- *Corresponding Author:
- Anudeep Balla
Department of pharmaceutics
Acharya & BM Reddy College of Pharmacy
Bengaluru, India
E-mail: anudeepballa@gmail.com
| Date of Received | 23 December 2019 |
| Date of Revision | 29 May 2020 |
| Date of Acceptance | 24 June 2020 |
| Indian J Pharm Sci 2020;82(4):622-631 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The current research was aimed at formulating brain targeting polymeric nanoparticles of a hydrophilic anti-parkinson agent (Ropinirole HCl) to improve the drug passage to brain. Two different approaches were used for the study. One was Chitosan nanoparticles prepared by ionic gelation method using tripolyphosphate as cross-linking agent and the other one was poly (lactic-co-glycolic acid)50:50 monomer ratio poly D,L-lactic-co-glycolic acid nanoparticles prepared by a modified two step nanoprecipitation method using polyvinyl alcohol as stabilizer. Polysorbate-80 was used to coat the nanoparticles to further improve its passage through brain. Minitab 17 statistical software was employed to create general full factorial design. Drug-excipient compatibility study by Fourier transform infrared spectroscopy and differential scanning calorimetry revealed no possible interactions. Kinetic modelling of in vitro release showed that poly D,L-lactic-co-glycolic acid nanoparticles were more linear towards Korsemeyer- Peppasmodel indicating drug release from the nanoparticles was by a combination of bulk degradation followed by diffusion (Fickian diffusion). The scanning electron microscopy studies showed that poly D,L-lactic-co-glycolic acid nanoparticles were found to be spherical with either smooth or rough at surface. Stability studies performed at refrigerator conditions (3-5±2º) showed no significant changes upon storage. Results of in vivo blood brain barrier crossing study showed that when compared with pure drug, the formulated nanoparticles carried the drug to brain effectively. Owing to the lower particle size and pharmacy and drug information, poly D,L-lactic-co-glycolic acid nanoparticles were able to pass through blood brain barrier effectively than chitosan nanoparticles. Also, the ability of poly D,L-lactic-co-glycolic acid to release drug in a sustained manner for weeks makes it a promising drug delivery system.
Keywords
Anti-parkinson, nanoparticles, Ropinirole HCl
Parkinson’s disease (PD) is one of the most common and severely debilitating neurodegenerative disorders. It is characterized by a progressive loss of dopamine neurons in the substantia nigra pars compacta of the brain and this results in inability to store and regulate the release of dopamine. The cause for neuron depletion remains largely unknow. Accordingly, striatal dopamine receptor activation becomes increasingly dependent on the peripheral availability of an exogenously administered dopaminergic agent. As the disease progresses, the patient begins to experience motor abnormalities such as akinesia, resting tremor, gait and balance problems. The advancement of the disease results in worsening of these symptoms [1].
Dopamine agonists can be used in the early stages of Parkinson’s disease to reduce symptoms as it can delay the need for levodopa, thus postponing the drug resistance and motor fluctuations associated with long term dopamine therapy. Dopamine agonists along with levodopa can also be given in the later stages when levodopa alone cannot control the symptoms. Ropinirole HCl (RH) is the selected dopamine agonist for the current study and is highly hydrophilic.
The brain is probably one of the least accessible organs for the delivery of drugs due to the presence of the blood-brain barrier (BBB) that controls the transport of endogenous and exogenous compounds, thus providing the neuroprotective function. The structural BBB is formed by the cerebral capillary endothelial cells that, in contrast to endothelial cells in capillary blood vessels in most other tissues, are closely joined to each other by tight junctions produced by the interaction of several transmembrane proteins. Moreover, these endothelial cells demonstrate very little fenestration and display only low pinocytic activity [2].
This physical barrier effectively abolishes any aqueous paracellular diffusional pathways between the extracellular fluid in the blood and brain. This diffusion is dependent on lipophilicity and molecular weight. Lipophilic drugs can diffuse across the BBB by direct permeation through the cell membrane if their molecular weight is not more than 500Da [3]. However, many of these lipophilic molecules will be actively removed from the cerebral compartment by the adenosine triphosphate binding cassette efflux transporters, such as P-glycoprotein (P-gp) or multidrug resistance proteins. Thus, many potential drugs with activity at a particular site or receptor in the brain have failed in the treatment of central nervous system (CNS) disorders. These drugs simply do not enter the CNS in sufficient quantities to be effective, which, consequently, diminishes their therapeutic value [4].
To bypass the BBB and to deliver therapeutics into the brain two different approaches are currently used – invasive & Noninvasive.
Delivering drugs to the CNS is impaired by the presence of the BBB that represents the main obstacle for CNS drug development, many hydrophilic drugs and neuropeptides etc. may have difficulty in crossing the BBB [5]. Especially coating of the nanoparticles with the polysorbate (Tween) surfactants resulted in transport of drugs across the blood brain barrier. The mechanism for transport was suggested to be endocytosis via the low density lipoprotein receptor of the endothelial cells after adsorption of lipoproteins form blood plasma to the nanoparticles. Investigations revealed the role of apolipoprotein-E for transport of drugs across the BBB. It is suggested that the recognition and interaction with lipoprotein receptors on brain capillary endothelial cells is responsible for the brain uptake of the drug. Passage of the BBB may also be achieved by masking certain drug characteristics preventing or limiting binding to cellular efflux systems like p-glycoprotein, a cellular transporter associated with drug removal from cells. P-glycoprotein is one of the ATP dependent efflux transporters that has an important physiological role in limiting drug entry into the brain [6].
