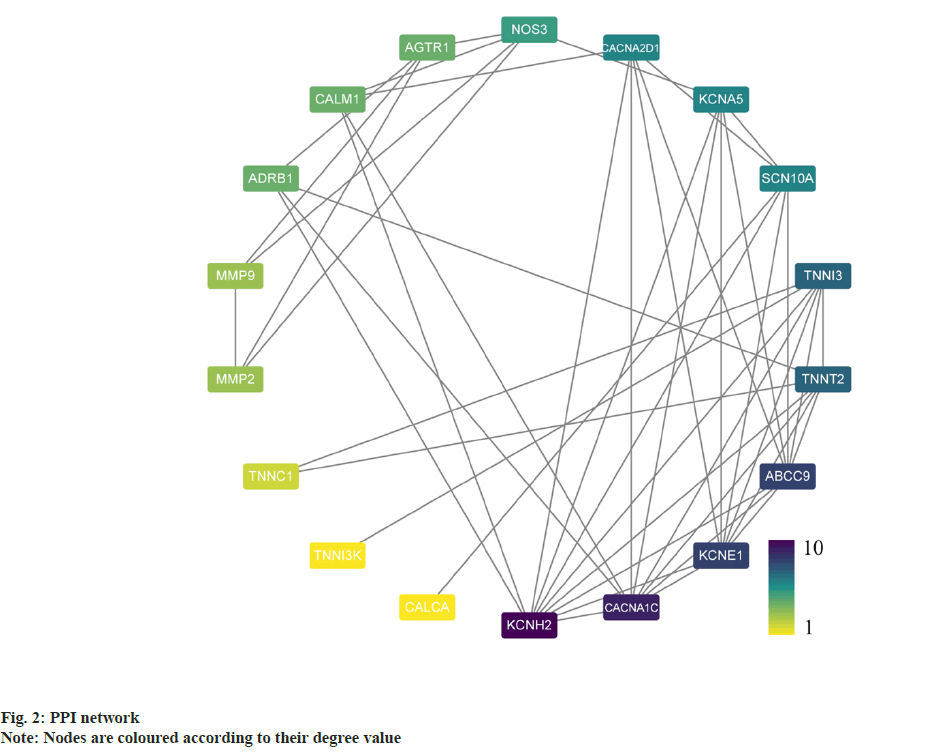
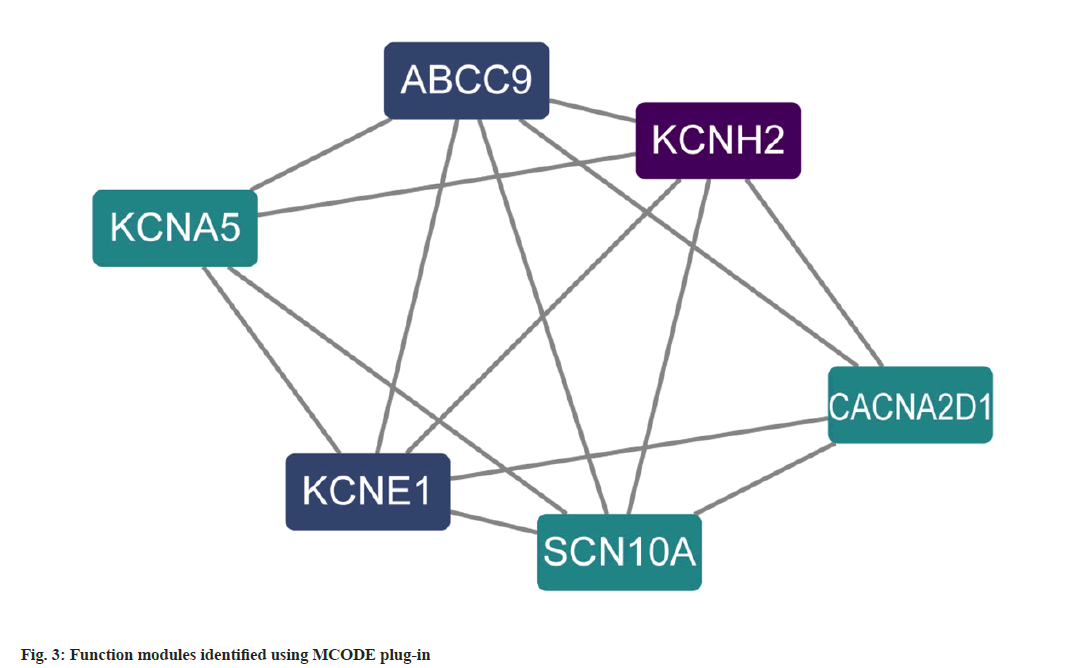
PPI analysis was carried out where a total of 18 potential targets for the intervention of atrial fibrillation were obtained. The relevant targets were uploaded to STRING for protein interaction analysis in order to construct a PPI network and the results were imported into Cytoscape, the degree values for each gene were calculated using the CytoNCA plugin, and a PPI network was constructed based on the degree values (fig. 2). Targets with degree values greater than median were defined as hub targets. Table 1 shows the names and topological parameters of the hub targets. Two potential protein complexes or functional modules were discovered simultaneously using the MCODE plug-in (fig. 3).
