- *Corresponding Author:
- G. Murtaza
Department of Pharmacy, COMSATS University Islamabad, Lahore Campus, Pakistan
E-mail: gmdogar356@gmail.com
| This article was originally published in a special issue, “Biomedical Research in Healthcare Setting” |
| Indian J Pharm Sci 2020:82(3)Spl issue5;56-65 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Long non-coding RNAs were analyzed using microarray testing in stable angina pectoris on a genomewide scale. According to the inclusion and exclusion criteria, long non-coding RNAs gene-chip was used to detect the expression of long non-coding RNAs in the stable angina pectoris and the healthy control group. Gene-chip analysis, signal pathway analysis and the predictive results of target genes were used. After amplification of the sample size, the expression of long non-coding RNAs in anticoagulated peripheral blood was screened using fluorescent quantitative real-time polymerase chain reaction. Statistical software SPSS 22.0 was employed for statistical data analysis using two independent samples of the t-test. The long non-coding RNA expression profiles of stable angina pectoris were established from the results of the long non-coding RNA gene chip. A total of 226 upregulated and 173 downregulated long non-coding RNAs were identified to be significantly and differentially expressed between the stable angina pectoris group and the healthy control group, and 106 upregulated and 346 downregulated mRNAs were identified. Differences in long non-coding RNAs ENST00000418539.1 and ENST00000589524.1 were verified using quantitative real-time polymerase chain reaction, and the results were consistent with the chip-test outcomes. The long non-coding RNA expression profile of stable angina pectoris was established by analysing the long non-coding RNAs gene-chip. The long non-coding RNA expression was consistent with the results of the microarray test and could have a potential role in the development of stable angina pectoris.
Keywords
Long non-coding RNA, stable angina pectoris, microarray
Stable angina pectoris (SAP) is one of the most commonly found clinical forms of coronary artery disease (CAD). Despite major advances in the management of CAD including percutaneous coronary intervention (PCI), coronary artery bypass grafting (CABG), and novel therapeutic techniques such as ivabradine, angina pain still persists in up to 14 % of SAP patients[1-3]. A number of alternative therapies, for instance, counter-pulsation, stem cell-based treatments and myocardial or intracoronary application of proteins are employed to improve angina symptoms and facilitate coronary perfusion in refractory angina SAP patients[4-6]. However, none of the mentioned strategies could succeed in achieving long-term positive results so far. On the other side, the advancements in genomics have significantly contributed to the development of several novel biomarkers with immense clinical significance in the management of CAD[7,8].
The long non-coding RNAs (lncRNAs) are fragments of RNA with a length of more than 200 basepairs that are transcribed from DNA but do not encode for any protein[9]. In recent years, lncRNAs have gained considerable importance because of their implications in gene expression and epigenetic regulation, pre/ post-translational mRNA processing, transcriptional control and pre/post-translational mRNA processing and their involvement in the initiation and development of CAD[10]. Hitherto, a number of these lncRNAs are clinically employed as biomarkers for diagnosis in many serious pathological conditions, for example, the lncRNA H19 in gastric cancer[11], the lncRNA PCA3 in prostate cancer[12] and the lncRNA LIPCAR in cardiac insufficiencies following myocardial infarction[13]. Despite the emerging clinical significance of lncRNAs, the role of these invaluable biomarkers in SAP patients remains obscure.
In order to fully understand the lncRNA biology including its role in SAP, it is necessary to characterize the expression pattern of lncRNA. In the present study, the main purpose is to investigate the expression of lncRNAs in peripheral blood cells of 5 SAP patients and 5 matched healthy control subjects by microarray analysis. The results were further validated by qRTPCR in 38 SAP patients and 38 healthy controls. The major focus was to identify differentially expressed lncRNAs and mRNAs throughout the process and finding potential markers that exhibited significant changes in expression.
Materials and Methods
Ethics:
The present research work was approved by the Xinjiang Medical University (XMU) Research Ethics Committee. The enrolment of the participants from XMU affiliated Hospitals was performed within 1 y starting from December 2014 to December 2015. The participants were well informed about the objectives of the experiments prior to the commencement of the study after getting their written consent.
Patients and sample collection:
The inclusion criteria of the study included patients with SAP who received treatment involving primary percutaneous coronary intervention. Those patients were considered for the study whose diagnostic conditions were in accordance with pre-stated SAP standards[14] and have undergone coronary angiography and percutaneous coronary stenting. The study did not include patients who suffered from cardiac failure, valvular issues, and acute coronary syndrome. Likewise, the patients who developed infections, chronic inflammatory complications or other severe systematic disorders such as renal and hepatic insufficiencies, malignancies and autoimmune reactions were excluded from the study. As mentioned earlier, only those patients who gave written consent to undergo the experiments were included. For the IncRNA chip analysis, 10 subjects were randomly selected using the coding system. Five of these subjects included SAP patients that were selected using SAP-H4, -H7, -H8, -H9, and -H10 coding whereas the remaining 5 subjects included normal volunteers who were selected using normal N-H5, -H6, -H7, -H8 and -H10 coding. Moreover, blood samples from 38 patients and 38 healthy volunteers were also included in the study to validate IncRNA expression using quantitative real-time PCR.