The upper limit of pore size in the BBB that enables passive flow of molecules across it is usually <1 nm; however, particles that have a diameter of several nanometers can also cross the BBB by carrier-mediated transport. Although very small nanoparticles (NPs) may just pass through the BBB, this uncontrolled passage into the brain may not be desirable and strategies are being developed for controlled passage as well as targeted drug delivery to the brain [7].
PLGA is a US FDA-approved copolymer that is used in a host of therapeutic devices, owing to its biodegradability and biocompatibility. PLGA is synthesized by the random ring-opening copolymerization of two different monomers. PLGA NPs deliver molecules considered too large and complex to be transported by known vectors. PLGA is nontoxic, does not illicit an immune response, causes comprehensive transfection, crosses the BBB and supports sustained drug release.
Polymer based nanoparticles are made from natural & biodegradable polymers such as Chitosan, Poly (D,L-Lactide- co Glycolide) (PLGA), polylactic acid (PLA), and polycyanoacrylate (PCA) etc. The mechanism for the transport across the BBB has been characterized as receptor-mediated endocytosis by the brain capillary endothelial cells. Transcytosis then occurs to transport the nanoparticles across the tight junction of endothelial cells and into the brain.
So, the current research is focused at developing a NPDDS of antiparkinson agent that can effectively carry the drug through BBB and target the brain providing sustained drug release with minimal side effects.
Materials and Methods
Materials:
Ropinirole Hydrochloride was purchased from Yarrow Chem Pvt. Ltd, Mumbai. PLGA was purchased from Lactel – Durect Corporation, USA. Chitosan was purchased from Sigma Aldrich, USA. PVA and Sodium Tripolyphosphate(TPP) was purchased from Central Drug House (p) ltd, New Delhi. HPLC grade Acetonitrile, Water, Methanol were purchased from S.D FineChem. HPLC grade Potassium Dihydrogen Phosphate and Orthophosphoric acid were purchased from Finar Chemicals, Ahmedabad. All other chemicalsused areof analytical grade.
Drug-excipient compatibility studies by Fourier transform infrared & Differential scanning calorimetry (FTIR &DSC) [8]:
Compatibility studies were carried out to find out any possible interactions between drug and excipients used in the formulations. FTIR was used to quantify the interaction between the drug and the carrier used in formulation. Spectra were recorded for pure drug and for drug and polymers (1:1) physical mixture, on Bruker tensor-27 spectrophotometer. The spectra were then compared with reference spectrum of drug. DSC is a technique in which determines the physical nature of a material by recording its endotherm or exotherm. By comparing the DSC curves of a pure drug sample with that of formulation determines the stability of physical form of a drug and any possible interaction with the excipients.
Preparation of chitosan nanoparticles (Ionic gelation method) [9,10]:
This method involves an ionic interaction between the positively charged amino groups of chitosan and the polyanion TPP, which acts as chitosan crosslinker. Chitosan solution (0.75%) was prepared using 2% Glacial Acetic Acid. 0.25% TPP solution was prepared in distilled water. Calculated amount of drug was dissolved in chitosan solution and added dropwise to TPP solution under continuous stirring. Nanoparticles were formed by ionic interaction and the solution becomes turbid after complete formation of nanoparticles. The ratio of Chitosan:TPP was varied in various formulations to optimize the best formulation. NPs were collected by centrifugation at 15,000 rpm for 45 min at 4º and the supernatant will be used to determine entrapment efficiency (EE) and loading capacity (LC). The resultant nanoparticles were then resuspended in water. The formulation chart was presented in Table 1.
| Formulation | Ratio | |
|---|---|---|
| Chitosan (0.75 %) |
Sodium Tripolyphosphate (0.25%) |
|
| CF1 | 1 | 1 |
| CF2 | 1 | 1.5 |
| CF3 | 1 | 2 |
| CF4 | 1.5 | 1 |
| CF5 | 1.5 | 1.5 |
| CF6 | 1.5 | 2 |
| CF7 | 2 | 1 |
| CF8 | 2 | 1.5 |
| CF9 | 2 | 2 |
Table 1: Formulation Chart for Chitosan NPs (CF1 – CF9)
Formulation of PLGA-PVA NPs by Modified nanoprecipitation method [11]:
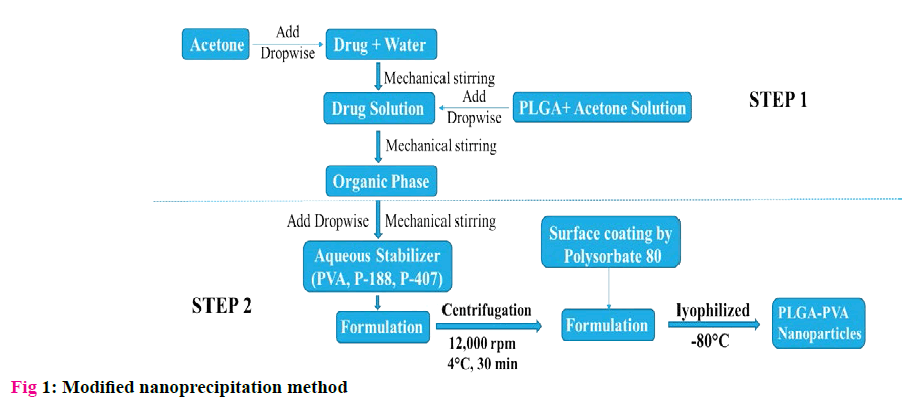
Nanoprecipitation had been proved to be an efficient technique in the preparation of PLGA NPs when compared to solvent evaporation or other techniques. However, it is mostly limited to hydrophobic drugs. But the drug used in the study is hydrophilic to the core with solubility in water (60 mg/ml), Ethanol (<1 mg/ ml) and practically insoluble in acetone. So, a modified two step Nanoprecipitation technique designed especially for hydrophilic drugs was used in preparing the nanoparticles. The method was represented in fig 1.
