- *Corresponding Author:
- X. K. Hu
Department of Radiotherapy and Oncology, The Second Affiliated Hospital of Harbin Medical University, Harbin, Heilongjiang 150086, China
E-mail: h06425@hrbmu.edu.cn
| This article was originally published in a special issue, “Drug Development in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(5) Spl Issue “55-66” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Colorectal cancer is one of the most commonly occurred cancers, although deubiquitination enzymes are related to tumor, the work of ubiquitin specific peptidase 5 in colorectal cancer is still not fully elucidated. Ubiquitin specific peptidase 5 expressions and prognosis was evaluated by analyzing a public dataset from the cancer genome atlas, gene expression omnibus database and a literature review. The biological function of ubiquitin specific peptidase 5 was investigated by gene set enrichment analysis. Ubiquitin specific peptidase 5 interaction proteins were shown by biological general repository for interaction datasets. Ubiquitin specific peptidase 5 is highly expressed and might be potentially used as a diagnostic and prognostic biomarker in colorectal cancer. Functional network analysis suggested that ubiquitin specific peptidase 5 is associated with proteasome, ribosome, deoxyribonucleic acid replication, cell cycle, spliceosome, etc. We discovered the correlation of ubiquitin specific peptidase 5 with several cancer related kinases, E2F transcription factors family and micro ribonucleic acids. Additionally, we presented about the ubiquitin specific peptidase 5 interaction proteins through protein-protein interaction network. Our results revealed information about ubiquitin specific peptidase 5 expression and its potential regulatory networks in colorectal cancer.
Keywords
Ubiquitin specific peptidase 5, colorectal cancer, gene function, gene set enrichment analysis
Colorectal Cancer (CRC) has been recognized as a cancer with high incidence and mortality[1,2]. In the past decades, several new therapeutic drugs were introduced to CRC, but prolonged survival is limited in advanced stage patients. Recently, increasing research evidence showed that multiple molecular pathways involved in the occurrence and development of CRC. Among which, Ubiquitin-Proteasome System (UPS) plays a significant role[3,4].
UPS is an important way for the degradation of proteins in eukaryotic cells. Ubiquitination enzymes and Deubiquitination (DUB) enzymes regulates targeted protein expression at post-transcriptional level. Ubiquitin Specific Peptidase (USP) family is an important member of DUBs. Numerous studies have shown that aberrantly expressed USP family are associated with tumor cell proliferation, migration and invasion[3,5]. USP5 is also known as a deubiquitinating enzyme belonging to the USP family, which has been confirmed to be associated with multiple physiological processes and pathological conditions[6]. USP5 plays pivotal roles by targeting substrates, such as p53[7], Forkhead box M1 (FOXM1)[8], c-Maf[9] and beta (β)-catenin[10]. Upregulation of USP5 expression has been found in a variety of cancers and related to poor prognosis[10-14]. Previous research studies mentioned that high USP5 expression related to tumor cell growth[15] and brigatinib[16] resistance in CRC. However, there are few articles and insufficient evidence about the role of USP5 in CRC.
In our study, we sought to unveil the clinical role of USP5 in CRC by bioinformatics analysis based on The Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and literature reviews. Then we used Gene Set Enrichment Analysis (GSEA) to explore the potential in depth mechanism of USP5, including Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), kinase targets, transcription factor targets and micro Ribonucleic Acid (miRNA) targets.
Materials and Methods
USP5 expression in CRC based on GEO datasets:
The microarray profiles of CRC were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/) from January 2010 to June 2020. The search strategy formulated was as follows: (colon OR rectal OR colorectal) AND (cancer OR tumor OR neoplasm OR malignancy OR carcinoma). Inclusion criteria were listed as Homo sapiens of CRC and normal tissues; the total number of cancer and healthy samples were >50; the expression data of USP5 could be provided or calculated in both CRC and normal tissues. Series which does not meet eligibility criteria were excluded.
USP5 expression levels were calculated in both CRC and normal tissues. Unpaired student’s t test in GraphPad prism v7.04 (GraphPad Software, California, USA) was performed to analyze the differences in USP5 expression. Receiver Operator Characteristic (ROC) curve were used to identify diagnostic value. Means and Standard Deviations (SD) were extracted to estimate USP5 expression level in cancer and control groups by using Review Manager 5.3. Heterogeneity across studies was analyzed by Chi-square (χ2) test and I2 statistics. I2>50 % or p<0.05 was considered heterogeneous and random-effects model was used.
USP5 messenger RNA (mRNA) expression in CRC based on TCGA datasets:
TCGA microarray data based on RNA-sequencing (RNA-seq) were obtained by The University of Alabama Cancer (UALCAN) data analysis portal (http://ualcan.path.uab.edu)[17]. We used this interactive web-portal to analyze the differential expression of USP5 in CRC and normal tissues.
Immunohistochemistry (IHC) of USP5 protein expression:
To show USP5 protein expression, IHC datasets were retrieved from The Human Protein Atlas database (https://www.proteinatlas.org/)[18].
Survival analysis:
TCGA survival data was downloaded by using OncoLnc (http://www.oncolnc.org/)[19]. GSE87211 microarray including survival data was obtained from GEO database. The best cut-off was analyzed by X-tile[20], which is a bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Kaplan-Meier survival curves were drafted by GraphPad.
Literature search and study for USP5 in CRC:
Related studies were selected by comprehensively searching on PubMed and Web of Science from January 2010 to June 2020. The search strategy was made as follows, (colon OR rectal OR colorectal) AND (cancer OR tumor OR neoplasm OR malignancy OR carcinoma) AND (USP5 OR ubiquitin specific peptidase 5). Publications fulfilled one of the following inclusion criteria were considered eligible, which includes the difference of USP5 expression between CRC and normal tissues were shown; USP5 expression and clinical data of colorectal patients were shown. Studies were excluded if they met any one of the following conditions which includes reviews, case report, conference abstracts, letters and comments; results based on TCGA data since we had evaluated USP5 expression in TCGA data by ourselves.
Bioinformatics analyses of USP5:
LinkedOmics (http://www.linkedomics.org/) is a publicly available portal including multi-omics data from TCGA, displaying correlated genes of USP5[21]. The website also used GSEA to perform analyses of GO, KEGG pathways, candidate target genes of protein kinases, transcriptional factors and miRNAs.
The Protein-Protein Interaction (PPI) network was constructed by Biological General Repository for Interaction Datasets (BioGRID) (https://thebiogrid.org/)[22]. BioGRID is a classic database in studying protein interactions by searching 72 408 publications for 1 871 024 protein and genetic interactions, 28 093 chemical associations and 874 796 post translational modifications from major model organism species.