| Gene | Degree | Betweenness | Closeness |
|---|---|---|---|
| KCNH2 | 10 | 38.386814 | 0.653846 |
| Calcium voltage-gated Channel subunit α 1C (CACNA1C) | 9 | 24.253479 | 0.607143 |
| KCNE1 | 8 | 12.682296 | 0.586207 |
| ABCC9 | 8 | 12.682296 | 0.586207 |
| Troponin T2 (TNNT2) | 7 | 23.965813 | 0.548387 |
| Troponin I-Interacting 3 (TNNI3) | 7 | 41.923077 | 0.5 |
| CACNA2D1 | 6 | 4.133333 | 0.515152 |
| Sodium (S) CN10 α | 6 | 32.4 | 0.515152 |
| KCNA5 | 6 | 41.062515 | 0.566667 |
| Nitric Oxide Synthase 3 (NOS3) | 5 | 47 | 0.472222 |
| Calmodulin 1 (CALM1) | 4 | 15.631258 | 0.515152 |
| Angiotensin II Receptor Type 1 (AGTR1) | 4 | 19.772894 | 0.425 |
| Adrenoceptor β 1 (ADRB1) | 4 | 32.106228 | 0.53125 |
| Matrix Metallopeptidase 9 (MMP9) | 3 | 0 | 0.361702 |
| MMP2 | 3 | 0 | 0.361702 |
| Troponin C Type 1 (TNNC1) | 2 | 0 | 0.377778 |
| TNNI3 Kinase (K) | 1 | 0 | 0.34 |
| CALCA | 1 | 0 | 0.346939 |
Table 1: Name and Topological Parameters of the Hub Target
Further, analysis of the correlation between potential intervention targets and atrial fibrillation was evaluated. 18 potential intervention targets were uploaded to VarElect, and the disease was searched with the keyword "atrial fibrillation". The correlation analysis was carried out to obtain the results, as shown in Table 2. All the 18 potential intervention targets were directly related to atrial fibrillation, and the correlation scores of Potassium voltagegated Channel subfamily A member 5 (KCNA5), Adenosine Triphosphate-Binding Cassette subfamily C member 9 (ABCC9), KCNE1, and KCNH2 were found to be the highest were compared with the hub target. Each active ingredient of YDD was molecularly docked with the corresponding target to verify their relationship.