RNA extraction, labelling and hybridization:
Blood samples with EDTA as anticoagulant were subjected to the total RNA extraction using Trizol reagent (Invitrogen) with subsequent purification using mirVana miRNA isolation kits as per manufacturer’s instructions (Ambion, Austin, TX, USA). The RNA was quantified and standardized using Nano Drop spectrophotometer by measuring OD (260/280 nm) whereas its integrity was evaluated by 1 % formaldehyde denaturing gel electrophoresis. Microarray-based gene expression analysis was performed for sample labelling and array hybridization as per the specified protocol (Agilent Technology). The complementary DNA (cDNA) was subjected to fluorescent labelling using Cy3 and Cy5- dCTP dyes through the RNA amplification method as described earlier[15]. The dsDNA sequences were synthesized from 1 μg total RNA by using CbcScript reverse transcriptase (Capitalbio) using T7 Oligo (dT/dN) primers[16]. Thereafter, PCR nucleospin extract II kit was used to purify dsDNA. It was further eluted with elution buffer (30 μl), concentrated to 15 μl through vacuum evaporation and later subjected to in vitro transcription reactions at 14 h at 37° temperature using a T7 Enzyme Mix.
After reverse transcription, Klenow enzyme labelling was employed with CbcScript II reverse transcriptase[17]. Briefly, 4 μg random nanomer was mixed with 2 μg of the amplified RNA with subsequent denaturation at 60° for 5 min followed by cooling on ice. Then, 2 μl of 0.1 M DTT was mixed with 1.5 μl CbcScript II reverse transcriptase and 5 μl of 4×first-strand buffers. The mixture was first incubated for 10 min at 25° followed by incubation for 90 min at 36°. As stated above, the PCR NucleoSpin extract II kit was used to purify the cDNA products that were further concentrated to 14 μl using a vacuum evaporator. After that, 4 μg random nanomer was mixed with cDNA. The mixture was heated for 3 min at 90° temperature followed by cooling on ice for 5 min. Cy5-dCTP/Cy3-dCTP along with Klenow buffer and dNTP were added to the mixture to make final concentrations of 40 μM Cy-dCTP, 120 μM dCTP, and 240 μM each of dATP, dTTP and dGTP.
A hybridization solution (80 μl) consisting of 0.2 % SDS, 3×SSC, 5×Denhardt’s solution and 25 % formamide was mixed with Cy3-dCTP and dCTP. The solution was incubated for 3 min at 90° temperature to denature its DNA content. The solution was loaded onto microarray using an Agilent Hybridization Oven for an overnight period at 40° temperature and 20 rpm rotation. The arrays were later washed for 5 min at 42° with 2×SSC and 0.2 % SDS followed by another washing at room temperature for 5 min with 0.2×SSC[18].
LncRNA and mRNA microarrays:
The total RNA (200 ng) from each sample was subjected to long non-coding RNA microarray by using Agilent lncRNA+mRNA array V 4.0 (4×180 K format). Each array contained probes that interrogated about 41 000 and 34 000 human lncRNAs and mRNAs, respectively following the Agilent protocol (Agilent, USA). The RNAs were detected by probes in replicate. The integration of the target sequences was performed by utilizing different database such as the Human LincRNA Catalog (14353 sequences), GENCODE/ ENSEMBL (23898 sequences), LNCipedia (21488 sequences), RefSeq (7760 sequences), NRED (13701 sequences), USCS (5627 sequences), Antisense ncRNA pipeline (1053 sequences), UCRs (962 sequences), Hox ncRNAs (407 sequences), LncRNAs-a (3019 sequences), H-InvDB (1038 sequences), Chen Ruisheng lab from the Institute of Biophysics, Chinese Academy of Science (848 sequences)[19].
Microarray imaging and data analysis:
The Gene Spring software (Agilent, V13.0) was used to summarize, normalize and analyse the lncRNA and mRNA array data for quality control. Volcano Plot filtering with hierarchical clustering was used to identify the statistically significant lncRNAs that were differentially expressed. The random variance model was used to identify the differentially expressed genes whereas the p-value was calculated by paired t-test. A fold change (FC) ≥2.0 was established and p-value <0.05 was required as the significance threshold for the up and down-regulated genes. Data were subjected to Log 2 transformation whereas the CLUSTER 3.0 software (Adjust Data function) was used for the median centring of genes. Moreover, Java Tree view software (Stanford University School of Medicine, Stanford, CA, USA) was used for tree visualization.
Functional and pathway assessment in relation to LncRNA:
The Kyoto Encyclopaedia of Genes and Genomes (KEGG) were used for the evaluation of the major pathway of the distinctively expressed genes, with a level of significance set at 0.05.
Quantitative real-time polymerase chain reaction (qRT-PCR):
The expression level of lncRNA was validated using qRT-PCR. Briefly, TRIZOL reagent was used to extract total RNA from monocytes which were later reverse transcribed by using cDNA reverse transcription kits following the manufacturer-advised protocol (Invitrogen, CA). Primer 6.0 software was used to design the primers for each lncRNA whereas the BLAST (basic local alignment search tool) from NCBI was used to compare these primers in order to confirm the uniqueness of their sequences. The primer sequences used are shown in Table 1. The PCR amplification was performed for 40 cycles at the temperature of 95° for 10 min, 60° for 15 s, and 60° for 60 s using ABI 7900HT RT-PCR System (Applied Biosystems, USA). The amplifications were conducted in a total volume of 10 μl that contained 0.5 μl of each of the primer mix and the genomic cDNA, 4 ul of nucleasefree water and 5 μl of Power SYBR Green PCR Master Mix (Applied Biosystems, USA). The PCR reactions were carried out in triplicate, and the comparative cycle threshold method (CT, 2−ΔΔCT) was used to calculate the relative expression of lncRNA, having β-action as the endogenous control. The level of each lncRNA in SAP patients was expressed as the fold changes against the averaged level of the same lncRNA in healthy controls.