Results and Discussion
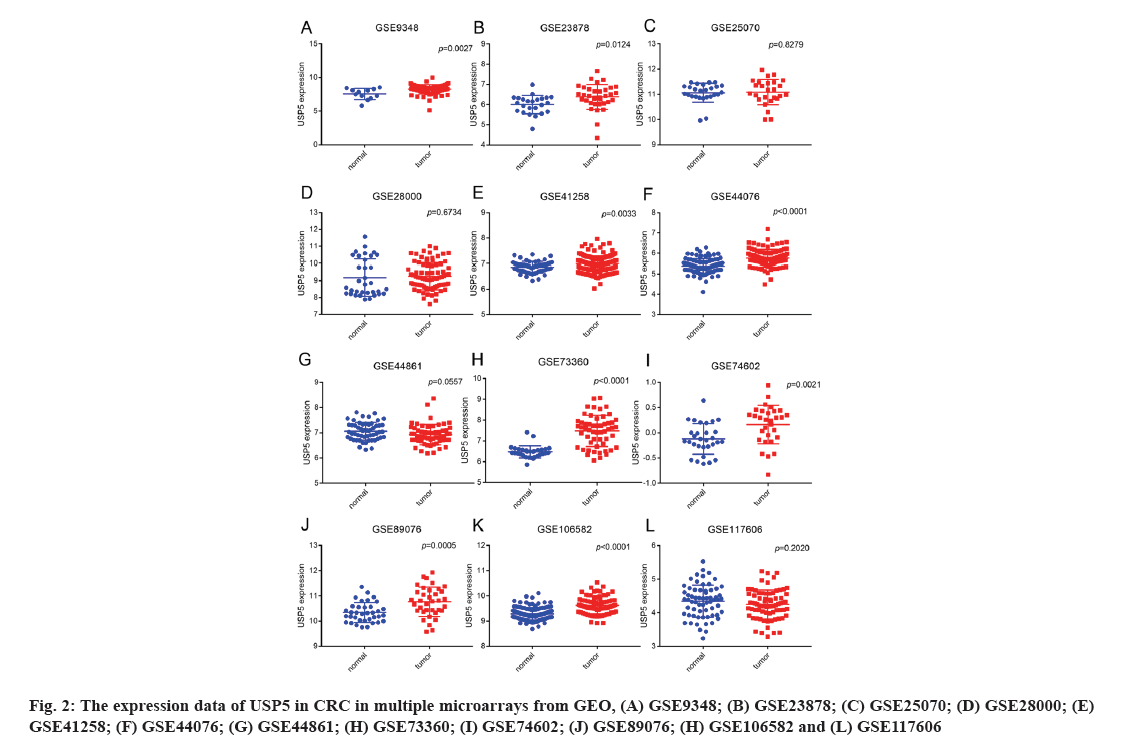
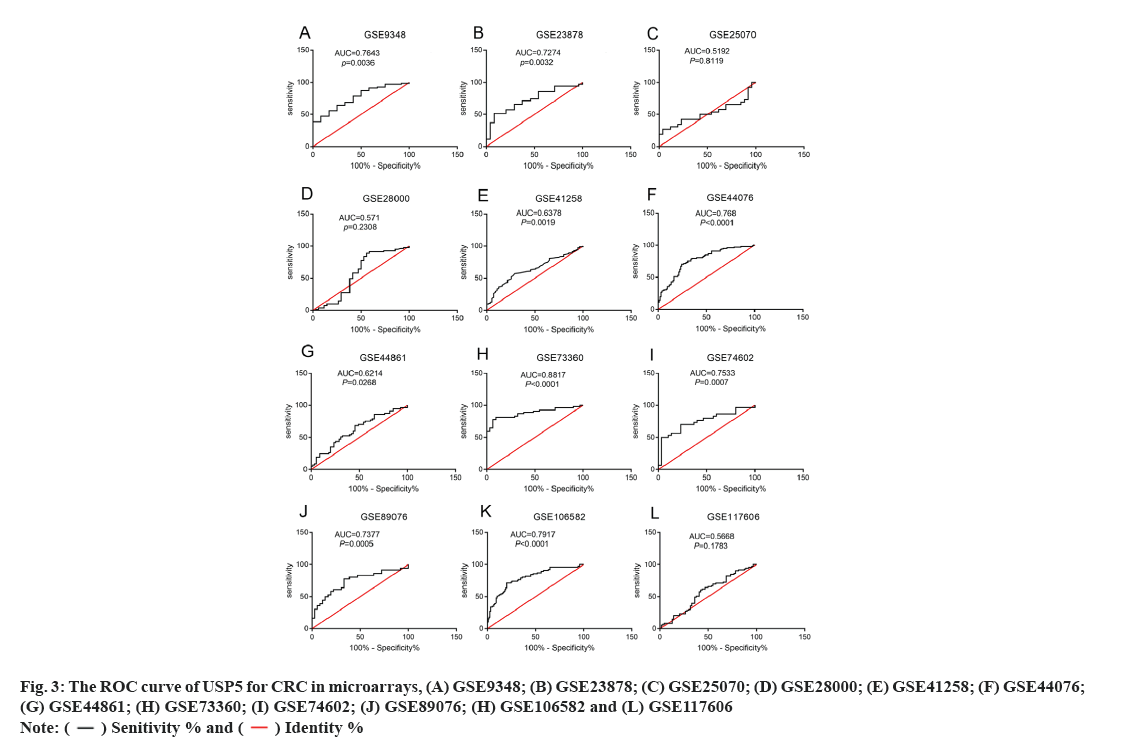
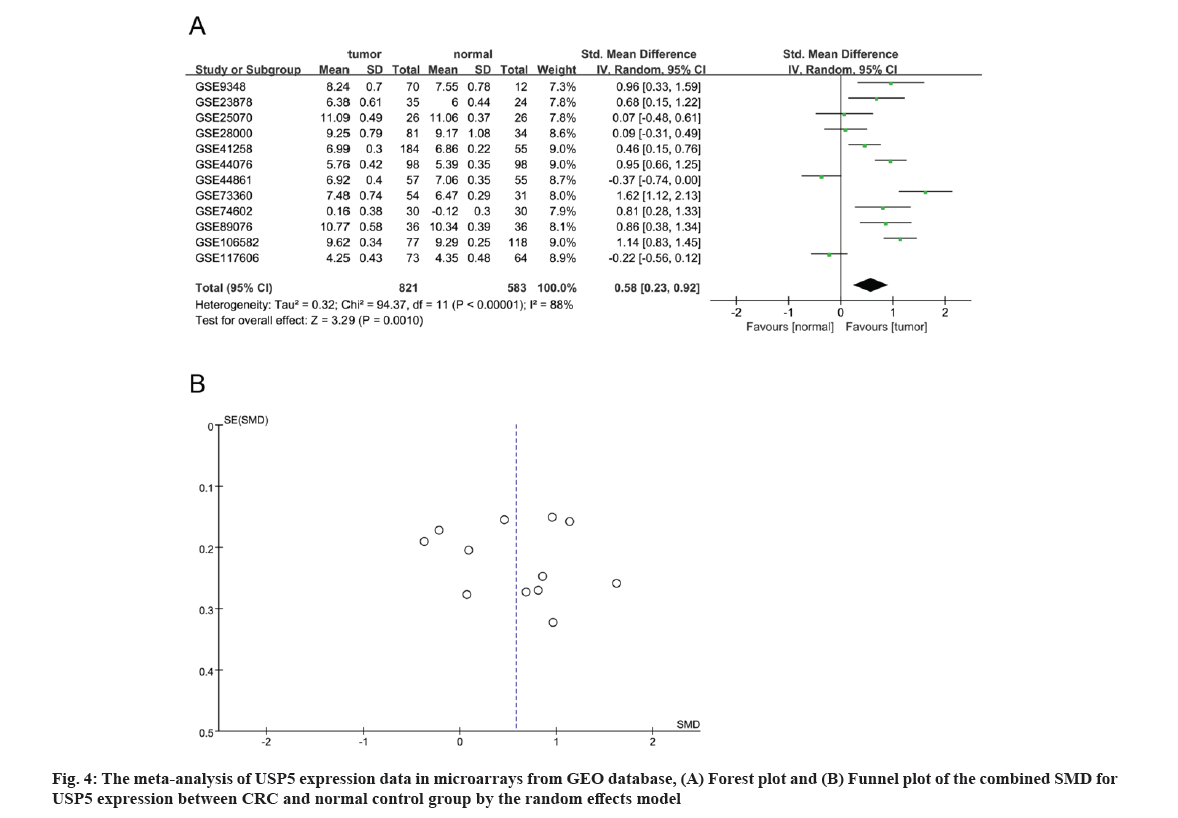
A total of 3068 microarrays from the GEO database were identified, among which 12 microarrays such as GSE9348, GSE23878, GSE25070, GSE28000, GSE41258, GSE44076, GSE44861, GSE73360, GSE74602, GSE89076, GSE106582 and GSE117606 met the entry criteria (fig. 1). Eight microarrays which include GSE9348, GSE23878, GSE41258, GSE44076, GSE73360, GSE74602, GSE89076 and GSE106582 demonstrated that the expression of USP5 was significantly higher in clinical CRC than normal tissues, whereas no significant difference was found in other four GEO datasets such as GSE25070, GSE28000, GSE44861 and GSE117606 (Table 1 and fig. 2). USP5 had shown a significant diagnostic value in seven microarrays which include GSE9348, GSE23878, GSE44076, GSE73360, GSE74602, GSE89076 and GSE106582 (fig. 3). 821 tumor samples and 583 normal samples were used in this meta-analysis. The pooled Standard Mean Difference (SMD) of USP5 was 0.58 with 95 % Confidence Interval (CI) (0.23-0.92) by random-effects model (p<0.0001, I2=88 %) (fig. 4A) and the results of funnel plot were shown in fig. 4B.