| Gene | Description | Score |
|---|---|---|
| KCNA5 | Potassium voltage-gated channel subfamily A member 5 | 124.02 |
| ABCC9 | ATP binding cassette subfamily C member 9 | 82.32 |
| KCNE1 | Potassium voltage-gated channel subfamily E regulatory subunit 1 | 63.03 |
| KCNH2 | Potassium voltage-gated channel subfamily H member 2 | 54.47 |
| CACNA1C | Calcium voltage-gated channel subunit α 1C | 53.3 |
| TNNI3K | Troponin I-interacting 3 kinase | 42.97 |
| TNNT2 | Troponin T2, cardiac type | 31.84 |
| TNNI3 | Troponin I3, cardiac type | 29.23 |
| SCN10A | Sodium voltage-gated channel α subunit 10 | 28.34 |
| CALM1 | Calmodulin 1 | 28.16 |
| CACNA2D1 | Calcium voltage-gated channel auxiliary subunit α 2 delta 1 | 25.39 |
| ADRB1 | Adrenoceptor β 1 | 16.41 |
| AGTR1 | Angiotensin II receptor type 1 | 16.28 |
| TNNC1 | Troponin C type 1 | 13.93 |
| NOS3 | Nitric oxide synthase 3 | 11.14 |
| MMP2 | Matrix metallopeptidase 2 | 10.43 |
| MMP9 | Matrix metallopeptidase 9 | 8.99 |
| CALCA | Calcitonin related polypeptide α | 8.6 |
Table 2: The Correlation Analysis of Genes
Enrichment analysis of potential intervention targets and functional modules was also studied. 18 potential intervention targets were uploaded to Metascape for GO-BPs, CC, MF, and KEGG pathway enrichment analysis, and where p<0.01 were exported to create a bubble map for enrichment analysis (fig. 4). The common targets mainly involved include regulation of blood circulation, myocardial contraction, heart rate regulation, and cardiac conduction in GO-BP (fig. 4a). GO-CCs are involved in voltage-gated potassium channel complexes, myofibrils, contractile fibers, cardiac troponin complexes, and cation channel complexes (fig. 4b). The KEGG pathway mainly focused on adrenergic signaling in cardiomyocytes, dilated cardiomyopathy, myocardial contraction, hypertrophic cardiomyopathy, calcium signaling pathway, cyclic Guanosine Monophosphate-Protein Kinase G (cGMP-PKG) signaling pathway, fluid shear stress and atherosclerosis, oxytocin signaling pathway, cyclic Adenosine 3′,5′-Monophosphate (cAMP) signaling pathway, Gonadotropin-Releasing Hormone (GnRH) signaling pathway, etc., (fig. 4c). The functions of GO molecules mainly involve voltage-gated single-atom cation channel activity, gated channel activity, voltage-gated potassium channel activity, cardiomyocyte action potential repolarization, passive transmembrane transporter activity, and inorganic cationic transmembrane transporter activity (fig. 4d).
In addition, GO and KEGG enrichment analysis was performed on the functional modules found in PPI, and the results were visualized using the ggplot package in R language (fig. 5), which showed that the modules could participate in the regulation of cardiac conduction, cardiac contraction, and blood circulation regulation of the transport of single atom ions across membranes, etc.
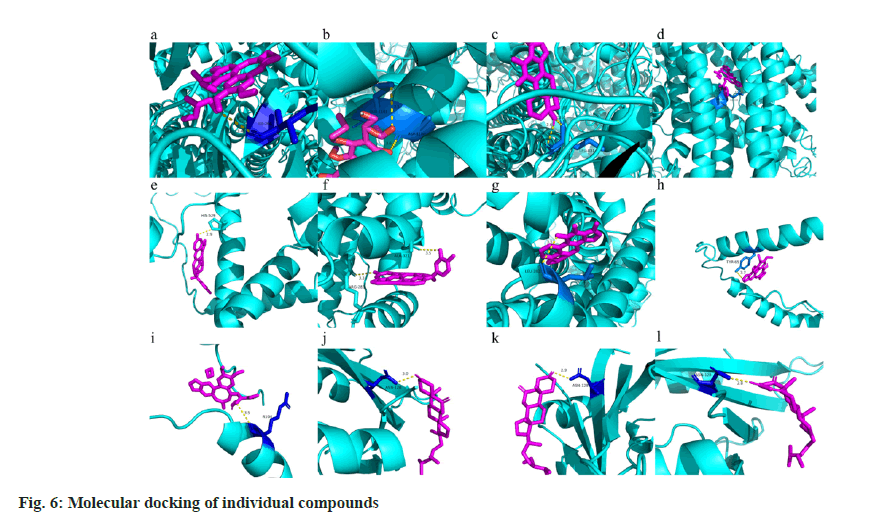
Molecular docking and validation was explained here. ABCC9, KCNA5, KCNH2 and KCNE1 were downloaded from the PDB and AlphaFold database, pretreated with ligands, and then was subjected to molecules using AutoDock Vina to export the results. The results were visualized using PyMOL (fig. 6). 4 genes which were closely related to atrial fibrillation were included in the molecular docking. The docking information of the active ingredients and targets is shown in Table 3. The results showed that the active ingredients of YDD could interact with the targets. A total of 12 composite structures with the lowest binding energy for docking were selected. Angeloyl gomisin P was linked to Isoleucine (Ile266) (fig. 6a); gomisin E was linked to Glutamine (Gln 1101) and Aspartic acid (ASP-1100) (fig. 6b); przewaquinone C with GLN-1101 and ASP-1100 (fig. 6c); schisantherin D with GLN-1101 and ASP- 1100 (fig. 6d) of ABCC9 via hydrogen bond linkage while schisanlactone A with Histidine (His-529) (fig. 6e); schisanlactone D with Alanine (Ala-321) and Arginine (Arg283) (fig. 6f); tanshinone IIA with Leucine (Leu-282) of KCNA5 (fig. 6g) of KCNA5 were linked via hydrogen bonding. Epigomisin O with Tyrosine (Tyr-65) (fig. 6h) and schisandrin B with Arg-104 (fig. 6i) of KCNE1 were linked via hydrogen bonding. Similarly in KCNH2, the 5 Alpha (α)-Stigmastan-3-6-dione core can form a hydrogen bond with residue Asparagine (Asn-128) (fig. 6j), the core of cadaverine can form a hydrogen bond with residue Asn-128 (fig. 6k) and the core of epigomisin O can form a hydrogen bond with residue Asn-128 (fig. 6l).