Six formulations (PF1 to PF6) were performed by using DOE of Minitab 17 with two levels of PLGA (20 mg/ ml, 50 mg/ml) and three levels of PVA (0.25%, 0.5%, 1%). The ratio of org phase: aqueous phase was set at 1:40. The drug concentration was kept constant in all formulations at 25 mg/ml. Table 2 represents the formulation chart.
| Formulation | PLGA(mg/ml) (Aqueous phase) |
PVA (%) (Organic phase) |
Ratio (Org:Aqueous) |
|---|---|---|---|
| PF1 | 50 | 0.5 | 1:40 |
| PF2 | 50 | 1 | 1:40 |
| PF3 | 20 | 1 | 1:40 |
| PF4 | 20 | 0.5 | 1:40 |
| PF5 | 50 | 0.25 | 1:40 |
| PF6 | 20 | 0.25 | 1:40 |
Table 2: Formulation Chart for PLGA-PVA NPs (PF1 – PF6)
Surface coating of PLGA nanoparticles [12]:
Polysorbate-80 coating was done to facilitate the transport of formulated nanoparticles through BBB to brain. The prepared nanoparticles were re-suspending nanoparticles in phosphate buffered saline under constant stirring. Relative to total suspension volume, polysorbate 80 was then added to give a final solution of 0.75% (m/v) polysorbate 80.
Characterization of nanoparticles:
%Entrapment efficiency [13]:
%Entrapment efficiency & loading capacity were determined by indirect estimation. RH-loaded nanoparticles were centrifuged at 15,000 rpm and 4º for 30 min using REMI Ultra Centrifuge. The nonentrapped drug (free drug) was determined in the supernatant solution using UV spectrophotometer or HPLC (Shimadzu LC-20AT) at 249 nm. The peak area was determined and amount of free drug is determined by extrapolating the calibration curve.
Particle size determination:
The particle size of the PLGA nanoparticles was determined by Malvern Zeta sizer ZS90. It performs size measurements using a process called dynamic light scattering (also known as PCS - photon correlation spectroscopy) measures brownian motion and relates this to the size of the particles.
Zeta potential measurement:
A potential exists between the particle surface and the dispersing liquid which varies according to the distance from the particle surface – this potential at the slipping plane is called the zeta potential. The Zetasizer Nano series measures zeta potential using a combination of the measurement techniques: electrophoresis and laser doppler velocimetry, sometimes called laser doppler electrophoresis.
In vitro diffusion studies [14]:
The in vitro release profile of RH NPs was performed using dialysis sacs. The drug loaded nanoparticulate formulation (containing about 5 mg of drug) was placed in pretreated dialysis sacs which were immersed into 100 ml of PBS, pH 7.4, at 37±0.5º and magnetically stirred at 50 rpm. Aliquots were withdrawn from the release medium at intervals 0.5, 1, 1.5, 2, 3, 4, 5, 6, 7, 8, 9, 10, 12 and 24 h and replaced with the same amount of phosphate buffer. The samples were analyzed at 249 nm.
Kinetic modeling of in vitro drug diffusion profiles [15,16]
The dissolution profiles of all formulations were fitted to zero order, first order, Higuchi and Korsmeyer– Peppas models to ascertain the kinetic modeling of the drug release. The methods were adopted for deciding the most appropriate model.
Scanning electron microscopy study [17]:
The surface morphology of nanoparticles was measured by scanning electron microscopy (SEM) (EM-LEO 435VP, Carl Zeiss SMT Inc., NY) equipped with 15 kv, SE detector with a collector bias of 300 V. The lyophilized sample was carefully mounted on an aluminum stub using a double stick carbon tape. Samplewas then introduced into an automated sputter coated with a very thin film of gold before scanning the sample under SEM.
In vivo blood brain barrier crossing study [18,19]:
Healthy adult Wistar rats weighing 180-220 g were used as animal model. The rats were randomly divided into different groups. Group 1 served as the control, Group 2 was injected with drug solution, Group 3 was intravenously injected with chitosan nanoparticles and Group 4 was intravenously injected with PLGA nanoparticles in tail vein. After time intervals of 0.5, 2, 4 and 8 h, 12 h they were sacrificed by decapitation. Brain was quickly dissected and stored at -20°. Internal standard is externally spiked to it before homogenization. The homogenate is centrifuged at 8000 rpm, 4° for 30 min (Methanol is added to precipitate the proteins and) and clear supernatant is collected for HPLC analysis. By estimating the amount of drug present in brain, the ability of formulated nanoparticles to pass BBB and target brain was estimated.