| GEO series | Contributor | Year | Samples | Normal (n) | Cancer (n) | Stage | Platform |
|---|---|---|---|---|---|---|---|
| GSE9348 | Hong et al.[23] | 2010 | Tissue | 12 | 70 | Early stage | GPL570 (HG-U133_Plus_2) Affymetrix Human Genome U133 Plus 2.0 Array |
| GSE23878 | Uddin et al.[24] | 2010 | 24 | 35 | |||
| GSE25070 | Hinoue et al.[25] | 2011 | 26 | 26 | GPL6883 Illumina HumanRef-8 v3.0 gene expression beadchip | ||
| GSE28000 | Jovov et al.[26] | 2012 | 34 | 81 | GPL1708 Agilent-012391 Whole Human Genome Oligo Microarray G4112A (Feature number version) | ||
| GPL4133 Agilent-014850 Whole Human Genome Microarray 4×44K G4112F (Feature number version) | |||||||
| GSE41258 | Sheffer et al.[27] | 2012 | 55 | 184 | I-IV | GPL96 (HG-U133A) Affymetrix Human Genome U133A Array | |
| GSE44076 | Sole et al.[28] | 2014 | 98 | 98 | II | GPL13667 (HG-U219) Affymetrix Human Genome U219 Array | |
| GSE44861 | Ryan et al.[29] | 2014 | 55 | 57 | GPL3921 (HT_HG-U133A) Affymetrix HT Human Genome U133A Array | ||
| GSE73360 | Condorelli et al.[30] | 2016 | 31 | 54 | GPL17586 (HTA-2_0) Affymetrix Human Transcriptome Array 2.0 (Transcript (gene) version) | ||
| GSE89076 | Satoh et al.[31] | 2017 | 36 | 36 | I-IV | GPL16699 Agilent-039494 SurePrint G3 Human GE v2 8×60K Microarray 039381 (Feature number version) |
Table 1: Characteristics of Studies Based on Geo Dataset
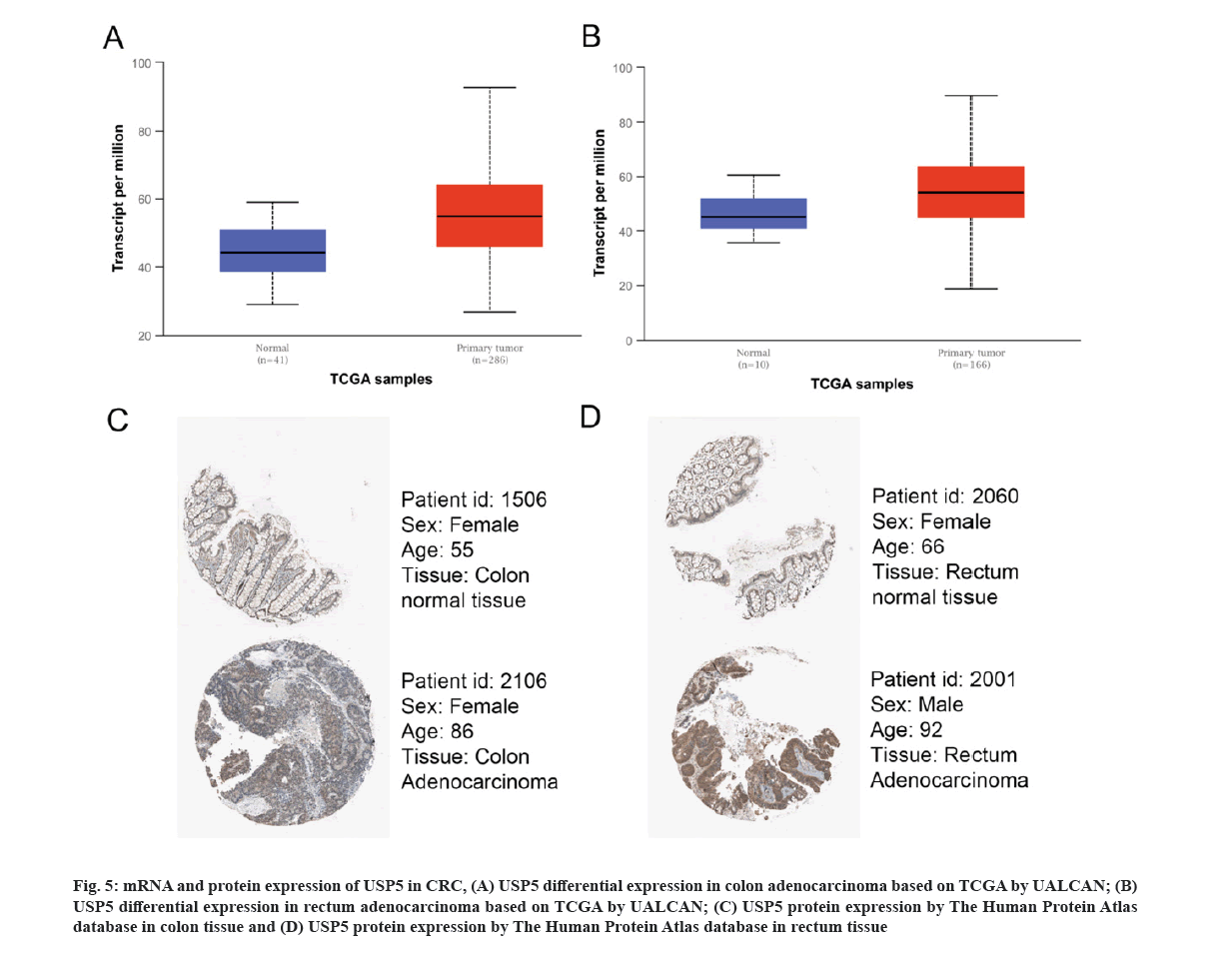
We next analyzed USP5 mRNA expression based on TCGA data by UALCAN. The differential expression in colon adenocarcinoma and rectum adenocarcinoma were shown respectively (fig. 5A and fig. 5B), consistent with GEO database results.