| Gene | Active ingredients | Affinity (kcal/mol) |
|---|---|---|
| ABCC9 | Angeloyl Gomisin P | -9.2 |
| ABCC9 | Gomisin E | -11.7 |
| ABCC9 | Przewaquinone C | -11.5 |
| ABCC9 | Schisantherin D | -10.9 |
| KCNA5 | Schisanlactone A | -12.3 |
| KCNA5 | Schisanlactone D | -12.8 |
| KCNA5 | Tanshinone iia | -10.1 |
| KCNE1 | Epigomisin O | -7.7 |
| KCNE1 | Schizandrer B | -7.1 |
| KCNH2 | 5 α Stigmastan 3,6 dione | -6.5 |
| KCNH2 | Cadaverine | -6.3 |
| KCNH2 | Epigomisin O | -6.2 |
Table 3: Target and Active Substance Docking Energy
Atrial fibrillation is the most common arrhythmia in clinical practice, and the incidence of atrial fibrillation is gradually increasing with the gradual aging of the world, which brings a great health burden to countries. YDD is a traditional Chinese medicine compound preparation, and long-term clinical studies have found that it has the efficacy in treating arrhythmia. This study aimed to use network pharmacology combined with molecular docking technology to predict the potential targets of YDD in the treatment of atrial fibrillation.
Through the collection and preliminary screening of the data, we identified 18 potential intervention targets, constructed a PPI network and H-C-T-D Venn diagram to evaluate their association. We intuitively expressed the relevant targets and pathways of each component of YDD acting on atrial fibrillation and found the potential functional group through the MCODE plug-in. The enrichment analysis of potential targets showed that YDD mainly regulates myocardial contraction, rhythm control, voltagegated potassium channel, etc., and found that the signal pathways of YDD decoction to intervene in atrial fibrillation mainly were involved in adrenergic signaling, dilated cardiomyopathy, myocardial contraction, hypertrophic cardiomyopathy, calcium signaling pathway, etc., through the analysis of potential functional modules.
It is found that the regulation of atrial fibrillation by the complex is through heart conduction, regulation of cardiac contraction, blood circulation and regulation of transmembrane transport of single atomic ions, etc. Adrenergic signaling has a huge impact on the electrical activity of cardiomyocytes[36], and when adrenergic signaling is activated, it activates adenylyl cyclase (through conjugated proteins cyclase), increases intracellular cAMP concentrations and thus activates cAMP-dependent Protein Kinase A (PKA), phosphorylates L-type calcium channels, leading to channel opening, increased intracellular calcium concentrations, and over activated adrenergic signaling resulting in intracellular calcium overload, thereby triggering increased sexual activities (such as early and delayed post-depolarization), and development of atrial fibrillation[37]. In addition, over activation of adrenergic receptors can also affect the depolarization and repolarization of heart cells, increasing the enrichment dispersion between the atrial muscles, resulting in the formation of fold back between the atria and myocardium, resulting in the occurrence of atrial fibrillation[38]. The same voltagegated potassium channel is also closely related to atrial fibrillation, and the voltage-gated potassium channel is a key ion channel for cardiomyocyte depolarization and repolarization, when the voltagegated potassium channel is over activated, Potassium (K+) ion outflow is significantly increased, and the myocardial repolarization time course is significantly shortened, which induces atrial fibrillation and promotes electrical remodeling[39].