| lncRNA ID | Forward primer | Reverse primer | Hybridization temperature (°) |
|---|---|---|---|
| ENST00000418539.1 | GGTGGTACCGGAGGGAATCT | CCACTAGATCGAGGTGCTTGG | 60 |
| ENST00000589524.1 | CGGGGATGGGGAAAGAACAG | CCCAGGCCCAGTTAGAAACTT | 60 |
| Beta-actin | CTCCATCCTGGCCTCGCTGT | GCTGTCACCTTCACCGTTCC | 60 |
Table 1: The Randomly Selected Lncrnas
Statistical analysis:
The data were statistically analyzed through SPSS software version 21.0 (IBM Inc, Chicago, Illinois, USA). An independent sample t-test was used to compare the differential expression levels of lncRNAs between groups whereas Fisher’s exact test was employed for GO and pathway analysis. The two-tailed statistical differences between groups were considered significant at p-value <0.05.
Results and Discussion
The study included a total of 38 SAP patients and 38 healthy individuals. The clinical parameters of the patients are presented in Table 2. The peripheral blood samples were utilized to extract of the total RNA. The lncRNA/mRNA expression of 5 SAP patients was compared with that of the healthy controls by microarray whereas the data from the remaining individuals were utilized for validation purposes. The expression of 41 000 lncRNA probes was determined. As per the GENCODE gene annotations, the tested lncRNAs were classified as bidirectional, intronic, intergenic, sense, antisense, and unknown[20]. The gene classification was based on the coding-gene locations with respect to the lncRNA.
| Variables | Controls (n=38) | SAP (n=38) | P |
|---|---|---|---|
| Age (y) | 62.11±8.90 | 58.87±7.90 | 0.099 |
| Male/female (number) | 17/21 | 25/13 | 0.135 |
| BMI | 25.55±2.59 | 24.83±2.76 | 0.488 |
| Active smoker (%) | 2 (5.2 %) | 7 (18.4 %) | 0.153 |
| Active drinker (%) | 1 (2.6 %) | 5 (13.1 %) | 0.200 |
| Diabetes, 𝑛 (%) | 7 (18.4 %) | 10 (26.3 %) | 0.583 |
| Hypertension (%) | 11 (28.9 %) | 28 (73.6 %) | 0.0002* |
| Systolic BP (mmHg) | 122.18±14.06 | 130.29±15.6 | 0.02* |
| Diastolic BP (mmHg) | 77.73±10.41 | 77.86±9.51 | 0.954 |
| FBS (mM) | 5.01±1.06 | 6.19±2.55 | 0.010* |
| TG (mM) | 1.46±0.92 | 1.67±0.96 | 0.346 |
| TC (mM) | 4.06±0.94 | 3.94±1.42 | 0.680 |
| HDL-C (mM) | 1.30±0.36 | 1.16±0.27 | 0.083 |
| LDC-C (mM) | 2.60±0.73 | 2.66±1.23 | 0.785 |
| TBIL (mM) | 13.19±18.40 | 12.04±4.89 | 0.722 |
| eGFR (ml/min) | 115.5±31.79 | 103.9±31.52 | 0.377 |
| LVEF (%) | 64.29±3.47 | 62.65±5.85 | 0.483 |
Table 2: Clinical Characteristics of the Patients
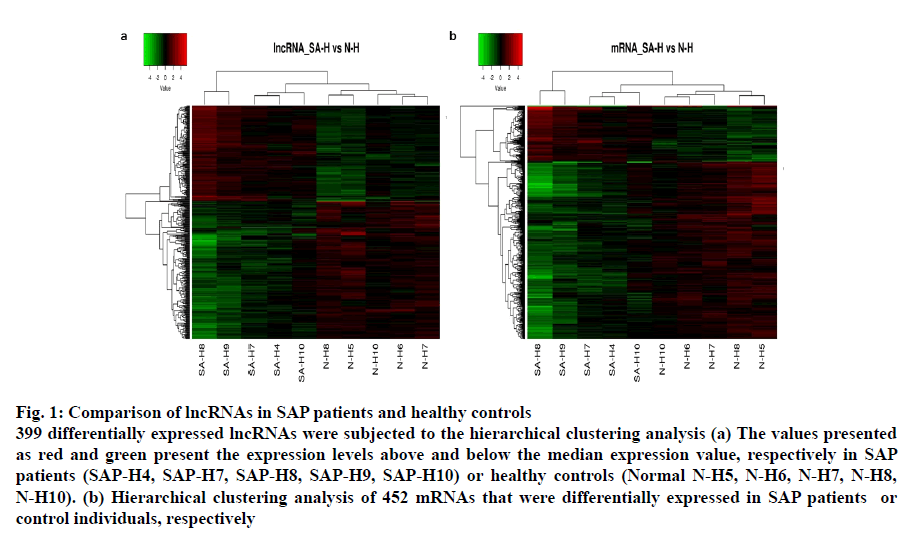
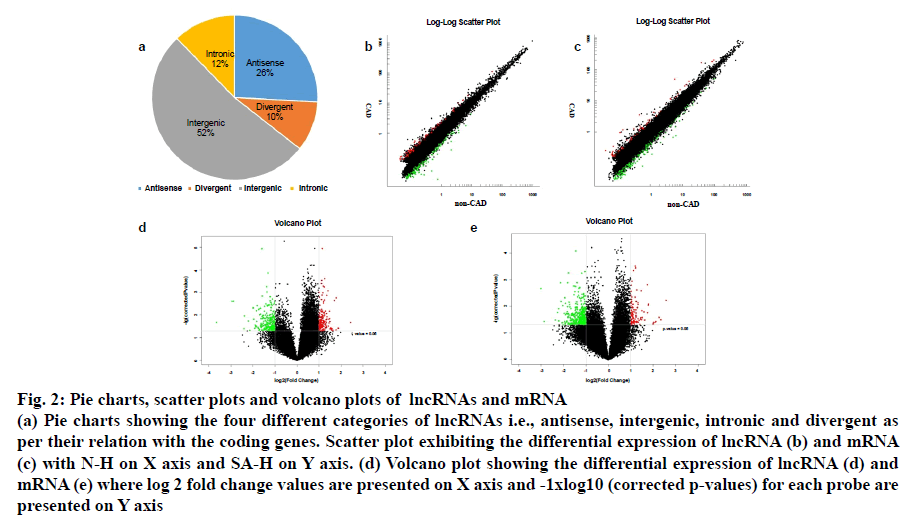
The lncRNA probes from the SAP patients’ and control samples (n=5) were analysed in a locus-by-locus fashion. The identification as well as the differential expression of 226 upregulated and 173 downregulated lncRNA was performed on the basis of fold change (FC) and corrected p-value (p<0.05; FC>2.0). Table 3 presents the top 20 distinctively expressed lncRNAs as per the FC values in SAP patients. Owing to its simple design and wide acceptability, the hierarchical clustering technique was employed for analysis of lncRNA and gene expression data in order to generate a hypothesis about the relationship among samples[21]. Present results exhibited distinguishable lncRNA expression profiles that are presented in fig. 1a. The variation of lncRNA expression between the diseased and control groups are presented in the scatter and volcano plots where each dot corresponds to a lncRNA (fig. 2b-2d). The green, red and black dots exhibit the downregulated (FC value <–2, p<0.05), upregulated (FC value >2, p<0.05) and the remaining lncRNAs (–2< FC value <2, p>0.05), respectively.