Fig 5: mRNA and protein expression of USP5 in CRC, (A) USP5 differential expression in colon adenocarcinoma based on TCGA by UALCAN; (B) USP5 differential expression in rectum adenocarcinoma based on TCGA by UALCAN; (C) USP5 protein expression by The Human Protein Atlas database in colon tissue and (D) USP5 protein expression by The Human Protein Atlas database in rectum tissue
The Human Protein Atlas database was used to present USP5 protein expression and IHC results revealed that USP5 is highly expressed in CRC (fig. 5C and fig. 5D).
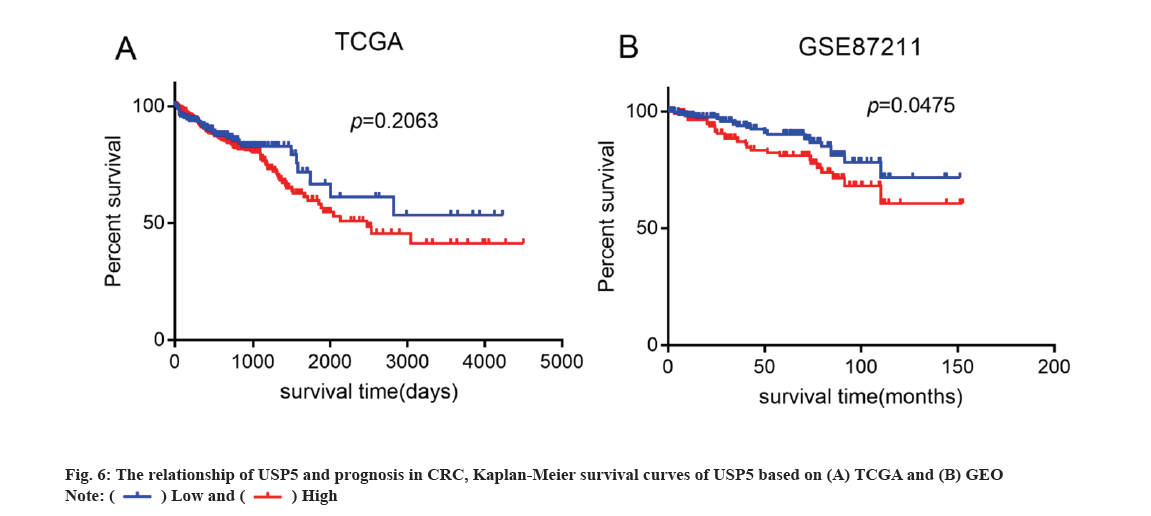
Kaplan-Meier survival analysis was performed by using TCGA and GEO database. As shown in fig. 6, USP5 high expression was correlated with poor Overall Survival (OS), which indicated that USP5 expression might be a potential indicator of poor clinical prognosis in patients with CRC.
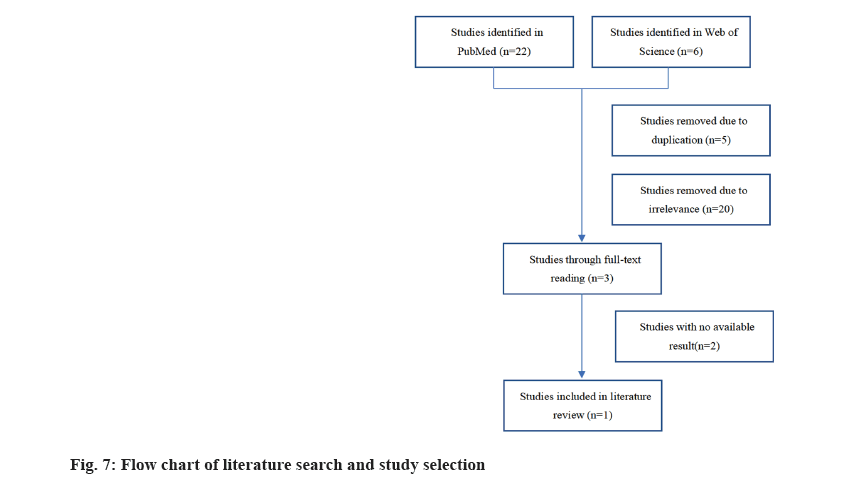
We explored USP5 expression in CRC based on literature data (fig. 7) where, only one study met the selection criteria. USP5 was considered highly expressed in CRC as a result of quantitative Reverse Transcriptase-Polymerase Chain Reaction (qRT-PCR), immunoblotting and immunohistochemistry of cell lines. Also, high USP5 expression was correlated with tumor stage and poor prognosis.