The 4 most closely related targets i.e., KCNA5, ABCC9, KCNE1, and KCNH2 and disease association analysis were obtained through PPI. KCNA5 gene encodes the Kv1.5 channel protein[40], a voltage-gated potassium channel. In the heart, Kv1.5 channels are mainly distributed in atrial myocytes, especially on the cell membranes of atrial myocytes[41]. Mutations or aberrant expression of the KCNA5 gene may alter the function of the Kv1.5 channel, thereby affecting the repolarization of atrial myocytes and increase the formation of reentrant circuits. Some studies have found mutations in KCNA5 gene among patients with familial atrial fibrillation, which further confirms the association between the KCNA5 gene and atrial fibrillation[42-44]. ABCC9 encodes Sulfonylurea Receptor 2 (SUR2)[45,46] which is a component of the ATP-sensitive potassium channel (KATP) channel and together with the Inward-Rectifying Potassium (Ikr) channel subunit forming a functional KATP channel[47].
Studies have found that certain variants in the ABCC9 gene are associated with an increased risk of atrial fibrillation[48]. KCNE1 is a gene encoding Beta (β) subunits of potassium channels that help maintain potential balance in cardiac muscle cells to maintain normal cardiac rhythm and function[49]. KCNE1 regulates the Delayed Rectifier Potassium (IKs) channels in the heart, and numerous studies have found that mutations in the KCNE1 gene can cause changes in cardiac repolarization leading to atrial fibrillation[50]. KCNH2 is a classic gene encoding a human Ether-a-go-go-Related Gene (hERG)/Ikr, which is part of an important potential-dependent potassium channel in cardiac myocytes[51-53]. A large number of studies have confirmed that KCNH2 gene mutations are associated with atrial fibrillation susceptibility[54,55], and KCNH2 gene mutations lead to a decrease in the encoding of IKr, and the action potential of IKr current is significantly reduced, which prolongs the repolarization time and induces atrial fibrillation. A recent study found that KCNH2-G628S gene therapy can successfully and safely reduce the risk of atrial fibrillation[56].
In this paper, network pharmacology, enrichment analysis, and molecular docking verification were performed. It is speculated that YDD can intervene in atrial fibrillation through a variety of mechanisms. This comprehensive regulatory effect of multiple targets is vividly reflected in the PPI, association, and enrichment analysis in this paper, which has incomparable advantages over existing drugs. This study promoted the explanation of the molecular mechanism of YDD in the intervention of atrial fibrillation, which can provide a comprehensive idea for further research. However, the data in the included databases are derived from different experimental studies, which still have some limitations. Analysis of the potential targets and mechanisms of YDD decoction in the intervention of atrial fibrillation by data mining and bioinformatics methods should be further explored through biological experiments and in the follow-up studies will be carried out based on this study to further elucidate the targets and mechanisms of YDD in the intervention of atrial fibrillation.
Conflict of interests:
The authors declared no conflict of interests.
References
- Hendriks JM, Gallagher C, Middeldorp ME, Lau DH, Sanders P. Risk factor management and atrial fibrillation. Europace 2021;23:52-60.
[Crossref] [Google Scholar] [PubMed]
- Hsieh JC, Shih H, Xin LL, Yang CC, Han CL. 12-lead ECG signal processing and atrial fibrillation prediction in clinical practice. Technol Health Care 2023;31(2):417-33.
[Crossref] [Google Scholar] [PubMed]
- Li H, Song X, Liang Y, Bai X, Liu-Huo WS, Tang C, et al. Global, regional, and national burden of disease study of atrial fibrillation/flutter, 1990-2019: Results from a global burden of disease study, 2019. BMC Public Health 2022;22(1):1-13.
[Crossref] [Google Scholar] [PubMed]
- Migdady I, Russman A, Buletko AB. Atrial fibrillation and ischemic stroke: A clinical review. Semin Neurol 2021;41(4):348-64.
[Crossref] [Google Scholar] [PubMed]
- Gutierrez C, Blanchard DG. Diagnosis and treatment of atrial fibrillation. Am Fam Physician 2016;94(6):442-52.