Fig. 1: Comparison of lncRNAs in SAP patients and healthy controls
399 differentially expressed lncRNAs were subjected to the hierarchical clustering analysis (a) The values presented as red and green present the expression levels above and below the median expression value, respectively in SAP
patients (SAP-H4, SAP-H7, SAP-H8, SAP-H9, SAP-H10) or healthy controls (Normal N-H5, N-H6, N-H7, N-H8,
N-H10). (b) Hierarchical clustering analysis of 452 mRNAs that were differentially expressed in SAP patients or
control individuals, respectively
Fig. 2: Pie charts, scatter plots and volcano plots of lncRNAs and mRNA
(a) Pie charts showing the four different categories of lncRNAs i.e., antisense, intergenic, intronic and divergent as per their relation with the coding genes. Scatter plot exhibiting the differential expression of lncRNA (b) and mRNA (c) with N-H on X axis and SA-H on Y axis. (d) Volcano plot showing the differential expression of lncRNA (d) and
mRNA (e) where log 2 fold change values are presented on X axis and -1xlog10 (corrected p-values) for each probe are
presented on Y axis
| lncRNA ID | Probe name | FC value | Regulation | class | database |
|---|---|---|---|---|---|
| ENST00000435749.1 | p15383 | 12.56803 | down | Intergenic | ENSEMBL |
| ENST00000429408.1 | p15381 | 7.353047 | down | Antisense | ENSEMBL |
| ENST00000448195.1 | p15382 | 5.201858 | down | Divergent | ENSEMBL |
| TCONS_00018817 | p18209 | 4.632731 | down | Intergenic | HumanLincRNACatalog |
| uc001vjh.1 | p25856 | 4.416233 | down | Intronic | UCSC |
| ENST00000417559.1 | p1657 | 3.976769 | down | Intergenic | ENSEMBL |
| ENST00000584157.1 | p6780 | 3.914835 | down | Intronic | ENSEMBL |
| ENST00000601079.1 | p34870_v4 | 3.73959 | down | Intergenic | ENSEMBL |
| ENST00000600225.1 | p34873_v4 | 3.693014 | down | Intergenic | ENSEMBL |
| ENST00000414106.1 | p1794 | 3.665167 | up | Antisense | ENSEMBL |
| ENST00000610044.1 | p37042_v4 | 3.654405 | down | unknown | ENSEMBL |
| ENST00000524978.1 | p15203 | 3.596647 | down | Divergent | ENSEMBL |
| ENST00000565674.1 | p6539 | 3.559462 | down | Intergenic | ENSEMBL |
| ENST00000512007.1 | p13944 | 3.440334 | up | Intergenic | ENSEMBL |
| ENST00000455929.1 | p261 | 3.424183 | up | Intronic | ENSEMBL |
| ENST00000565633.1 | P6583 | 3.354947 | down | Divergent | ENSEMBL |
| ENSG00000263072.1 | P5841 | 3.34036 | down | Antisense | ENSEMBL |
| XR_426740.1 | p39378 | 3.279137 | down | unknown | ENSEMBL |
| ENST00000549616.1 | p3742 | 3.26393 | down | Antisense | ENSEMBL |
| uc.209- | p27676 | 3.259788 | down | Intronic | UCR |
Table 3: Top 20 of Distinctively Expressed Lncrnas According to the Fc Values in Sap Patients Compared with that in Healthy Controls
The distinctive lncRNAs were further analyzed as per their categorizations, despite the fact that a considerable proportion of lncRNAs (62.9 %) was not categorized. As it is evident from fig. 2a, the classified lncRNAs were dominated by the intergenic and antisense (52 and 26 % respectively) versus the divergent and intronic (10 and 12 % respectively).
