- *Corresponding Author:
- V. B. Tatipamula
Pharmaceutical Chemistry Department, AU College of Pharmaceutical Sciences, Andhra University, Visakhapatnam-530 003, India
E-mail: tvinaybharadwaj@andhrauniversity.edu.in
| Date of Submission | 29 October 2018 |
| Date of Revision | 28 December 2018 |
| Date of Acceptance | 27 April 2019 |
| Indian J Pharm Sci 2019;81(3):569-574 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Considering the antidiabetic potentiality of the moss Taxithelium napalense, the present study was undertaken to explore the chemical constituents that are present in this species. The entire moss Taxithelium napalense was extracted by ethanol and the extract obtained was subjected to gas chromatography-mass spectrometry. Sixty nine compounds were identified, which are reported for the first time from this species. From the gas chromatography-mass spectrometry analysis, it was determined that the octadecanoic acid methyl ester was the chief component present in Taxithelium napalense. In addition, the total phenolic and flavonoid contents of ethanol extract of Taxithelium napalense were found to be 101.43±0.38 mg Q/g and 229.73±3.07 mg GA/g, respectively. Additionally, the inhibitory concentration 50 of the ethanol extract of Taxithelium napalense on α-glucosidase was found to be 34.5 µg/ml, while acarbose value was 29.5 µg/ml. This is the first report on the chemical investigation of the moss Taxithelium napalense.
Keywords
Moss, chemical constituents, gas chromatography, mass spectroscopy, metabolites
Mosses are non-vascular plants with rhizoids. They are grouped into the division of Bryophyte under the subdivision Musci. About 17 000 species of moss falling in 89 families and ca. 898 genera are distributed across the world and in India, about 1786 moss species are recorded till now [1]. Generally, mosses are habituated in wet environments such as alpine and rainforest ecosystems, in addition, mosses require fresh water to their reproduction [1,2]. Due to their unique habitat, they are scrutinized for biological evaluations because of their nutritional contents [2], for instance, Sphagnum palustre moss has renoprotective effects [3] and aromatase inhibitory activity [4].
Taxithelium napalense (Schwaerg) Broth, family Sematophyllaceae is an epiphytic moss. T. napalense is habituated on twigs and branches of other plants in semi evergreen forests found in palaeotropical region [5]. Earlier, the phytochemical, antioxidant and in vivo antidiabetic potential of the ethanol extract of T. napalense was reported [6] . In the present study, it was aimed to identify the active metabolites present in this moss using gas chromatography-mass spectrometry (GC-MS) analysis. Additionally, the total flavonoid and phenolic contents along with its α-glucosidase inhibitory activity was also determined.
The specimens of Taxithelium nepalense (Schwaerg) Broth was collected from barks of mangrove, Rhizophora species from Bhitarkanika island (20°72’ N latitude and 86°87’ E Longitude) Orissa, India in March 2016. The moss T. nepalense was authenticated in the Council of Scientific and Industrial Research (CSIR)-National Botanical Research Institute (NBRI), Lucknow and deposited at Bryophyte Herbarium, CSIR-NBRI, Lucknow, India with an accession number LWG-5/VB-Orissa-2016 [1].
The moss specimens collected from the mangrove plants were shade-dried. The dried lichen material was powdered using a blender and about 12 g of moss material was exhaustively extracted thrice with ethanol at room temperature. The mixture was filtered through muslin cloth and evaporated under reduced pressure to obtain ethanol extract (1.21 g, 10.08 % w/w) as a dark greenish solid, which were stored in amber colored vials and preserved at 4° till further use.
All the chemicals and reagents used in the present study were of analytical grade. p-nitrophenyl-α-Dglucopyranoside, rat intestinal acetone powder, sucrose and acarbose, were procured from Sigma-Aldrich (Mumbai, India). Glibenclamide was purchased from Aventis Pharma Ltd. (Mumbai, India).