[Google Scholar] [PubMed]
- Shi R, Norman M, Chen Z, Wong T. Individualized ablation strategy guided by live simultaneous global mapping to treat persistent atrial fibrillation. Future Cardiol 2018;14(3):237-49.
[Crossref] [Google Scholar] [PubMed]
- Wang XY, Hu HY, Ji ZC, Zhai JB, Liu CX, Zhang JH. Systematic review and Meta-analysis of efficacy and safety of Yangxin Dingji capsules in treatment of arrhythmia. Zhongguo Zhong Yao Za Zhi 2021;46(20):5418-27.
[Crossref] [Google Scholar] [PubMed]
- Liang Y, Liang B, Wu XR, Chen W, Zhao LZ. Network pharmacology-based systematic analysis of molecular mechanisms of Dingji Fumai decoction for ventricular arrhythmia. Evid Based Complement Alternat Med 2021:1-12.
[Crossref] [Google Scholar] [PubMed]
- Zhao L, Zhang H, Li N, Chen J, Xu H, Wang Y, et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J Ethnopharmacol 2023;309:116306.
[Crossref] [Google Scholar] [PubMed]
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:1-6.
[Crossref] [Google Scholar] [PubMed]
- Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Res 2019;47:D976-82.
[Crossref] [Google Scholar] [PubMed]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res 2021;49(D1):D1388-95.
[Crossref] [Google Scholar] [PubMed]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: A major update to the drug bank database for 2018. Nucleic Acids Res 2018;46(D1):D1074-82.
[Crossref] [Google Scholar] [PubMed]
- Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F, et al. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res 2022;50:D1398-407.
[Crossref] [Google Scholar] [PubMed]
- Rappaport N, Nativ N, Stelzer G, Twik M, Guan-Golan Y, Iny Stein T, et al. MalaCards: An integrated compendium for diseases and their annotation. Database 2013:1-14.
[Crossref] [Google Scholar] [PubMed]
- Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45(D1):D833-9.
[Crossref] [Google Scholar] [PubMed]
- Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): A knowledgebase of human genes and genetic phenotypes. Curr Protoc Bioinformatics 2017;58:1-12.
[Crossref] [Google Scholar] [PubMed]
- The UniProt Consortium. UniProt: The universal protein knowledgebase in 2023. Nucleic Acids Res 2023;51:D523-31.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google Scholar] [PubMed]
- Lin JS, Lai EM. Protein-protein interactions: Co-immunoprecipitation. Methods Mol Biol 2017;1615:211-19.
[Crossref] [Google Scholar] [PubMed]
- Tang H, Bi H, Liu B, Lou S, Song Y, Tong S, et al. WRKY33 interacts with WRKY12 protein to up-regulate RAP2.2 during submergence induced hypoxia response in Arabidopsis thaliana. New Phytol 2021;229(1):106-25.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 2021;49:D605-12.
[Crossref] [Google Scholar] [PubMed]
- Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72.
[Crossref] [Google Scholar] [PubMed]
- Xie R, Li B, Jia L, Li Y. Identification of core genes and pathways in melanoma metastasis via bioinformatics analysis. Int J Mol Sci 2022;23(2):1-14.
[Crossref] [Google Scholar] [PubMed]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016;54:1-3.
[Crossref] [Google Scholar] [PubMed]
- Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucleic Acids Res 2015;43:D1049-56.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000;28(1):27-30.
[Crossref] [Google Scholar] [PubMed]
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10(1):1-10.
[Crossref] [Google Scholar] [PubMed]
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open babel: An open chemical toolbox. J Cheminform 2011;3:1-14.
[Crossref] [Google Scholar] [PubMed]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res 2000;28(1):235-42.
[Crossref] [Google Scholar] [PubMed]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold protein structure database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 2022;50:D439-44.
[Crossref] [Google Scholar] [PubMed]
- Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and AutoDock/Vina. J Comput Aided Mol Des 2010;24(5):417-22.
[Crossref] [Google Scholar] [PubMed]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, et al. AutoDock4 and AutoDock tools4: Automated docking with selective receptor flexibility. J Comput Chem 2009;30(16):2785-91.
[Crossref] [Google Scholar] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31(2):455-61.
[Crossref] [Google Scholar] [PubMed]
- Saikia S, Bordoloi M. Molecular docking: Challenges, advances and its use in drug discovery perspective. Curr Drug Targets 2019;20(5):501-21.