A total of 452 out of the 34 000 total detected mRNA probes were found statistically significant and were differentially expressed as presented in fig. 1b. 106 probes out of the mentioned 452 were up-regulated whereas 346 were down-regulated. As per the FC values in diseased individuals, the top 20 of distinctively expressed mRNAs are presented in Table 4.
| Probe name | P value | FC value | Regulation | Gene Symbol | Ensembl ID |
|---|---|---|---|---|---|
| A_24_P102053 | 0.031056 | 23.32903 | down | OCLN | ENST00000617792 |
| A_23_P92672 | 0.027624 | 23.27681 | down | OCLN | ENST00000622858 |
| A_23_P143526 | 0.002113 | 8.010953 | down | S100B | ENST00000291700 |
| A_33_P3249364 | 0.036857 | 7.317906 | down | TMTC1 | ENST00000553189 |
| A_24_P289709 | 0.005901 | 5.97426 | up | unknown | ENST00000623781 |
| A_33_P3360412 | 0.032299 | 5.141089 | down | PIGQ | ENST00000409439 |
| A_32_P385587 | 0.031513 | 5.080685 | up | ALAS2 | ENST00000330807 |
| A_23_P22800 | 0.025994 | 4.812657 | up | TSPY3 | ENST00000623101 |
| A_24_P339126 | 0.01698 | 4.788326 | down | PRSS21 | ENST00000575199 |
| A_33_P3260373 | 0.012049 | 4.647984 | down | ISY1-RAB43 | ENST00000561790 |
| A_21_P0000664 | 0.034943 | 4.632324 | down | CD4 | ENST00000544344 |
| A_23_P385500 | 0.03479 | 4.631608 | down | RNF166 | ENST00000541206 |
| A_23_P61551 | 0.001302 | 4.410628 | down | CD2BP2 | ENST00000569466 |
| A_23_P135486 | 0.036884 | 4.334105 | up | AHSP | ENST00000302312 |
| A_33_P3292531 | 0.006511 | 4.333651 | down | TMEM201 | ENST00000508400 |
| A_23_P254944 | 0.038887 | 4.277977 | up | GSTT1 | ENST00000621513 |
| A_23_P392076 | 0.041157 | 4.210579 | down | STRN3 | ENST00000555358 |
| A_33_P3316403 | 0.040835 | 4.11949 | down | MAN2A2 | ENST00000560534 |
| A_33_P3412613 | 0.00949 | 4.105124 | down | OCLN | ENST00000393053 |
| A_33_P3355954 | 0.040041 | 4.061537 | up | OCLN | unknown |
Table 4: Top 20 of Distinctively Expressed Mrnas According to the Fc Values in Sap Patients Compared with that in Healthy Controls
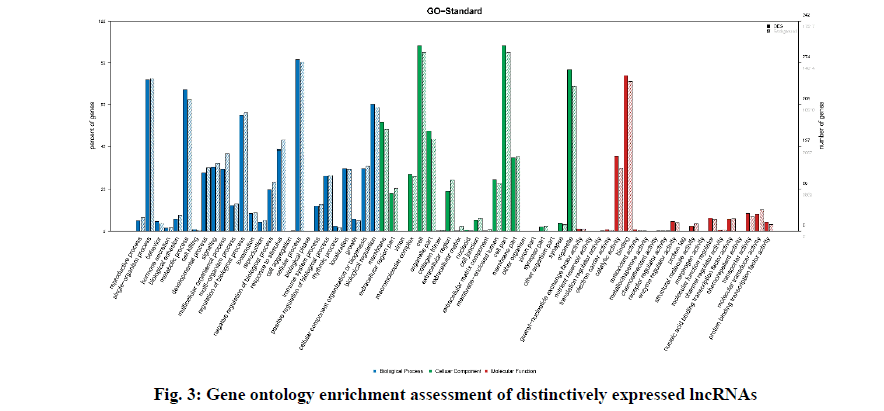
Fig. 3 describes the gene ontology (GO) terms that were associated with different biological processes in SAP patients. Fig. 3c-3e exhibits clear segregation of mRNAs between the control group and SAP patients as evident from the scatter and volcano plots where every dot corresponds to a differentially expressed mRNA. The green, red and black dots in the plot indicate the downregulated (FC value <–2, p<0.05), upregulated (FC value >2, p<0.05) and the remaining mRNAs (–2<FC value <2, p>0.05), respectively. The results exhibited a significant difference in mRNA expression between the diseased and healthy subjects.