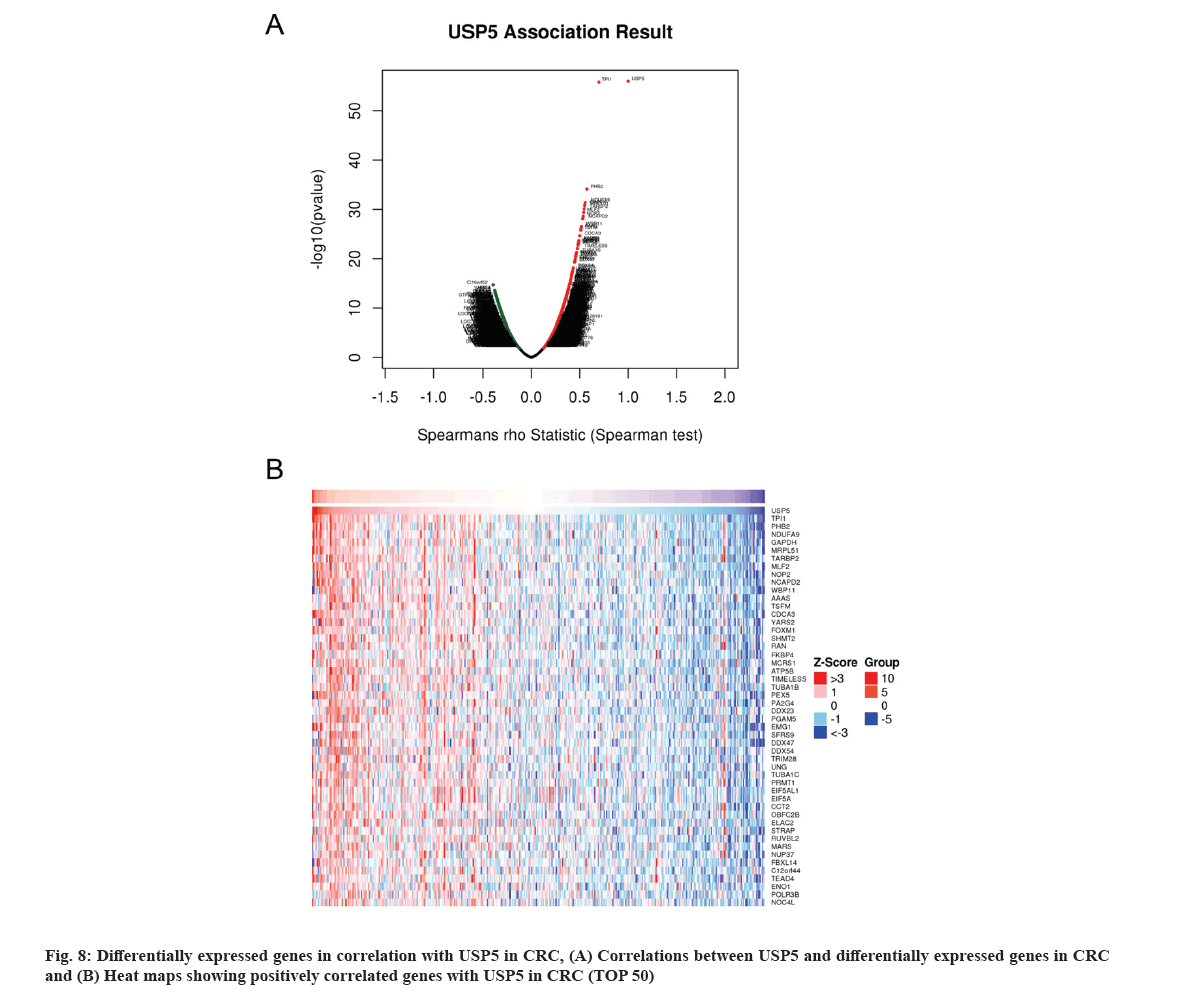
USP5 co-expression genes were obtained based on LinkedOmics (fig. 8A) and a total of 5699 genes co-expressed with USP5 were statistically significant with False Discovery Rate (FDR)<0.05. Top 50 positively correlated significant genes were displayed in fig. 8B. USP5 expression presented a significant correlation with Triosephosphate Isomerase 1 (TPI1), Prohibitin-2 (PHB2) and Nicotinamide Adenine Dinucleotide (NADH) Ubiquinone oxidoreductase subunit A9 (NDUFA9).
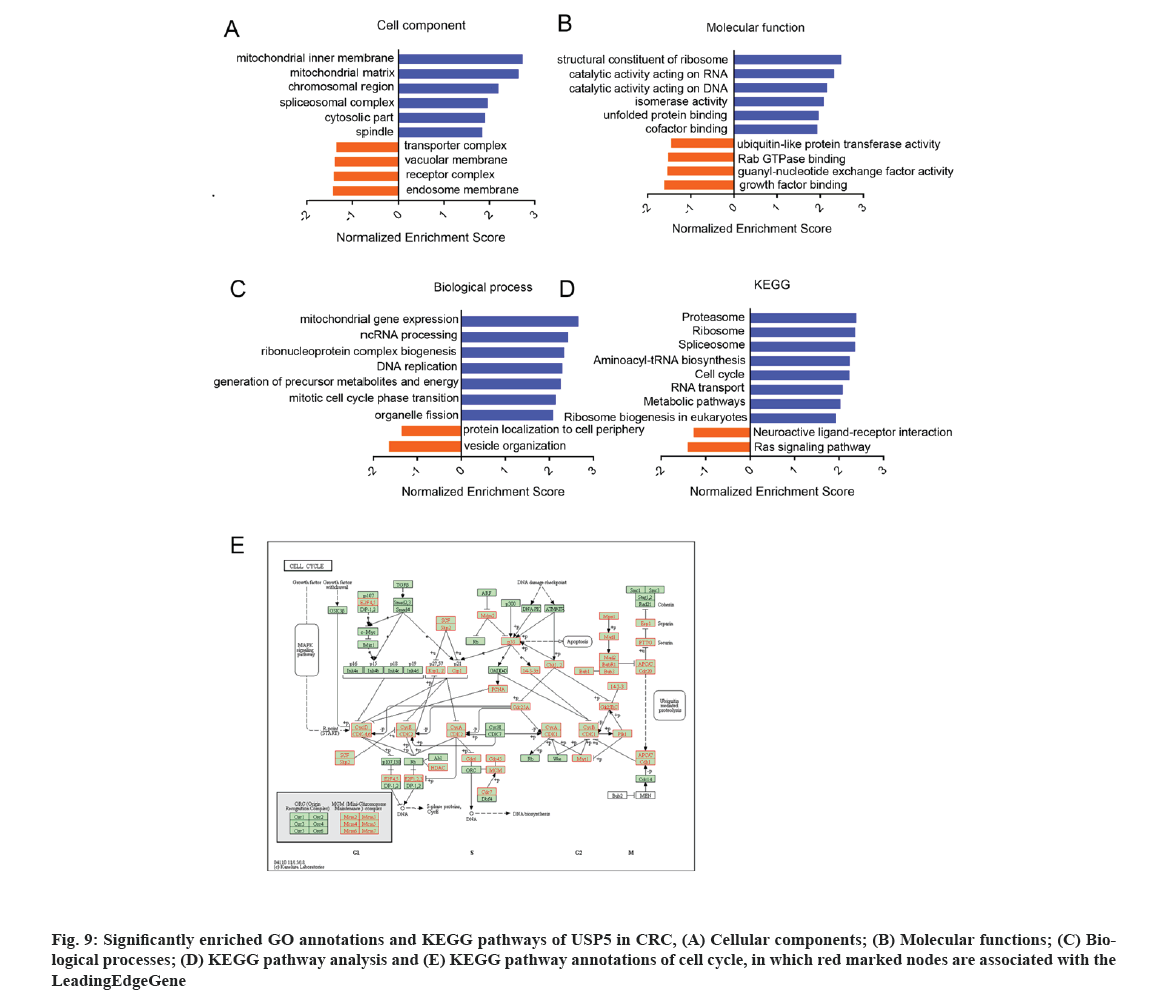
GO analysis showed that USP5 expression correlated genes located mainly in mitochondrial, ribosomal, chromosomal and spliceosomal complexes. They participated in cell cycle regulation, Deoxyribonucleic Acid (DNA) replication and RNA processing (fig. 9A-fig. 9C). KEGG pathway analysis showed enrichment in proteasome, ribosome, spliceosome, cell cycle and several metabolic pathways (fig. 9D and fig. 9E).