The phytochemical investigation of ethanol extract of T. nepalense was performed on GC-MS equipment (GCMS-QP2010 Plus, Shimadzu, Europe). Experimental parameters of GC-MS system were, column oven temperature: 50°, injection temperature: 250°, injection mode: split; flow control mode: linear velocity; pressure: 29.7 kPa; total flow: 10.9 ml/min; column flow: 0.72 ml/min; linear velocity: 30.7 cm/s; purge flow: 3.0 ml/min; split ratio: 10.0. Oven temperature program was 50° hold time for 2 min, at 200° hold time for 0 min and 280° hold time for 2 min. The GC program was ion-source temperate at 210°, interface temperature at 250°, solvent cut time: 3 min, detector gain: 1.07 kV+0.20 kV and threshold of 1000.
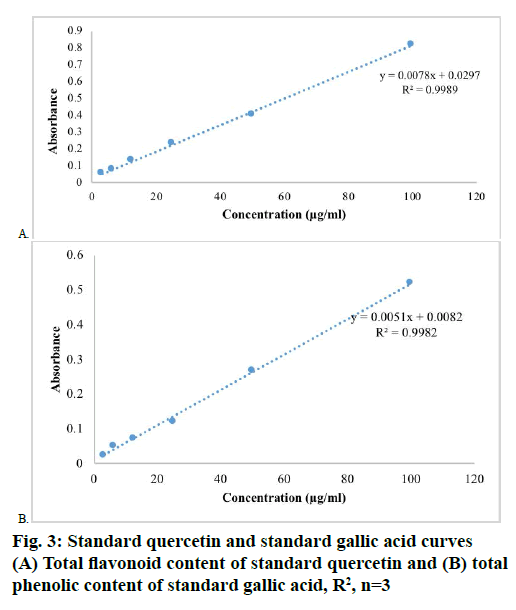
The total flavonoid content [7] of the extracts were determined by using aluminium chloride spectrophotometric method, in which AlCl3 forms complex with hydroxyl groups of flavonoids exist in the testing sample. To the extract (1 mg/ml) or standard quercetin solution (3.125, 6.25, 12.5, 25, 50, 100 μg/ml), added 3 ml of methanol, 1 ml of 2 % AlCl3 solution, 200 μl of 1 M potassium acetate and made up to 10 ml with distilled water and incubated for 60 min at room temperature, whereas the blank contained only reagents and the absorbance was noted at 415 nm. Based on the measured absorbance of the test sample, the total flavonoid content was read on the calibration curve and the total flavonoid content was expressed in terms of quercetin equivalent (mg of Q/g of extract).
The total phenolic content [7] was estimated by Folin- Ciocalteu’s method. To the extract (1 mg/ml) or standard gallic acid solution (3.125, 6.25, 12.5, 25, 50, 100 μg/ml), added 0.5 ml of Folin-Ciocalteau reagent, 1.5 ml of sodium carbonate (20 %) solution and volume made up to 10 ml with distilled water and incubated at room temperature for 120 min and the absorbance was measured at 750 nm using spectrophotometer against blank (contained only reagents). Based on the measured absorbance of the test sample, the total phenolic content was read on the calibration curve and the total phenolic content was expressed in terms of gallic acid equivalent (mg of GA/g of extract).
The assay of α-glucosidase inhibitory activity was estimated using the method reported by Tatipamula et al. [8] in triplicate (n=3). Two microlitres of α-glucosidase from rat intestine acetone powder solution (a stock solution of 1.0 mg/ml in 10 mM phosphate buffer, pH 6.8, diluted 40-fold with phosphate buffer) was mixed with 20 μl of the samples at different concentrations (25, 50, 75 and 100 μg/ml solubilized in dimethyl sulfoxide) to which 100 μl of 50 mM phosphate buffer (pH 6.8) was added in a 96 well microtitre plate and incubated for 5 min at 37°. After incubation, 50 μl of substrate (5 mM of p-nitrophenyl- α-D-glucopyranoside prepared in 50 mM of phosphate buffer, pH 6.8) was added and the entire reaction mixture was again incubated for 20 min at 37°. Thereafter the reaction was terminated by adding 50 μl of Na2CO3 (1 M) and the final volume was made up to 150 μl. The amount of p-nitrophenol released from substrate was noted at 405 nm spectrophotometrically (Spectra MAX plus 384, Molecular Devices Corporation, Sunnyvale, CA, USA). Dimethyl sulfoxide and acarbose were used as control and standard, respectively. Percent enzyme inhibition was calculated using the Eqn., percent inhibition = (C–S)/C×100, where C is the absorbance of the control, S is the absorbance of sample. IC50 values of the samples were determined by plotting percent inhibition against concentrations.