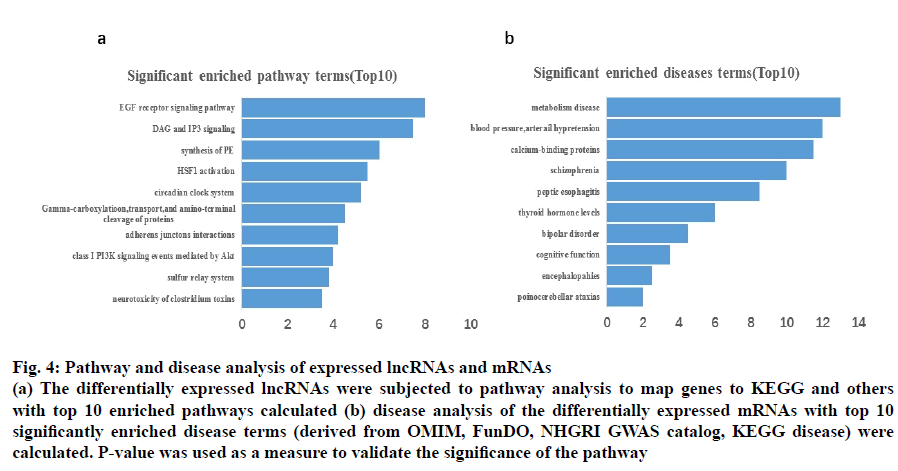
Six different databases including the Kyoto Encyclopaedia of Genes and Genomes pathway analysis KEGG, Biocarta, BioCyc, PANTHER, PID and Reactome were used to acquire 30 significantly enriched pathways. Fig. 4b shows the top 10 significant enriched disease terms that include, metabolic disease, arterial hypertension, calcium-binding proteins, schizophrenia, peptic esophagitis, thyroid hormone levels, bipolar disorder, cognitive function, encephalopathies and pontocerebellar ataxias.
Fig. 4: Pathway and disease analysis of expressed lncRNAs and mRNAs
(a) The differentially expressed lncRNAs were subjected to pathway analysis to map genes to KEGG and others
with top 10 enriched pathways calculated (b) disease analysis of the differentially expressed mRNAs with top 10
significantly enriched disease terms (derived from OMIM, FunDO, NHGRI GWAS catalog, KEGG disease) were
calculated. P-value was used as a measure to validate the significance of the pathway
The suspected genetic modifications that correspond to the aberrantly expressed lncRNA were further evaluated by the microarray analysis for functional linking studies. The results suggested that the top 10 lncRNAs were related to EGF receptor signalling pathway, DAG and IP3 signalling, synthesis of PE, HSF1 activation, circadian clock system, gammacarboxylation, transport, and amino-terminal cleavage of proteins, Adherens junction interactions, class I PI3K signaling events mediated by Akt, sulfur relay system, and neurotoxicity of clostridium toxins (fig. 4a).
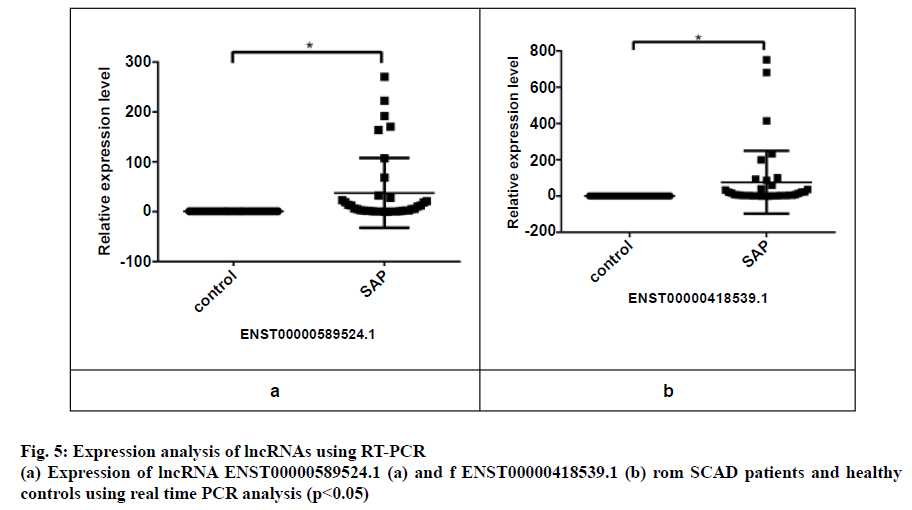
Three peripheral blood samples from each of the 38 SAP patients and healthy controls were randomly selected to evaluate lncRNA expression by using qRT-PCR. The PCR primers employed to detect the gene expression are presented in Table 1. The details of the 2 upregulated lncRNAs are presented in Table 5, where the comparison of the lncRNAs expression in patients and healthy controls is shown in fig. 5. It is evident from the results that the expression of lncRNA ENST00000418539.1 and lncRNA ENST00000589524.1 was significantly higher in the SAP patients when compared with the healthy controls (p<0.05). These findings were in close agreement with the microarray analysis.
| Gene name | p-value | Fold change | Regulation | Probe | Chromosome | Start | End | Class |
|---|---|---|---|---|---|---|---|---|
| ENST00000418539.1 | 0.030583 | 1.836234 | up | P9782 | 2 | 47558198 | 47571656 | Intergenic |
| ENST00000589524.1 | 0.008033 | 1.895215 | up | P8706 | 19 | 36440385 | 36442421 | Intergenic |
Table 5: The Randomly Selected Lncrnas
Congestive heart disease (CHD) in one of the most common cardiovascular ailments with a significantly higher mortality rate worldwide[22]. Among CHD, a substantial proportion of patients comes from SAP, the incidence of death and myocardial infarction was 2.3/100 patient-years[23-25], making an early diagnosis of SAP remained dismal. Identification of novel biological information has been recognized as better stratification and management of patients with angina pectoris syndrome. Recently, many authoritative lncRNAs not only are implicated in the expression of genes at epigenetics, transcriptional, and post-transcriptional levels but also are involved in the pathophysiology of cardiovascular diseases, including heart failure, cardiac hypertrophy, cardio-metabolic diseases, myocardial infarction[26-30].