Results and Discussion
FTIR study showed that all the characteristic peaks of drug were present in the spectra of physical mixture of between drug and excipients thus indicating there was no interaction between them. Results of the compatibility studies are shown in the Table 3.
| Peak No | FREQUENCY(cm-1) | ||||
|---|---|---|---|---|---|
| Ropinirole HCl |
Drug+ Chitosan+ TPP | Drug+ PLGA | Standard frequency range |
Description | |
| 1 | 3415 | 3466 | 3436 | 3500-3100 | N-H Stretch (2°Amine) |
| 2 | 3067 | 3072 | 3034 | 3100-3000 | C=C stretch |
| 3 | 2974 2886 |
2939 2887 |
2918 | 3000-2850 | Alkyl CH Stretch |
| 4 | 1710 | 1711 | 1690 | 1755-1650 | C=O (Ketone) |
| 5 | 1603 | 1606 | 1558 | 1700-1500 | Aromatic C=C bending |
| 6 | 1240 | 1229 | 1293 | 1340-1020 | C-N Stretch |
Table 3: Drug-Excipient Compatibility Study by FTIR-PEAK Picking
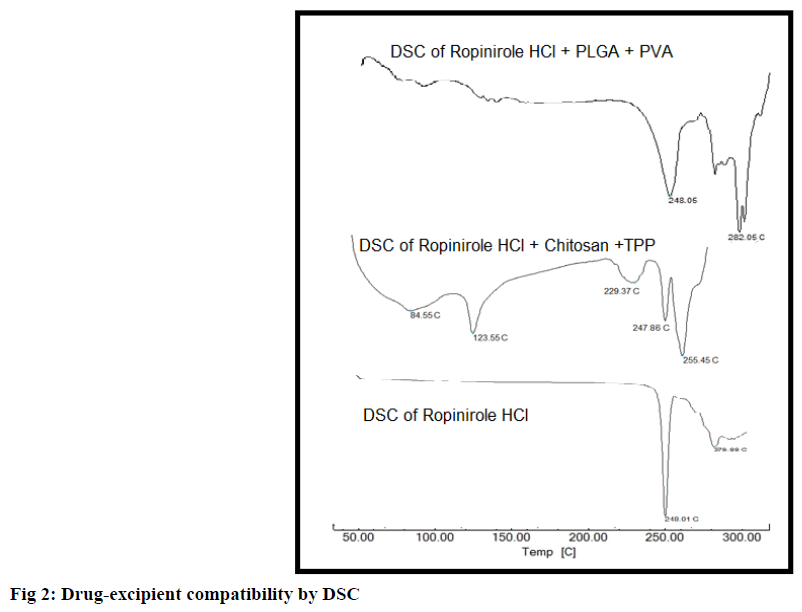
DSC analysis of RH-chitosan physical mixture revealed no possible interaction between them with drug endotherm reported at 247.86º. DSC analysis of RHPLGA physical mixture revealed no possible interaction between them with drug endotherm reported at 248.05º. Results are shown in fig 2. Standard melting point of drug is 243-250º.
From the results, it was observed that Chitosan NPs with high TPP showed higher entrapment due to improved crosslinking resulting in tighter polymer matrix. Formulations with lower chitosan concentration resulted in low entrapment, but highest chitosan concentration didn’t offer best entrapment either. It may be because concentration of TPP was inadequate to crosslink the higher chitosan amounts. Ideal concentration of polymer coupled with good crosslinking helps in tighter polymer matrix and higher entrapment. CF6 with 1.5:2 ratio of Chitosan:STPP showed higher entrapment efficiency of 61.37±0.21%. The results are shown in Table 4.
| Formulation Code | % Encapsulation efficiency | Particle size (nm) | Polydispersity index | Zeta potential (mV) |
|---|---|---|---|---|
| CF1 | 31.53±1.25 | 305.50±13.08 | 0.174±0.004 | 21.61±0.45 |
| CF2 | 29.51±0.74 | 304.03±5.37 | 0.141±0.010 | 19.11±0.32 |
| CF3 | 36.22±0.26 | 290.73±21.60 | 0.171±0.007 | 16.11±1.47 |
| CF4 | 48.92±1.38 | 348.70±21.30 | 0.138±0.031 | 26.47±0.87 |
| CF5 | 52.77±0.87 | 321.73±19.71 | 0.172±0.019 | 23.22±0.96 |
| CF6 | 61.37±0.21 | 299.51±11.17 | 0.147±0.021 | 20.77±0.99 |
| CF7 | 37.41±0.09 | 549.63±29.55 | 0.283±0.043 | 29.13±1.13 |
| CF8 | 42.32±0.26 | 576.77±51.46 | 0.289±0.057 | 25.86±2.01 |
| CF9 | 51.35±2.67 | 522.30±29.19 | 0.271±0.037 | 23.31±0.88 |
All the values are Mean of 3 values ±SD
Table 4: Characterization of Chitosan Nanoparticles
From the results, it was observed that the amount of PLGA and PVA had a significant impact on %EE and loading capacity. An increase in %EE & loading capacity was observed with the increase in PLGA amount. Whereas, concentration of PVA showed an inverse relationship. NPs with high amount of PLGA and low amount of PVA showed better entrapment of 83.30% as seen in PF5 (50 mg/ml PLGA, 0.25% PVA). The volume of nanosuspension was kept constant. The increased %EE with increase in PLGA concentration was due to increase in viscosity of organic phase, which avoids diffusion of drug from organic to aqueous phase. The increased viscosity also increases particles size which accommodates more drug with in NPs. As the concentration of PVA increases, the particle size decreases which results in poor %EE [20]. The results are shown in Table 5.