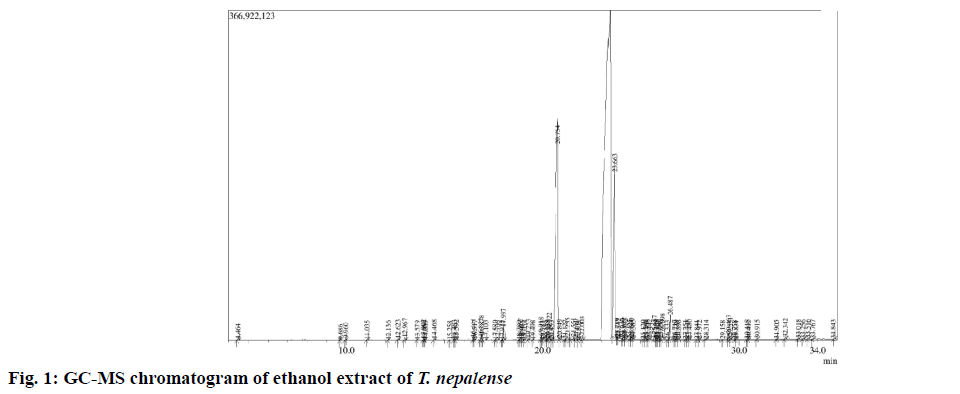
After successful extraction of the whole moss material, the dried extract was subjected to GC-MS analysis. The GC-MS spectrum revealed the presence of various chemical components with different retention times (fig. 1), whereas the MS analyzes the compounds eluted at different times to detect its nature and structure of the compounds. The results pertaining to GC-MS analysis of the ethanol extract of T. nepalense lead to the identification of a number of compounds. The various compounds present in the entire stressed moss of ethanol extract of T. nepalense along with their retention time, molecular formula, molecular weight, peak area (%) and nature of the compound that were detected by the GC-MS were represented in Table 1. A total of 69 compounds were identified by GC-MS analysis (Table 1). In addition, the composition determined for the ethanol extract of T. nepalense corresponds to 63.59 % of the entire GC-MS chromatogram.
| RT (min) | Name of the compound | Molecular formula | Molecular weight (g/mol) |
Peak area (%) | Nature of the compound |
|---|---|---|---|---|---|
| 4.464 | Hexanal | C6H12O | 100.16 | 0.10 | Aliphatic aldehyde |
| 9.686 | Nonanal | C9H18O | 142.24 | 0.02 | Aliphatic aldehyde |
| 9.960 | Octanoic acid | C8H16O2 | 144.21 | 0.15 | Aliphatic carboxylic acid |
| 11.035 | 1-Dodecene | C12H24 | 168.32 | 0.05 | Aliphatic alkene |
| 12.136 | 2-Decenal | C10H18O | 154.25 | 0.03 | Aliphatic alkene |
| 12.623 | (Z)-2,4-Decadienal | C10H16O | 152.24 | 0.13 | Aliphatic alkene |
| 12.967 | (E,E)-2,4-Decadienal | C10H16O | 152.24 | 0.27 | Aliphatic alkene |
| 13.579 | (E,E)-2-Undecenal | C11H20O | 168.28 | 0.02 | Aliphatic alkene |
| 13.902 | 1-Pentadecene | C15H30 | 210.41 | 0.10 | Aliphatic alkene |
| 13.950 | 2-Decanone | C10H20O | 156.27 | 0.01 | Aliphatic ketone |
| 14.005 | Tetradecane | C14H30 | 198.39 | 0.03 | Aliphatic alkane |
| 14.468 | Nonanoic acid | C9H18O2 | 158.24 | 0.13 | Aliphatic carboxylic acid |
| 15.258 | 2-Tridecanone | C13H26O | 198.35 | 0.02 | Aliphatic ketone |
| 16.441 | 1-Hexadecene | C16H32 | 224.43 | 0.19 | Aliphatic alkene |
| 16.517 | Hexadecane | C16H34 | 226.45 | 0.10 | Aliphatic alkane |
| 16.878 | 4,5-Dimethoxy benzophenone | C15H14O4 | 258.27 | 0.50 | Aromatic ketone |
| 17.580 | 2,4-Di-t-butyl-6-nitro-6-dodecanone | C20H40NO3 | 342.54 | 0.05 | Aliphatic ketone |
| 17.922 | Hexadecanal | C16H32O | 240.43 | 0.03 | Aliphatic aldehyde |