By using GSEA, we analyzed kinase, transcription factor targets and miRNA networks of related genes in order to further explore the targets of USP5 in CRC. The top 5 most significant target networks were listed in Table 2. The kinase target networks included Polo Like Kinase 1 (PLK1), Cyclin Dependent Kinase 1 (CDK1), Cyclin Dependent Kinase 2 (CDK2), Aurora Kinase A (AURKA) and Aurora Kinase B (AURKB). The transcription factor targets network mainly correlated to E2F Transcription Factor 1 (E2F) family and Forkhead-Related Activator 2 (FREAC2). miR-493 (ATGTACA), miR-369-3P (GTATTAT), miR-18A, miR-18B (GCACCTT), miR-448 (ATATGCA) and miR-129 (GCAAAAA) were potentially related to USP5.
| Enriched category | GeneSet | LeadingEdgeNum | p value | FDR |
|---|---|---|---|---|
| Kinase target | Kinase_PLK1 | 42 | 0 | 0 |
| Kinase_CDK1 | 74 | 0 | 0 | |
| Kinase_CDK2 | 96 | 0 | 0 | |
| Kinase_AURKA | 16 | 0 | 0 | |
| Kinase_AURKB | 35 | 0 | 4.84E-04 | |
| Transcription factor target | V$E2F_Q6 | 72 | 0 | 0 |
| V$E2F_Q4 | 73 | 0 | 0 | |
| V$E2F_Q4_01 | 66 | 0 | 0 | |
| V$E2F1_Q6 | 67 | 0 | 0 | |
| V$FREAC2_01 | 92 | 0 | 0 | |
| miRNA target | ATGTACA, miR-493 | 137 | 0 | 0 |
| GTATTAT, miR-369-3P | 89 | 0 | 0 | |
| GCACCTT, miR-18A, miR-18B | 56 | 0 | 0 | |
| ATATGCA, miR-448 | 96 | 0 | 0 | |
| GCAAAAA, miR-129 | 86 | 0 | 2.83E-04 |
Note: LeadingEdgeNum: The number of leading edge genes and V$: The annotation found in Molecular Signatures Database (MSigDB) for transcription factors
Table 2: The Kinase, Transcription Factor-Target and miRNA Networks of USP5 in CRC
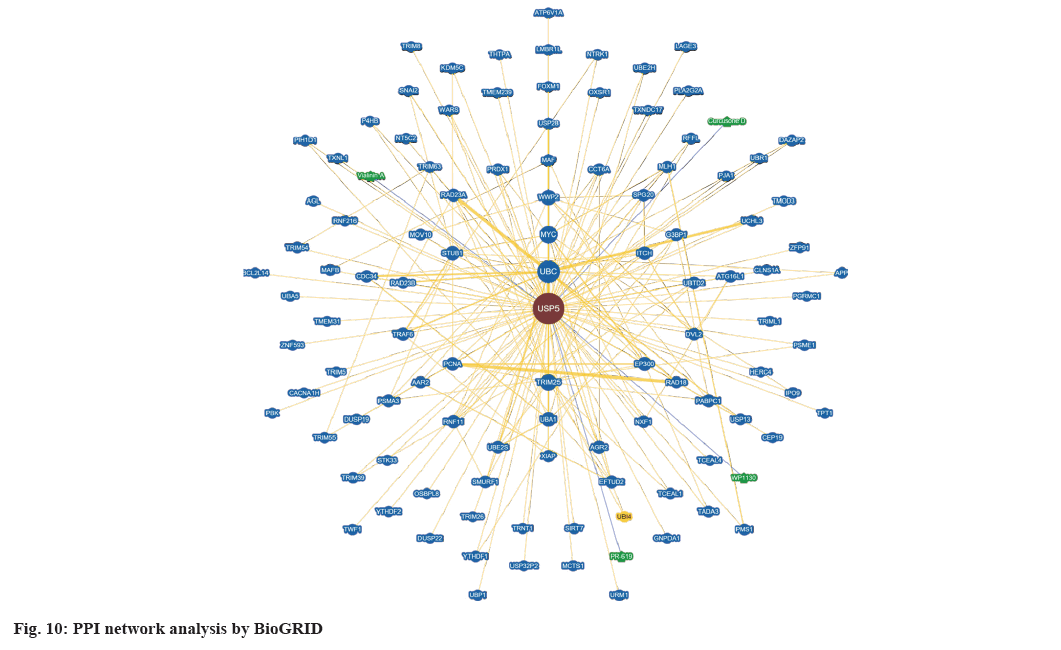
We constructed PPI network by BioGRID (fig. 10) and a total of 105 interactors and 155 published interactions were found. In addition, 4 unique small molecule chemical associations were displayed in network, including vialinin A, degrasyn (WP1130), curcusone D and 2,6-Diaminopyridine-3,5-bis(thiocyanate) (PR-619).
Emerging evidence showed that UPS were characterized in promoting tumorigenesis and tumor progression[32]. Multiple DUBs has been confirmed as novel promising biomarkers and therapeutic targets for cancer, in which USP5 is one of the potential deubiquitinated proteins[6]. The present study aimed at analyzing the relationship between USP5 expression, prognosis and the potential functional role of USP5 in CRC.
In this study, we identified the aberrantly highly expressed USP5 in tumor tissues compared with normal tissues based on GEO datasets. A large cohort of 821 CRC samples and 583 normal samples were involved in the comprehensive meta-analysis. TCGA and The Human Protien Atlas results drew the same conclusion. Additionally, we found diagnostic values of USP5 high expression. Considering the early detection problem, USP5 deserves further clinical validation as a potential diagnostic marker. Kaplan-Meier survival analysis showed that high level of expression of USP5 results in shorter survival rate in CRC patients and the above conclusions are consistent with literature review[15].
Since USP5 has been involved in several significant physiological functions, we need to figure out USP5 co-expression genes and its enrichment by GSEA. USP5 neighboring gene networks generally showed different degrees of amplification in CRC, which indicated that USP5 might involve in proteasome, ribosome, DNA replication, cell cycle, spliceosome and several metabolic pathways. As a classic deubiquitinated protein, USP5 definitely plays a role in proteasome. USP5 in regulating RNA splicing[13] and DNA damage repair[33] are consistent with the former studies. In pancreatic cancer, USP5 can promote tumor progression by regulating cell cycle[11]. But whether USP5 can promote cell cycle in CRC still needs further research.
Next, we used GSEA enrichment analysis to reveal the target kinase, transcription factors and miRNA. We found that USP5 is associated with a network of kinases including PLK1, CDK1, CDK2, AURKA and AURKB in CRC. These kinases regulate tumor cell proliferation and cell cycle[34-37]. In fact, PLK1 is an essential gene for the correct execution of cell division[38]. AUR/PLK1 axis directly regulates mitosis and other non-canonical targets, including cellular-myelocytomatosis (c-myc)[39-41]. Although there are some surprisingly debated topics about whether PLK1 is an oncogene or a tumor suppressor gene, this does not affect the use of PLK1 inhibitors as an anticancer drug during last decades[42,43].
The transcription factor targets of USP5 mainly correlated to E2F family. E2F family, as an important transcriptional factors family, has been implicated in the regulation of many cell possesses related to cell proliferation, differentiation, cell cycle, apoptosis and DNA repair[44]. Our analysis suggests that E2F family might be an important target of USP5 and regulate cell proliferation and cycle through USP5. Further studies need to test this hypothesis.
Several miRNAs were found related to USP5 in our study. miR-493, miR-129 and miR-448 suppresses proliferation and invasion in cancer[45-47], while miR-18a, miR-18b and miR-369 has the opposite effect[48-50]. Although these miRNAs have different degrees of relationship with tumor occurrence and development, their relevance to USP5 needs further experimental research.