[Crossref] [Google Scholar] [PubMed]
- Grandi E, Ripplinger CM. Antiarrhythmic mechanisms of beta blocker therapy. Pharmacol Res 2019;146:1-21.
[Crossref] [Google Scholar] [PubMed]
- Reinhardt F, Beneke K, Pavlidou NG, Conradi L, Reichenspurner H, Hove-Madsen L, et al. Abnormal calcium handling in atrial fibrillation is linked to changes in cyclic AMP dependent signaling. Cells 2021;10(11):1-17.
[Crossref] [Google Scholar] [PubMed]
- Grant AO. Mechanisms of atrial fibrillation and action of drugs used in its management. Am J Cardiol 1998;82:43N-9.
[Crossref] [Google Scholar] [PubMed]
- Geng L, Wang S, Zhang F, Xiong K, Huang J, Zhao T, et al. SNX17 (Sorting Nexin 17) mediates atrial fibrillation onset through endocytic trafficking of the Kv1.5 (Potassium voltage-gated channel subfamily a member 5) channel. Circ Arrhythm Electrophysiol 2019;12(4):1-12.
[Crossref] [Google Scholar] [PubMed]
- Ahmed M, Fezai M, Uzcategui NL, Hosseinzadeh Z, Lang F. SGK3 sensitivity of voltage gated K+ channel Kv1.5 (KCNA5). Cell Physiol Biochem 2016;38(1):359-67.
[Crossref] [Google Scholar] [PubMed]
- Yang Z, Browning CF, Hallaq H, Yermalitskaya L, Esker J, Hall MR, et al. Four and a half LIM protein 1: A partner for KCNA5 in human atrium. Cardiovasc Res 2008;78(3):449-57.
[Crossref] [Google Scholar] [PubMed]
- Olson TM, Alekseev AE, Liu XK, Park S, Zingman LV, Bienengraeber M, et al. Kv1.5 channelopathy due to KCNA5 loss-of-function mutation causes human atrial fibrillation. Hum Mol Genet 2006;15(14):2185-91.
[Crossref] [Google Scholar] [PubMed]
- Tian L, Liu G, Wang L, Zheng M, Li Y. KCNA5 gene polymorphism associate with idiopathic atrial fibrillation. Int J Clin Exp Med 2015;8(6):9890-6.
[Google Scholar] [PubMed]
- Wang K, Zhao J, Guo Z. Interaction of KCNA5, CX43, and CX40 proteins in the atrial muscle of patients with atrial fibrillation. Cell Biol Int 2022;46(11):1834-40.
[Crossref] [Google Scholar] [PubMed]
- McClenaghan C, Hanson A, Sala-Rabanal M, Roessler HI, Josifova D, Grange DK, et al. Cantu syndrome-associated SUR2 (ABCC9) mutations in distinct structural domains result in KATP channel gain-of-function by differential mechanisms. J Biol Chem 2018;293(6):2041-52.
[Crossref] [Google Scholar] [PubMed]
- Park S, Lim BB, Perez-Terzic C, Mer G, Terzic A. Interaction of asymmetric ABCC9-encoded nucleotide binding domains determines KATP channel SUR2A catalytic activity. J Proteome Res 2008;7(4):1721-8.
[Crossref] [Google Scholar] [PubMed]
- Shindo T, Yamada M, Isomoto S, Horio Y, Kurachi Y. SUR2 subtype (A and B)-dependent differential activation of the cloned ATP-sensitive K+ channels by pinacidil and nicorandil. Br J Pharmacol 1998;124(5):985-91.
[Crossref] [Google Scholar] [PubMed]
- Olson TM, Alekseev AE, Moreau C, Liu XK, Zingman LV, Miki T, et al. KATP channel mutation confers risk for vein of Marshall adrenergic atrial fibrillation. Nat Clin Pract Cardiovasc Med 2007;4(2):110-6.
[Crossref] [Google Scholar] [PubMed]
- Van Horn WD, Vanoye CG, Sanders CR. Working model for the structural basis for KCNE1 modulation of the KCNQ1 potassium channel. Curr Opin Struct Biol 2011;21(2):283-91.
[Crossref] [Google Scholar] [PubMed]
- Ehmke H. Physiological functions of the regulatory potassium channel subunit KCNE1. Am J Physiol Regul Integr Comp Physiol 2002;282(3):R637-8.