| 17.997 | Methyl tetradecanoate | C15H30O2 | 242.40 | 1.14 | Aliphatic ester |
| 18.780 | Heptadecanoic acid | C17H34O2 | 270.46 | 0.02 | Fatty acid |
| 18.857 | 16-Methyl-1-octadecene | C19H38 | 265.51 | 0.26 | Aliphatic alkene |
| 18.933 | Heneicosane | C21H44 | 296.58 | 0.11 | Aliphatic alkane |
| 19.054 | 5-Octadecenoic acid | C18H34O2 | 282.47 | 0.06 | Fatty acid |
| 19.498 | Pentadecanoic acid | C15H30O2 | 242.40 | 0.17 | Fatty acid |
| 19.918 | 2-Undecanone | C11H22O | 170.30 | 0.07 | Aliphatic ketone |
| 19.983 | 6,10-Dimethyl-9-heptadecanone | C19H38O | 282.51 | 0.36 | Aliphatic ketone |
| 20.159 | 10-Methyl-E-11-tridece-1-ol-acetate | C16H30O2 | 254.41 | 0.02 | Aliphatic ester |
| 20.322 | 9-Hexadecenoic acid | C16H30O2 | 254.41 | 1.01 | Fatty acid |
| 20.450 | 9-Hexadecenoic acid methyl ester | C16H30O2 | 268.44 | 0.05 | Aliphatic ester |
| 20.754 | Octadecanoic acid methyl ester | C19H38O2 | 298.51 | 52.09 | Aliphatic ester |
| 20.849 | 3-Hydroxy cyclodecene | C10H18O | 154.25 | 0.12 | Cyclic hydroxyl compound |
| 21.680 | Cyclopropaneoctanoic acid | C11H20O2 | 184.28 | 0.28 | Fatty acid |
| 22.003 | Heptadecanoic acid methyl ester | C18H36O2 | 248.48 | 0.50 | Aliphatic ester |
| 23.789 | Methyl octadec-9-enoate | C19H36O2 | 297.487 | 0.11 | Aliphatic ester |
| 23.842 | Methyl 9-cis,11-trans-octadecadienoate | C19H34O2 | 294.48 | 0.08 | Aliphatic ester |
| 24.040 | Ethyl (9Z,12Z)-9,12-octadecadienoate | C20H36O2 | 308.51 | 0.18 | Aliphatic ester |
| 24.122 | (E)-9-Octadecenoic acid ethyl ester | C20H38O2 | 310.52 | 0.40 | Aliphatic ester |
| 24.425 | 9-Cis,11-trans-octadecadienoate-1-docosene | C18H32O2 | 280.45 | 0.07 | Fatty acid |
| 25.246 | Behenic alcohol | C22H46O | 326.61 | 0.08 | Aliphatic hydroxyl compound |
| 25.442 | 9,12,15-Octadecatrienoic acid | C18H30O2 | 278.44 | 0.17 | Fatty acid |
| 25.828 | 9,12,15-Octadecatrienoic acid methyl ester | C19H32O2 | 292.46 | 0.11 | Aliphatic ester |
| 25.890 | 6,9,12-Octadecatrienoic acid | C18H30O2 | 278.44 | 0.11 | Fatty acid |
| 25.996 | Oxiraneoctanoic acid 3-octyl- methyl ester | C19H36O3 | 312.49 | 0.11 | Aliphatic ester |
| 26.098 | Cyclopropaneoctanoic acid methyl ester | C12H22O2 | 198.31 | 0.94 | Aliphatic ester |
| 26.333 | 8-Hexadecenal | C16H30O | 238.42 | 0.04 | Aliphatic aldehyde |
| 26.717 | (9E,12E)-9,12-Octadecadienoyl chloride | C18H31ClO | 298.90 | 0.07 | Chlorinated aliphatic ketone |