According to the results of PPI network data, we found lots of genes were correlated with USP5. Some of them had already been verified, like FOXM1, Suppressor of Mothers against Decapentaplegic (SMAD) Ubiquitination Regulatory Factor 1 (SMURF1) and Tumor Necrosis Factor Receptor (TNFR)-Associated Factor 6 (TRAF6)[51-53]. Others might be a hotspot in the future researches. Four inhibitors such as WP1130, vialinin A, curcusone D and PR-619 might inhibit USP5, in which the first two inhibitors have been used and confirmed previously[9,54].
Cell cycle disorder resulted in tumor cell aberrant proliferation is one of the 10 tumor hallmarks[55]. The above evidence indicated that USP5 may be related to CRC cell cycle regulation, so we verified in our experiments. Overexpression of USP5 can change the expression of cyclin protein marker, confirming that USP5 can indeed regulate the cell cycle in CRC. The mechanism might be USP5 regulates the deubiquitination of many key proteins in cell cycle signaling pathways.
In conclusion, this study mainly used meta-analysis and bioinformatics methods to improve the expression and functional analysis of USP5 in CRC. Our results still need to be confirmed by further experimental studies.
Conflict of interests:
The authors declared no conflict of interests.
References
- Siegel RL, Miller KD, Sauer AG, Fedewa SA, Butterly LF, Anderson JC,
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70(1):7-30.
- Zhong JL, Huang CZ. Ubiquitin proteasome system research in gastrointestinal cancer. World J Gastrointest Oncol 2016;8(2):198-206.
[Crossref] [Google scholar] [PubMed]
- Roeten MS, Cloos J, Jansen G. Positioning of proteasome inhibitors in therapy of solid malignancies. Cancer Chemother Pharmacol 2018;81(2):227-43.
[Crossref] [Google scholar] [PubMed]
- Chen YJ, Wu H, Shen XZ. The ubiquitin-proteasome system and its potential application in hepatocellular carcinoma therapy. Cancer Lett 2016;379(2):245-52.
[Crossref] [Google scholar] [PubMed]
- Ning F, Xin H, Liu J, Lv C, Xu X, Wang M, et al. Structure and function of USP5: Insight into physiological and pathophysiological roles. Pharmacol Res 2020;157:1-27.
[Crossref] [Google scholar] [PubMed]
- Liu Y, Wang WM, Zou LY, Li L, Feng LU, Pan MZ, et al. Ubiquitin specific peptidase 5 mediates histidine‐rich protein Hpn induced cell apoptosis in hepatocellular carcinoma through P14‐P53 signaling. Proteomics 2017;17(12):1-31.
[Crossref] [Google scholar] [PubMed]
- Li XY, Wu HY, Mao XF, Jiang LX, Wang YX. USP5 promotes tumorigenesis and progression of pancreatic cancer by stabilizing FoxM1 protein. Biochem Biophys Res Commun 2017;492(1):48-54.
[Crossref] [Google scholar] [PubMed]
- Wang S, Juan J, Zhang Z, Du Y, Xu Y, Tong J, et al. Inhibition of the deubiquitinase USP5 leads to c-Maf protein degradation and myeloma cell apoptosis. Cell Death Dis 2017;8(9):1-13.
[Crossref] [Google scholar] [PubMed]
- Ma X, Qi W, Pan H, Yang F, Deng J. Overexpression of USP5 contributes to tumorigenesis in non-small cell lung cancer via the stabilization of β-catenin protein. Am J Cancer Res 2018;8(11):2284-95.
[Google scholar] [PubMed]
- Kaistha BP, Krattenmacher A, Fredebohm J, Schmidt H, Behrens D, Widder M, et al. The deubiquitinating enzyme USP5 promotes pancreatic cancer via modulating cell cycle regulators. Oncotarget 2017;8(39):66215-25.
[Crossref] [Google scholar] [PubMed]
- Du Y, Lin J, Zhang R, Yang W, Quan H, Zang L, et al. Ubiquitin specific peptidase 5 promotes ovarian cancer cell proliferation through deubiquitinating HDAC2. Aging 2019;11(21):9778-93.
[Crossref] [Google scholar] [PubMed]
- Izaguirre DI, Zhu W, Hai T, Cheung HC, Krahe R, Cote GJ. PTBP1‐dependent regulation of USP5 alternative RNA splicing plays a role in glioblastoma tumorigenesis. Mol Carcinog 2012;51(11):895-906.
[Crossref] [Google scholar] [PubMed]
- Meng J, Ai X, Lei Y, Zhong W, Qian B, Qiao K, et al. USP5 promotes epithelial-mesenchymal transition by stabilizing SLUG in hepatocellular carcinoma. Theranostics 2019;9(2):573-87.
[Crossref] [Google scholar] [PubMed]
- Xu X, Huang A, Cui X, Han K, Hou X, Wang Q, et al. Ubiquitin specific peptidase 5 regulates colorectal cancer cell growth by stabilizing Tu translation elongation factor. Theranostics 2019;9(14):4208-20.
[Crossref] [Google scholar] [PubMed]
- Zhang Z, Gao W, Zhou LI, Chen Y, Qin S, Zhang LU, et al. Repurposing brigatinib for the treatment of colorectal cancer based on inhibition of ER-phagy. Theranostics 2019;9(17):4878-92.
[Crossref] [Google scholar] [PubMed]
- Chandrashekar DS, Bashel B, Balasubramanya SA, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BV, et al. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017;19(8):649-58.
[Crossref] [Google scholar] [PubMed]
- Thul PJ, Lindskog C. The human protein atlas: A spatial map of the human proteome. Protein Sci 2018;27(1):233-44.
[Crossref] [Google scholar] [PubMed]
- Anaya J. OncoLnc: Linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. Peer J Comput Sci 2016;2:1-13.
- Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res 2004;10(21):7252-9.
[Crossref] [Google scholar] [PubMed]
- Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res 2018;46(1):956-63.
[Crossref] [Google scholar] [PubMed]
- Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, et al. The BioGRID interaction database: 2019 update. Nucleic Acids Res 2019;47(1):529-41.
[Crossref] [Google scholar] [PubMed]
- Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis 2010;27(2):83-90.
[Crossref] [Google scholar] [PubMed]
- Uddin S, Ahmed M, Hussain A, Abubaker J, Al-Sanea N, Jabbar A, et al. Genome-wide expression analysis of Middle Eastern colorectal cancer reveals FOXM1 as a novel target for cancer therapy. Am J Pathol 2011;178(2):537-47.
[Crossref] [Google scholar] [PubMed]
- Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, van den Berg D, et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012;22(2):271-82.
[Crossref] [Google scholar] [PubMed]
- Jovov B, Araujo-Perez F, Sigel CS, Stratford JK, McCoy AN, Yeh JJ, et al. Differential gene expression between African American and European American colorectal cancer patients. PLoS One 2012;7(1):1-9.
[Crossref] [Google scholar] [PubMed]
- Sheffer M, Bacolod MD, Zuk O, Giardina SF, Pincas H, Barany F, et al. Association of survival and disease progression with chromosomal instability: A genomic exploration of colorectal cancer. Proc Natl Acad Sci U S A 2009;106(17):7131-6.
[Crossref] [Google scholar] [PubMed]
- Sole X, Crous-Bou M, Cordero D, Olivares D, Guinó E, Sanz-Pamplona R, et al. Discovery and validation of new potential biomarkers for early detection of colon cancer. PLoS One 2014;9(9):1-11.