[Crossref] [Google Scholar] [PubMed]
- Chan PJ, Osteen JD, Xiong D, Bohnen MS, Doshi D, Sampson KJ, et al. Characterization of KCNQ1 atrial fibrillation mutations reveals distinct dependence on KCNE1. J Gen Physiol 2012;139(2):135-44.
[Crossref] [Google Scholar] [PubMed]
- Jiang YF, Chen M, Zhang NN, Yang HJ, Xu LB, Rui Q, et al. Association between KCNE1 G38S gene polymorphism and risk of atrial fibrillation: A PRISMA-compliant meta-analysis. Medicine 2017;96(25):1-8.
[Crossref] [Google Scholar] [PubMed]
- Temple J, Frias P, Rottman J, Yang T, Wu Y, Verheijck EE, et al. Atrial fibrillation in KCNE1-null mice. Circ Res 2005;97(1):62-9.
[Crossref] [Google Scholar] [PubMed]
- Wang QS, Wang XF, Chen XD, Yu JF, Wang J, Sun J, et al. Genetic polymorphism of KCNH2 confers predisposition of acquired atrial fibrillation in Chinese. J Cardiovasc Electrophysiol 2009;20(10):1158-62.
[Crossref] [Google Scholar] [PubMed]
- Sinner MF, Pfeufer A, Akyol M, Beckmann BM, Hinterseer M, Wacker A, et al. The non-synonymous coding IKr-channel variant KCNH2-K897T is associated with atrial fibrillation: Results from a systematic candidate gene-based analysis of KCNH2 (HERG). Eur Heart J 2008;29(7):907-14.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Hutt JA, Rajeshkumar B, Azuma Y, Duan KL, Donahue JK. Preclinical efficacy and safety of KCNH2-G628S gene therapy for postoperative atrial fibrillation. J Thorac Cardiovasc Surg 2017;154(5):1644-58.
[Crossref] [Google Scholar] [PubMed]
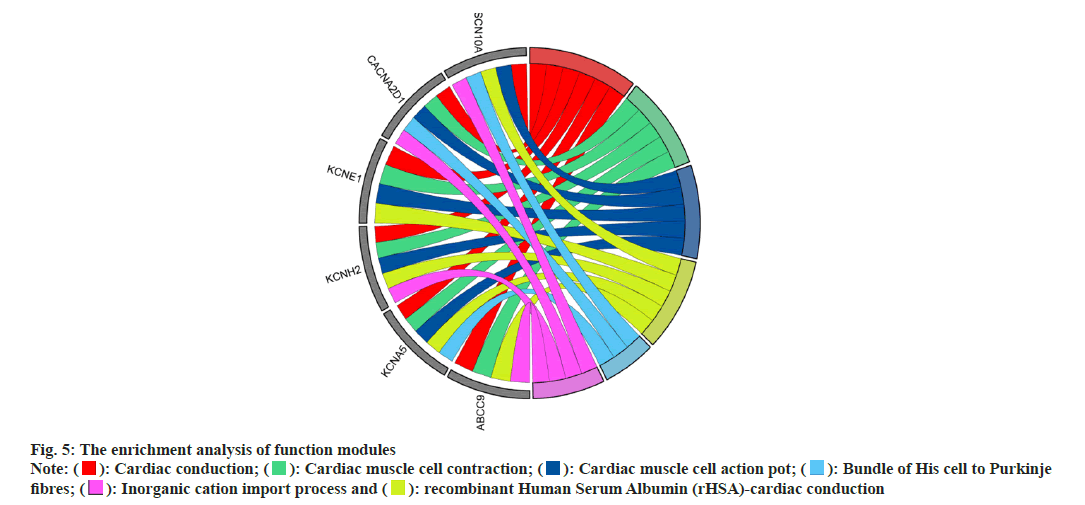
 ): Cardiac conduction; (
): Cardiac conduction; ( ): Cardiac muscle cell contraction; (
): Cardiac muscle cell contraction; ( ): Cardiac muscle cell action pot; (
): Cardiac muscle cell action pot; ( ): Bundle of His cell to Purkinje
fibres; (
): Bundle of His cell to Purkinje
fibres; ( ): Inorganic cation import process and (
): Inorganic cation import process and ( ): recombinant Human Serum Albumin (rHSA)-cardiac conduction
): recombinant Human Serum Albumin (rHSA)-cardiac conduction