| 26.779 | 4,4',6,6'-Tetra-tert-butyl-2,2'-biphenyldiol | C28H42O2 | 410.64 | 0.05 | Biphenolic compound |
| 26.898 | 2-Butyl-5-methyl-3-(2-methylprop-2-enyl)cyclohexanone | C15H26O | 222.37 | 0.06 | Cyclic ketone |
| 27.251 | 9-Octadecenoic acid methyl ester | C9H36O2 | 296.50 | 0.05 | Aliphatic ester |
| 27.348 | (Z,Z)-6,9-cis-3,4-epoxynonadecadiene | C19H34O | 278.48 | 0.11 | Aliphatic alkene |
| 27.480 | 1-Docosene | C22H44 | 308.59 | 0.12 | Aliphatic alkene |
| 27.841 | 1-(2,2-Dimethylcyclopentyl) ethanone | C9H16O | 140.23 | 0.06 | Cyclic ketone |
| 27.972 | Heneicosanoic acid | C21H42O2 | 326.57 | 0.07 | Fatty acid |
| 28.314 | 7,10-Hexadecadienoic acid | C16H28O2 | 252.40 | 0.10 | Fatty acid |
| 29.158 | 9-Heptadecanol | C17H36O | 256.47 | 0.02 | Aliphatic hydroxyl compound |
| 29.463 | Docosanoic acid | C22H44O2 | 340.59 | 0.97 | Fatty acid |
| 29.531 | 1,2-Benzenedicarboxylic acid | C8H6O4 | 166.13 | 0.40 | Aromatic acid |
| 29.758 | 9,11-Octadecadienoic acid | C18H32O2 | 280.45 | 0.04 | Fatty acid |
| 29.831 | 9-Octadecenoic acid | C18H34O2 | 282.47 | 0.04 | Aliphatic acid |
| 30.418 | 1-Hexadecanol | C16H34O | 242.45 | 0.10 | Aliphatic hydroxyl compound |
| 30.492 | Octadecane | C18H38 | 254.50 | 0.03 | Aliphatic alkane |
| 30.915 | Tricosanoic acid | C23H46O2 | 354.62 | 0.10 | Aliphatic acid |
| 31.905 | Pentatriacontane | C35H72 | 492.96 | 0.04 | Aliphatic alkane |
| 32.342 | Tetracosanoic acid | C24H48O2 | 368.65 | 0.22 | Fatty acid |
| 33.018 | Geranylgeraniol | C20H34O | 290.49 | 0.08 | Aliphatic hydroxyl compound |
| 33.256 | 1-Heptacosanol | C27H56O | 396.74 | 0.08 | Aliphatic hydroxyl compound |
| 33.516 | Squalene | C30H50 | 410.73 | 0.09 | Aliphatic alkane |
| 33.767 | Pentacosanoic acid methyl ester | C26H52O2 | 396.70 | 0.06 | Aliphatic ester |
| 34.843 | Heptacosane | C27H56 | 380.75 | 0.04 | Aliphatic alkane |
Table 1: Compounds identified from GC-MS analysis of the ethanol extract of Taxithelium napalense
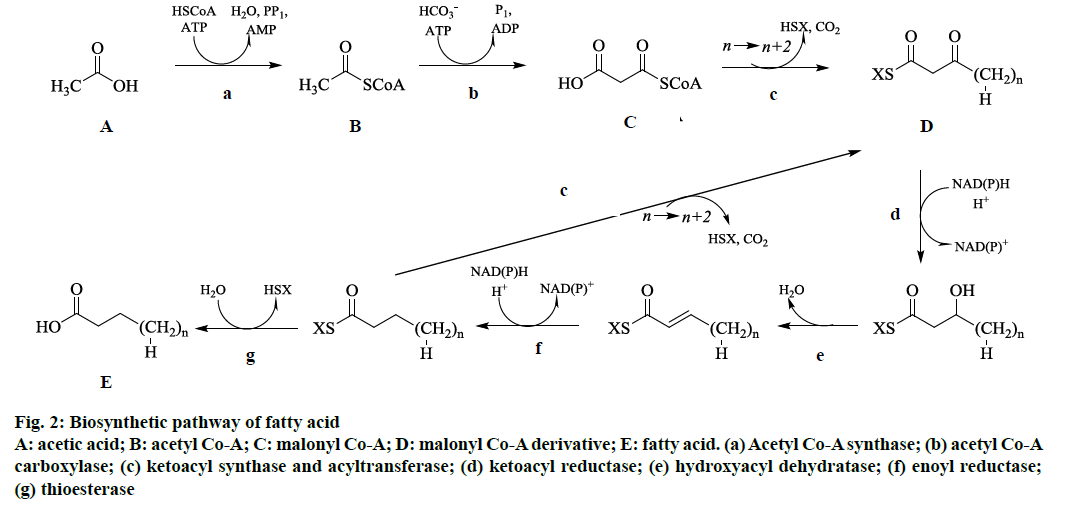
From the GC-MS analysis, it was observed that this moss species contained mostly aliphatic fatty acids and its derivatives. Therefore, the standard way for cells to synthesize fatty acids is through the fatty acid synthesis cycle as shown in fig. 2. This cycle of eight enzymes (acyl-CoA synthase, acyl-CoA carboxylase, acyltransferase, ketoacyl synthase, ketoacyl reductase, hydroxyacyl dehydratase, enoyl reductase, and thioesterase) and acyl carrier protein) was