The correlation between CAD and the expression pattern of lncRNAs is well-established with a number of studies published on the subject. Cai et al. had reported 51 up and 35 down-regulated lncRNAs with a total of 86 probes in CAD patients. They also reported 50 up and 60 down-regulated mRNAs in both PBMCs and plasma[31]. Accordingly, Ounzain et al. had reported the correlation of the lncRNA expression profile with myocardial infarction[32]. Owing to the rapidly emerging importance of lncRNA expression in the mitigation of cardiovascular ailments, lncRNA and mRNA genome-wide expression profile in 5 SAP patients and 5 healthy volunteers via microarray were examined. The differential expression of a total of 226 up and 173 downregulated lncRNA probes was verified in SAP patients with respect to the healthy controls. Likewise, 106 mRNAs were found upregulated with 346 down-regulated in SAP patients.
Furthermore, the findings have predicted the potential signalling pathways that could be related to the dysregulation of lncRNA with their co-expressed mRNAs in SAP patients. These pathways include nuclear factor kappa B (NFκB), mitogen-activated protein kinase (MAPK), p53 as well as the Bcl/caspase apoptotic pathways. Based upon the gene ontology and KEGG analysis, the likelihood of a coordinated pattern between lncRNA/mRNA expression and SAP pathology could be claimed. However, further investigations are required to ascertain this claim.
Recently, several studies have shown that some lncRNAs are involved in the development of various types of cardiovascular disease. The expression of lncRNA ANRIL is correlated with atherosclerosis severity which may be important in its pathogenesis[33]. Similarly, MIAT (myocardial infarction associated transcript/ Gomafu) and LIPCAR (lnc RNA predicting cardiac remodelling) which are associated with ventricular mass and output ratio have been reported as a predictor of left ventricular remodelling following acute myocardial infarction[34]. Urothelial cancer associated 1 (UCA1) is another lncRNA whose level is found associated with acute myocardial infarction. The reports have revealed a fall in UCAS1 levels in the early stages of infarction with a persistent increase from 3rd d onwards following an acute infarction attack[35]. The other infarction associated lncRNAs include ZFAS1 and CDR1-AS whose blood levels are correlated in a negative and positive manner respectively[36]. The findings of the present study stand in close commitment with the previous reports, concluding a positive correlation in the expression levels of lncRNA ENST00000418539.1 and ENST00000589524.1 with the prevalence of stable angina pectoris. The detection of these lncRNAs could be helpful in the early diagnosis of SAP patients and may provide in-depth information about the severity of the disease.
The sampling was limited to a single centre. It is wellknown that there are genetic differences in patients. Hence, our findings should be validated further by randomly choosing larger samples from a relatively heterogeneous population. Microarray technologies have been employed to study the expression profile of lncRNA in the peripheral blood samples of the SAP patients with subsequent comparison of these levels to the healthy controls. It was observed for the first time that the expression pattern of the lncRNA ENST00000418539.1 and ENST00000589524.1 are positively correlated with the prevalence and severity of stable angina pectoris in patients. These findings could be useful in future SAP management strategies.
Acknowledgments
This study was supported by grants from the Natural Science Foundation of China (ID 81660058) and Special Funds for the Key Laboratory of Xinjiang Autonomous Region in China (ID 2018D04029).
Conflict of interest
All authors report no conflicts of interest in this work.
References
- Williams B, Menon M, Satran D, Hayward D, Hodges JS, Burke MN, et al. Patients with coronary artery disease not amenable to traditional revascularization: prevalence and 3 y mortality. Cardiovasc Interv 2010;75:886-91.
- Mannheimer C, Camici P, Chester MR, Collins A, DeJongste M, Eliasson T, et al. The problem of chronic refractory angina; report from the ESC Joint Study Group on the Treatment of Refractory Angina. Eur Heart J 2002;23:355-70.
- Wilson SR, Scirica BM, Braunwald E, Murphy SA, Karwatowska-Prokopczuk E, Buros JL, et al. Efficacy of ranolazine in patients with chronic angina observations from the randomized, double-blind, placebo-controlled MERLIN-TIMI (Metabolic Efficiency With Ranolazine for Less Ischemia in Non-ST-Segment Elevation Acute Coronary Syndromes) 36 Trial. J Am Coll Cardiol 2009;53:1510-6.
- Tardif JC, Ponikowski P, Kahan T. Efficacy of the I(f) current inhibitor ivabradine in patients with chronic stable angina receiving beta-blocker therapy: a 4 mon, randomized, placebo-controlled trial. Eur Heart J 2009;30:540-8.
- Zipes DP, Svorkdal N, Berman D, Boortz‐Marx R, Henry T, Lerman A, et al. Spinal cord stimulation therapy for patients with refractory angina who are not candidates for revascularization. Neuromodulation 2012;15:550-9.
- Burkhoff D, Schmidt S, Schulman SP, Myers J, Resar J, Becker LC, et al. Transmyocardial laser revascularisation compared with continued medical therapy for treatment of refractory angina pectoris: a prospective randomised trial. ATLANTIC Investigators. Angina Treatments-Lasers and Normal Therapies in Comparison. Lancet 1999;354:885-90.
- Ruth McPherson MD. Chromosome 9p21 and Coronary Artery Disease. N Engl J Med 2010;362:1736-7.
- Ardissino D, Berzuini C, Merlini PA, Mannucci PM, Surti A, Burtt N, et al. Influence of 9p21.3 genetic variants on clinical and angiographic outcomes in early-onset myocardial infarction. J Am Coll Cardiol 2011;58:426-34.