| Formulation Code | Encapsulation efficiency (%) | Particle size (nm) | Polydispersity index | Zeta potential (mV) |
|---|---|---|---|---|
| PF1 | 75.35±1.79 | 308.37±23.02 | 0.152±0.023 | -0.61±0.13 |
| PF2 | 70.71±0.44 | 345.50±42.83 | 0.196±0.028 | -5.30±0.21 |
| PF3 | 67.47±1.04 | 264.67±11.40 | 0.115±0.035 | -6.00±0.42 |
| PF4 | 64.97±0.91 | 212.10±3.03 | 0.135±0.040 | -12.47±0.90 |
| PF5 | 83.30±0.61 | 351.97±20.94 | 0.140±0.043 | -7.25±0.75 |
| PF6 | 79.39±0.73 | 196.4±8.68 | 0.059±0.037 | -10.35±0.81 |
All the values are Mean of 3 values ±SD
Table 5: Characterization of PLGA Nanoparticles
The particle size of formulations was determined by Malvern Zetasizer -Nano ZS 90. The size of the chitosan NPs ranged between 275.5 to 576.7 nm. CF3 showed good particle size of 290.73±21.60 nm. The mean particle size increased with the increase in chitosan concentration [21]. Polydispersity index ranged from 0.141 to 0.289. All formulations were found to be moderate to less polydisperse (more homogenous) with values below 0.3. The results are shown in Table 4. The size of the particles ranged between 196.4nm to 351.97 nm. PF6 showed lowest particle size of 196.4±8.68nm. From the results, an increase particle size was observed with the increase in PLGA amount. Whereas, concentration of PVA showed an inverse relationship. Polydispersity index (PDI) ranged from 0.059 to 0.196. PDI values for all formulation were closer to 0 indicating less polydisperse nature. PF6 with a PDI value 0.059±0.037 was found to be less polydisperse. The results are shown in Table 5.
The increased particle size with increase in PLGA concentration was due to increase in viscosity of organic phase resulting in reduction of both net shear stress and dispersion of organic phase into the aqueous phase, resulting in the larger size of NPs. The final size particles depend on net shear stress available for the droplet breakdown [22]. As concentration of PVA increased, interfacial tension decreased, resulting in increased net shear stress and smaller NPs formation during the emulsification. Also, high concentration of PVA stabilized the formed NPs and prevented the coalescence resulting in smaller NPs [20]. At very high concentrations of PVA, size of NPs increased due to increment in viscosity of aqueous phase which results in reduction of net shear stress available for droplet breakdown [22].
Zeta potential for formulations was determined by Malvern Zetasizer -Nano ZS 90. Zeta potential values of Chitosan NP’s were ranging from 16.11 to 29.13 mV indicating moderate to good stability. It was observed that at constant TTP concentration, increase in chitosan concentration shifted the zeta potential value to positive side due to cationic nature of chitosan. Zeta potential decreased with increase in TPP at constant chitosan concentration due to negative charge of TPP. The results are shown in Table 4.
From the results, zeta potential was found to be negative ranging from -0.61 to -12.47 mV indicating poor to moderate stability. PF1 showed poor stability and PF4 showed moderate stability with zeta potential values of -0.61±0.13 mV and -12.47±0.90 mV respectively.
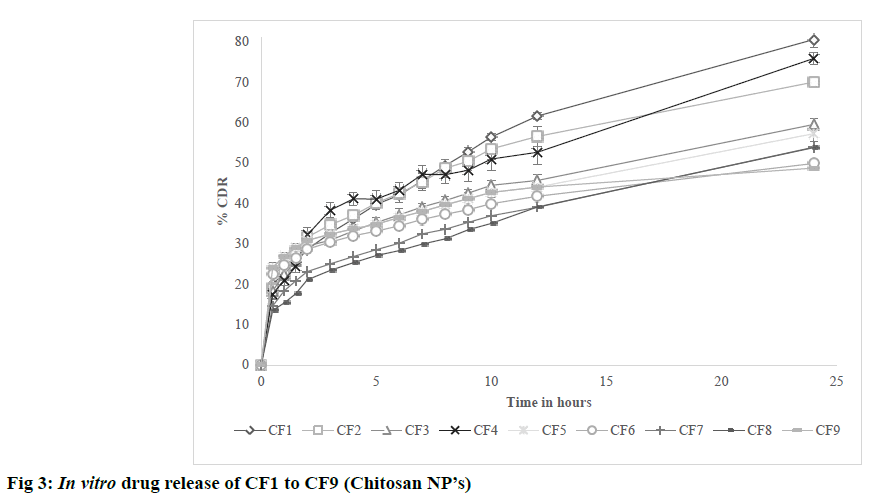
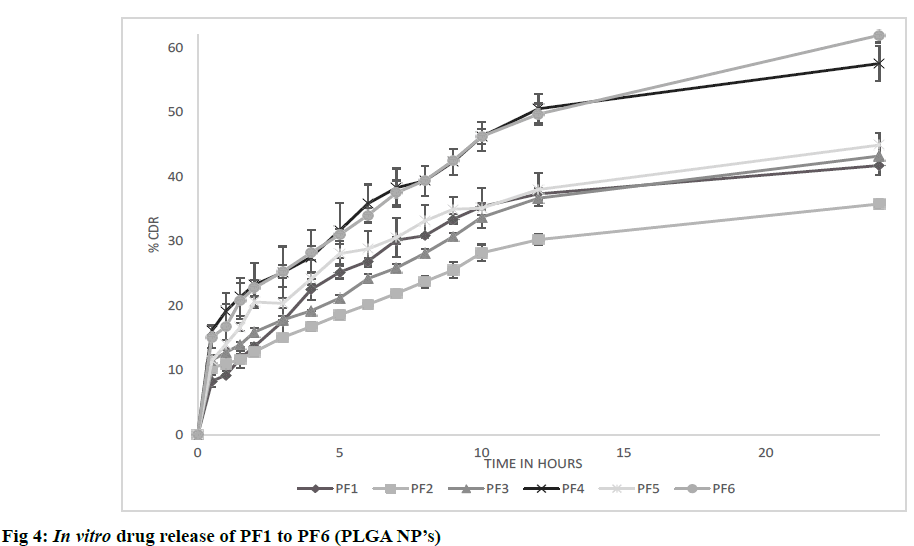
The possible reason for poor zeta potential values for PVA formulations may be due to presence of residual PVA on surface of nanoparticles which was found to act as shield between nanoparticles and its surrounding medium by masking the charged groups present on the surface. The results are shown in Table 5. In vitro diffusion was performed for formulations CF1 to CF9 in PBS pH 7.4 at 50 rpm and 37±0.5º for 24 h. All formulations showed initial burst release due to release of drug that is present on the surface of nanoparticles. CF1 showed higher release of 80.56% after 24 h. From the observed results, it was found that the nature of crosslinking directly affected the drug release from nanoparticles. Higher the cross linking, tighter the polymeric matrix and slower was the drug release as found