[Crossref] [Google scholar] [PubMed]
- Ryan BM, Zanetti KA, Robles AI, Schetter AJ, Goodman J, Hayes RB, et al. Germline variation in NCF4, an innate immunity gene, is associated with an increased risk of colorectal cancer. Int J Cancer 2014;134(6):1399-407.
[Crossref] [Google scholar] [PubMed]
- Condorelli DF, Spampinato G, Valenti G, Musso N, Castorina S, Barresi V. Positive caricature transcriptomic effects associated with broad genomic aberrations in colorectal cancer. Scientific Reports. 2018;8(1):1-16.
[Crossref] [Google scholar] [PubMed]
- Satoh K, Yachida S, Sugimoto M, Oshima M, Nakagawa T, Akamoto S, et al. Global metabolic reprogramming of colorectal cancer occurs at adenoma stage and is induced by MYC. Proc Natl Acad Sci U S A 2017;114(37):E7697-706.
[Crossref] [Google scholar] [PubMed]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell 2005;123(5):773-86.
[Crossref] [Google scholar] [PubMed]
- Nakajima S, Lan L, Wei L, Hsieh CL, Rapić-Otrin V, Yasui A, et al. Ubiquitin-specific protease 5 is required for the efficient repair of DNA double-strand breaks. PLoS One 2014;9(1):1-11.
[Crossref] [Google scholar] [PubMed]
- Bertolin G, Tramier M. Insights into the non-mitotic functions of Aurora kinase A: More than just cell division. Cell Mol Life Sci 2020;77(6):1031-47.
[Crossref] [Google scholar] [PubMed]
- Goldenson B, Crispino JD. The aurora kinases in cell cycle and leukemia. Oncogene 2015;34(5):537-45.
[Crossref] [Google scholar] [PubMed]
- Xie B, Wang S, Jiang N, Li JJ. Cyclin B1/CDK1-regulated mitochondrial bioenergetics in cell cycle progression and tumor resistance. Cancer Lett 2019;443:56-66.
[Crossref] [Google scholar] [PubMed]
- Tadesse S, Anshabo AT, Portman N, Lim E, Tilley W, Caldon CE, et al. Targeting CDK2 in cancer: Challenges and opportunities for therapy. Drug Discov Today 2020;25(2):406-13.
[Crossref] [Google scholar] [PubMed]
- Colicino EG, Hehnly H. Regulating a key mitotic regulator, polo‐like kinase 1 (PLK1). Cytoskeleton 2018;75(11):481-94.
[Crossref] [Google scholar] [PubMed]
- Macůrek L, Lindqvist A, Lim D, Lampson MA, Klompmaker R, Freire R, et al. Polo-like kinase-1 is activated by aurora A to promote checkpoint recovery. Nature 2008;455(7209):119-23.
[Crossref] [Google scholar] [PubMed]
- Joukov V, de Nicolo A. Aurora-PLK1 cascades as key signaling modules in the regulation of mitosis. Sci Signal 2018;11(543):1-25.
[Crossref] [Google scholar] [PubMed]
- Murga-Zamalloa C, Polk A, Hanel W, Chowdhury P, Brown N, Hristov AC, et al. Polo-like-kinase 1 (PLK-1) and c-myc inhibition with the dual kinase-bromodomain inhibitor volasertib in aggressive lymphomas. Oncotarget 2017;8(70):114474-80.
[Crossref] [Google scholar] [PubMed]
- Elsayed I, Wang X. PLK1 inhibition in cancer therapy: Potentials and challenges. Future Med Chem 2019;11(12):1383-6.
[Crossref] [Google scholar] [PubMed]
- de Cárcer G. The mitotic cancer target polo-like kinase 1: Oncogene or tumor suppressor?. Genes 2019;10(3):1-14.
[Crossref] [Google scholar] [PubMed]
- Lammens T, Li J, Leone G, de Veylder L. Atypical E2Fs: New players in the E2F transcription factor family. Trends Cell Biol 2009;19(3):111-8.
[Crossref] [Google scholar] [PubMed]
- Wang G, Fang X, Han M, Wang X, Huang Q. microRNA-493-5p promotes apoptosis and suppresses proliferation and invasion in liver cancer cells by targeting VAMP2. Int J Mol Med 2018;41(3):1740-8.
[Crossref] [Google scholar] [PubMed]
- Feng J, Guo J, Wang JP, Chai BF. miR-129-5p inhibits proliferation of gastric cancer cells through targeted inhibition on HMGB1 expression. Eur Rev Med Pharmacol Sci 2020;24(7):3665-73.
[Crossref] [Google scholar] [PubMed]
- Liu HY, Chang J, Li GD, Zhang ZH, Tian J, Mu YS. microRNA-448/EPHA7 axis regulates cell proliferation, invasion and migration via regulation of PI3K/AKT signaling pathway and epithelial-to-mesenchymal transition in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2020;24(11):6139-49.
[Crossref] [Google scholar] [PubMed]
- Al‑Kafaji G, Al-Naieb ZT, Bakhiet M. Increased oncogenic microRNA-18a expression in the peripheral blood of patients with prostate cancer: A potential novel non-invasive biomarker. Oncol Lett 2016;11(2):1201-6.
[Crossref] [Google scholar] [PubMed]
- Egeland NG, Jonsdottir K, Aure MR, Sahlberg K, Kristensen VN, Cronin-Fenton D, et al. miR-18a and miR-18b are expressed in the stroma of oestrogen receptor alpha negative breast cancers. BMC Cancer 2020;20(1):1-4.
[Crossref] [Google scholar] [PubMed]
- Guo L, Li B, Yang J, Shen J, Ji J, Miao M. Fibroblast‑derived exosomal microRNA‑369 potentiates migration and invasion of lung squamous cell carcinoma cells via NF1‑mediated MAPK signaling pathway. Int J Mol Med 2020;46(2):595-608.
[Crossref] [Google scholar] [PubMed]
- Qian G, Ren Y, Zuo Y, Yuan Y, Zhao P, Wang X, et al. Smurf1 represses TNF-α production through ubiquitination and destabilization of USP5. Biochem Biophys Res Commun 2016;474(3):491-6.
[Crossref] [Google scholar] [PubMed]
- Chen Y, Li Y, Xue J, Gong A, Yu G, Zhou A, et al. Wnt‐induced deubiquitination FoxM1 ensures nucleus β‐catenin transactivation. EMBO J 2016;35(6):668-84.
[Crossref] [Google scholar] [PubMed]
- Luo XB, Xi JC, Liu Z, Long Y, Li LT, Luo ZP, et al. Proinflammatory effects of ubiquitin-specific protease 5 (USP5) in rheumatoid arthritis fibroblast-like synoviocytes. Mediators Inflamm 2020;2020:1-9.
[Crossref] [Google scholar] [PubMed]
- Yoshioka Y, Ye YQ, Okada K, Taniguchi K, Yoshida A, Sugaya K, et al. Ubiquitin-specific peptidase 5, a target molecule of vialinin A, is a key molecule of TNF-α production in RBL-2H3 cells. PLoS One 2013;8(12):1-6.
[Crossref] [Google scholar] [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell 2011;144(5):646-74.
[Crossref] [Google scholar] [PubMed]