- Ma Y, Ma W, Huang L, Feng D, Cai B. Long non-coding RNAs, a new important regulator of cardiovascular physiology and pathology. Int J Cardiol 2015;188:105-10.
- Busch A, Eken SM, Maegdefessel L. Prospective and therapeutic screening value of non-coding RNA as biomarkers in cardiovascular disease. Ann Transl Med 2016;4: 236.
- Arita T, Ichikawa D, Konishi H, Komatsu S, Shiozaki A, Shoda K, et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res 2013;33:3185-93.
- Reis EM, Verjovski-Almeida S. Verjovski-Almeida, Perspectives of Long Non-Coding RNAs in Cancer Diagnostics. Front Genet 2012;3:32.
- Kumarswamy R, Bauters C, Volkmann I, Maury F, Fetisch J, Holzmann A, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 2014;114:1569-75.
- Montalescot G, Sechtem U, Achenbach S, Andreotti F, Arden C, Budaj A, et al. Comments on the 2013 ESC Guidelines on the Management of Stable Coronary Artery Disease. Revista Espa de Cardiologia 2014;67:80-6.
- Sun J, Wu J. Expression profiling of long noncoding RNAs in neonatal and adult mouse testis. Data in Brief 2015;4:322-27.
- Gao W, Hillwig ML, Huang L, Cui G, Wang X, Kong J, et al. A functional genomics approach to tanshinone biosynthesis provides stereo chemical insights. Org Lett 2009.11:5170-3.
- Li H, Li H, Yue H, Wang W, Yu L, Cao Y, et al. Comparison between smaller ruptured intracranial aneurysm and larger un-ruptured intracranial aneurysm: gene expression profile analysis. Neurosurg Rev 2017;40:419-25.
- Zhai H, Li XM, Liu F, Chen BD, Zheng H, Wang XM, et al. Expression pattern of genome-scale long noncoding RNA following acute myocardial infarction in Chinese Uyghur patients. Oncotarget 2017;8:31449-64.
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010;143:46-58.
- Mathiyalagan P, Keating ST, Du XJ, El-Osta A. Interplay of chromatin modifications and non-coding RNAs in the heart. Epigenetics 2014;9:101-12.
- Wang H, Zheng H, Azuaje F. Poisson-based self-organizing feature maps and hierarchical clustering for serial analysis of gene expression data. IEEE/ACM Trans Comput Biol Bioinform 2007;4:163-75.
- Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, et al. Heart disease and stroke statistics--2015 update: a report from the American Heart Association. Circulation 2015;131:e322-9.
- Gibbons RJ, Abrams J, Chatterjee K, Daley J, Deedwania PC, Douglas JS, et al. ACC/AHA 2002 guideline update for the management of patients with chronic stable angina--summary article: a report of the American College of Cardiology/American Heart Association Task Force on practice guidelines (Committee on the Management of Patients With Chronic Stable Angina). J Am Coll Cardiol 2003;41:159-68.
- Rapsomaniki E, Shah A, Perel P, Denaxas S, George J, Nicholas O, et al. Prognostic models for stable coronary artery disease based on electronic health record cohort of 102 023 patients. Eur Heart J 2014;35:844-52.
- Thom TA. Report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation 2006;113:e85-e151.
- Lindsay MA, Griffiths-Jones S, Young RS, Ponting CP. Identification and function of long non-coding RNAs. Essays Biochem 2013;54:113-26.
- Han P, Li W, Lin CH, Yang J, Shang C, Nurnberg ST, et al. A long noncoding RNA protects the heart from pathological hypertrophy. Nature 2014;514:102-6.
- Wang K, Liu F, Zhou LY, Long B, Yuan SM, Wang Y, et al. The long noncoding RNA CHRF regulates cardiac hypertrophy by targeting miR-489. Circ Res 2014;114:1377-88.
- Liu Y, Ferguson JF, Xue C, Ballantyne RL, Silverman IM, Gosai SJ, et al. Tissue-specific RNA-Seq in human evoked inflammation identifies blood and adipose LincRNA signatures of cardiometabolic diseases. Arterioscler Thromb Vasc Biol 2014;34:902-12.
- Ishii N, Ozaki K, Sato H, Mizuno H, Saito S, Takahashi A, et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 2006;51:1087-99.
- Cai Y, Yang Y, Chen X, Wu G, Zhang X, Liu Y, et al. Circulating 'lncRNA OTTHUMT00000387022' from monocytes as a novel biomarker for coronary artery disease. Cardiovasc Res 2016;112:714-724.
- Ounzain S, Micheletti R, Beckmann T, Schroen B, Alexanian M, Pezzuto I, et al. Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 2015;36:353-68a.
- Holdt LM, Hoffmann S, Sass K, Langenberger D, Scholz M, Krohn K, et al. Elements in ANRIL Non-Coding RNA at Chromosome 9p21 Modulate Atherogenic Cell Functions through Trans-Regulation of Gene Networks. PLOS Genetics 2013;9:e1003588.
- de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, Van Der Meer RW, Rijzewijk LJ, et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep 2016;6:37354.
- Yan Y, Zhang B, Liu N, Qi C, Xiao Y, Tian X, et al. Circulating Long Noncoding RNA UCA1 as a Novel Biomarker of Acute Myocardial Infarction. Biomed Res Int 2016;2016:7.
- Zhang Y, Sun L, Xuan L, Pan Z, Li K, Liu S, et al. Reciprocal Changes of Circulating Long Non-Coding RNAs ZFAS1 and CDR1AS Predict Acute Myocardial Infarction. Sci Rep 2016;6:22384.