in CF6 [23]. The results are shown in fig. 3. In vitro diffusion was performed for formulations PF1 to PF6 in PBS pH 7.4 at 37±0.5 for 72 h. A biphasic release was observed in all formulations with initial burst release due to free drug present on surface of nanoparticles) followed by sustained release. Among all PF6 showed higher release of 61.42% after 24 h. After initial burst release, the drug was released by bulk degradation followed by diffusion. PLGA undergoes bulk degradation in which random scission of ester bonds in the polymer backbone forms acidic monomers (lactic acid and glycolic acid) and oligomers which further catalyze the degradation of the parent polymer, a process known as autocatalysis [24]. From the observed results, it was found that higher the concentration of PLGA slower was the drug release due to its hydrophobic nature which delayed the degradation. It was also observed that formulation with high concentrations of PVA resulted in slowest drug release after 24 h as found in PF2. The possible reason can be due to formation of hydrogel barrier by PVA present on surface which may retard the release [24]. The results are shown in fig. 4.
The drug diffusion profiles of all formulations were fitted into various kinetic models. Both chitosan NP’s and PLGA NP’s were found to follow 1st order kinetics. From the results, it was also evident that both chitosan NP’s PLGA NP’s were more linear towards Korsemeyer- Peppas model with R2 value ranging from 0.979 to 0.996 and 0.973 to 0.989 respectively indicating that drug release mechanism was by diffusion. In Korsemeyer- Peppas plot the release component (n value) for all formulations was found to be less than 0.45 indicating fickian diffusion. The drug release from the nanoparticles was by a combination of bulk degradation followed by diffusion which involves the simultaneous penetration of the surrounding liquid into polymer matrix, dissolution of the drug, and leaching out of the drug through interstitial channels or pores [25]. The results are shown in Tables 6,7.
| Formulation | Model | ||||
|---|---|---|---|---|---|
| Zero order | 1st order | Higuchi | Korsemeyer- Peppas | ||
| R2 | R2 | R2 | R2 | n value | |
| CF1 | 0.939 | 0.809 | 0.994 | 0.979 | 0.38 |
| CF2 | 0.879 | 0.926 | 0.993 | 0.995 | 0.33 |
| CF3 | 0.900 | 0.961 | 0.974 | 0.991 | 0.32 |
| CF4 | 0.897 | 0.908 | 0.988 | 0.995 | 0.34 |
| CF5 | 0.835 | 0.968 | 0.974 | 0.991 | 0.19 |
| CF6 | 0.878 | 0.882 | 0.990 | 0.992 | 0.29 |
| CF7 | 0.878 | 0.939 | 0.988 | 0.996 | 0.30 |
| CF8 | 0.839 | 0.950 | 0.969 | 0.993 | 0.23 |
| CF9 | 0.903 | 0.972 | 0.967 | 0.986 | 0.18 |
Table 6: Kinetic Modelling of CF1 – CF9
| Formulation | Model | ||||
|---|---|---|---|---|---|
| Zero order | 1st order | Higuchi | Korsemeyer- Peppas | ||
| R2 | R2 | R2 | R2 | n value | |
| PF1 | 0.785 | 0.889 | 0.936 | 0.960 | 0.44 |
| PF2 | 0.903 | 0.957 | 0.988 | 0.973 | 0.38 |
| PF3 | 0.893 | 0.961 | 0.983 | 0.977 | 0.38 |
| PF4 | 0.792 | 0.916 | 0.944 | 0.978 | 0.34 |
| PF5 | 0.846 | 0.936 | 0.971 | 0.989 | 0.36 |
| PF6 | 0.819 | 0.948 | 0.961 | 0.985 | 0.37 |
Table 7: Kinetic Modelling of PF1 – PF6
The characterization data of all formulations was analyzed with Minitab 17 response optimizer. Entrapment efficiency, Particle size, Zeta potential and in vitro drug release were selected as responses. d-value (individual desirability) on a scale of 0 to 1 indicates the possibility of obtaining the desired results with the selected combination. Values closer to 1 indicates good desirability and vice versa. Generally same formulation cannot score high d value with all responses as evident from the results that different formulations were having good desirability with different responses. D value (composite desirability) was calculated taking all individual responses in to consideration. Higher the value of D, higher was the possibility for optimal results. From the results CF4 with D value of 0.519 and PF6 with D value of 0.653 were selected for in vivo studies. The results are shown in Tables 8,9.
| Formulation | d-value (individual desirability or goal) | Composite Desirability | |||
|---|---|---|---|---|---|
| Encapsulation Efficiency |
Particle Size | Zeta Potential | In vitro release |
||
| CF1 | 0.028 | 0.947 | 0.385 | 0.635 | 0.284 |
| CF2 | 0 | 0.951 | 0.231 | 0.346 | 0 |
| CF3 | 0.098 | 1 | 0 | 0.258 | 0 |
| CF4 | 0.267 | 0.797 | 0.769 | 0.442 | 0.519 |
| CF5 | 0.323 | 0.891 | 0.539 | 0.077 | 0.337 |
| CF6 | 0.451 | 0.968 | 0.308 | 0 | 0 |
| CF7 | 0.113 | 0.094 | 1 | 0.077 | 0.169 |
| CF8 | 0.183 | 0 | 0.692 | 0.115 | 0 |
| CF9 | 0.310 | 0.188 | 0.538 | 0.019 | 0.157 |
Table 8: Response Optimization of CF1 to CF9 by Minitab 17
| Formulation | d-value (individual desirability or goal) | Composite Desirability | |||
|---|---|---|---|---|---|
| Encapsulation Efficiency | Particle Size | Zeta Potential | In vitro release |
||
| PF1 | 0.296 | 0.312 | 0 | 0.133 | 0 |
| PF2 | 0.164 | 0.046 | 0.395 | 0 | 0 |
| PF3 | 0.071 | 0.624 | 0.454 | 0.258 | 0.269 |
| PF4 | 0 | 0.816 | 1 | 0.436 | 0 |
| PF5 | 0.523 | 0 | 0.559 | 0.219 | 0 |
| PF6 | 0.412 | 0.962 | 0.821 | 0.559 | 0.653 |
Table 9: Response Optimization of PF1 to PF6 by Minitab 17
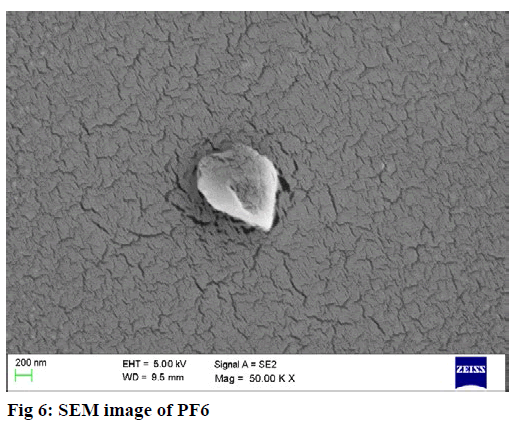
SEM analysis revealed PLGA NP’s (PF6) were found to be spherical and smooth whereas chitosan NP’s (CF4) were near spherical and rough in texture. The results are shown in fig 5, 6.
When compared with pure drug, the formulated nanoparticles carried the drug to brain effectively. A two-way ANOVA was employed for statistical analysis of Blood Brain Barrier crossing study. When compared with pure drug at different time intervals, PF6 with a p value<0.001 and CF4 with p value<0.05 (*) are found to be statistically significant. The results are shown in Table 10.
| Frequency | Pure drug | Formulation | |
|---|---|---|---|
| CF4 | PF6 | ||
| 30 min | 2.48±0.82 | 9.44±1.39 | 18.44±1.75 |
| 3 hours | 4.77±1.15 | 26.48±3.71 | 43.18±3.25 |
| 6 hours | 8.32±1.32 | 39.92±4.88 | 64.35±2.76 |
| 12 hours | 5.81±1.08 | 27.35±4.11 | 51.52±2.31 |
Table 10: BBB Crossing Study (% of Drug Present in Brain after I.V)
Acknowledgements:
The authors thank Shri. B. M. Reddy, President of JMJ Education Society & Shri. B. Premnath Reddy, Chairman and Smt. Shalini Reddy, Executive director of Acharya Institutes for providing the facilities to complete the research work successfully.
References
- Neha S, Viness P, Yahya E. Advances in the treatment of Parkinson’s disease. Prog Neurobio 2007;81(1):29-44.
- Ram B Gupta, Uday B. Kompella. Nanoparticle technology for drug delivery. New York: Taylor & Francis Group; 2006: p. 273-4.
- Gordon EM, Cornelio GH, Lorenzo CC III. First clinical experience using a ‘pathotropic’ injectable retroviral vector (Rexin-G) as intervention for stage IV pancreatic cancer. Int J Oncol 2004;24:177-85.
- Rockall AG, Sohaib SA, Harisinghani M. Diagnostic performance of nanoparticle-enhanced magnetic resonance imaging in the diagnosis of lymph node metastases in patients with endometrial and cervical cancer. J Clin Oncol 2005;23:2813-21.
- Sunitha R, Harika D, Kumar AP, Prabha KS, Prasanna PM. A review: nanoparticles as specified carriers in targeted brain drug delivery system. Am J Pharm Res 2011;1(2): 121-34.
- Amrita D, Madgulkar AR, Sanjay JK, Bhalekar MR, Andeep SC. Targeted drug delivery to CNS using nanoparticles. J Adv Pharm Sci 2012;2(1):179-91.
- Kewal KJ. Nanobiotechnology-based strategies for crossing the blood–brain barrier. Nanomed 2012;7(8):1225-33.
- Moji Christianah Adeyeye. Ed. Preformulation in solid dosage form development. Drugs and Pharmaceutical Sciences. Informa Healthcare; Newyork;2008. p. 2.
- Fazil Md, Sadab Md, Shabdul H, Manish K, Sanjula B. Jasjeet KS et al., Development and evaluation of Rivastigmine loaded chitosan nanoparticles for brain targeting. Eur J Pharm Sci 2012;47(1):6-15.
- Nagavarma BN, Hemant KS, Ayaz A, Vasudha LS, Shivakumar HG. Different techniques for preparation of polymeric nanoparticles- A review. Asian J Pharm Clin Res 2012;5(3):16-23.
- Moraima MC, Giselle MFF, Myreisa MC, Elsie AO, Jose ARM, Mercedes R et al. Two-step nanoprecipitation for the production of protein-loaded nanospheres. Results Pharm Sci 2012;2:79-85.
- Eameema M, Raju D, Anjali J, Venu G, Shaheen M, Wahid Khan. Intranasal delivery of nanoparticle encapsulated tarenflurbil: A potential brain targeting strategy for Alzheimer’s disease. Eur J Pharm Sci 2016;92:224-34.
- Werner JG, David DA, Jeffrey RB. Novel models for assessing blood-brain barrier drug permeation. Expert Ofin Drug Metab Toxico 2012;6:647-53.
- Nagavarma BVN, Hemant KS, Ayaz A, Vasudha LS, Shivakumar HG. Different techniques for preparation of polymeric nanoparticles- A review. Asian J Pharm Clin Res 2012;5(3):16-23.
- Shoaib MN, Tazeen J, Hamid AM, Rabia IY. Evaluation of drug release kinetics from ibuprofen matrix tablets using HPMC. Pak J Pharm Sci 2006;19(2):119-24.
- Gautam Singhvi, Mahaveer Singh. In vitro drug release characterization models. Int J Pharm Res 2011;2(1):77
- Rajan A, Mukerjee, A, Helson L, Vishwanatha J. Scale up, optimization and stability analysis of curcumin C3 complex loaded nanoparticles for cancer therapy. J Nanobiotech. 2012;10: 1-18.
- Fazil Md, Sadab Md, Shabdul H, Manish kumar, Sanjula B. Jasjeet KS et al. Development and evaluation of Rivastigmine loaded chitosan nanoparticles for brain targeting. Eur J Pharm Sci 2012;47(1):6-15.
- Wilson B, Samanta M, Santhi K, Kumar KP, Param KN, Suresh B. Targeted delivery of Tarine into the brain with poly(n-butyl cyanoacrylate) nanoparticles. Eur J Pharm Biopharm 2008;70:75-84.
- Mohammed H, Urszula D. PLGA biodegradable nanoparticles containing perphenazine or chlorpromazine hydrochloride: Effect of formulation and release. Int J Mol Sci 2014;15(12):23909-23.
- Sruthi S, Francisco MG, Bruno MM, Gustavo RRR. Parameters infuencing the size of chitosan-TPP nano- and microparticles. Sci Reports 2018;8:4695. DOI:10.1038/s41598-018-23064-4.
- Sarvesh B, Vibha C, Archna P. Polymeric nanoparticles containing diazepam: preparation, optimization, characterization, in vitro drug release and release kinetic study. Nano Convergence 2016;3(3):1-7.
- Vineetha P, Rao Vadaparthia, Kumara K, Dileep BJ, Veerabhadra A, Suresh K. Influence of organic solvents on nanoparticle formation and surfactants on release behaviour in vitro using costunolide as model anticancer agent. Int J Pharm Pharm Sci 2014;6(4):638-45.
- Zahra S, Soliman M, Hashem M, Elham K. Nanoparticles of chitosan loaded ciprofloxacin: fabrication and antimicrobial activity. Adv Pharm Bull 2017;7(3):427-32.
- Gautam Singhvi, Mahaveer Singh. In vitro drug release characterization models. Int J Pharm Res 2011;2(1):77.