initiated with acetic acid, CoA, and ATP by using acyl-CoA synthase as catalyst to make acetyl-CoA (fig. 2). By using another ATP and bicarbonate ion catalyzed by acyl- CoA carboxylase, which yields malonyl-CoA (fig. 2). The obtained malonyl-CoA was added to an acyl chain by catalysis with ketoacyl synthase and acyltransferase, which were usually activated with acyl carrier protein, to make an acyl chain two methylene groups longer (fig. 2). Additionally, reduction, dehydration, and reduction with ketoacyl synthase, hydroxyacyl dehydratase, and enoyl reductase catalysis, respectively, leads to a saturated and unhydroxylated acyl chain activated with acyl carrier protein (fig. 2). If the chain is of appropriate length, it was attacked by thioesterase to release acyl carrier protein, yielding the finished fatty acid (fig. 2). The various fatty acids derived by this pathway was subjected to further reduction, dehydration, and reduction reactions in the cells to yield respective derivatives namely aliphatic aldehydes, aliphatic hydroxyl, aliphatic ketones and aliphatic esters.
Figure 2: Biosynthetic pathway of fatty acid
A: acetic acid; B: acetyl Co-A; C: malonyl Co-A; D: malonyl Co-A derivative; E: fatty acid. (a) Acetyl Co-A synthase; (b) acetyl Co-A
carboxylase; (c) ketoacyl synthase and acyltransferase; (d) ketoacyl reductase; (e) hydroxyacyl dehydratase; (f) enoyl reductase;
(g) thioesterase
The results of total flavonoid content [7] was expressed in terms of mg of Q/g of extract by using standard calibration line Eqn., y = 0.0078x+0.0297 R²=0.9989 (fig. 3A), similarly, the total phenolic content were expressed in terms of mg of GA/g of extract by using standard calibration line Eqn., y = 0.0051x+0.0082 R²=0.9982 (fig. 3B). The ethanol extract of T. nepalense contained more total phenolic content than total flavonoid content. The total phenolic content [7] was found to be 229.73±3.07 mg GA/g, whereas the total flavonoid content was 101.43±0.38 mg Q/g. In addition, the GC-MS analysis also depicted the presence of higher amounts of phenolic compounds in ethanol extract of T. nepalense.
The in vitro antidiabetic activity was assessed by α-glucosidase inhibitory assay [8] using acarbose as standard and p-nitrophenyl-α-D-glucopyranoside as a substrate. From the inhibitory assay, it was estimated that the ethanol extract of T. nepalense exhibited significant inhibition of α-glucosidase enzyme with IC50 values of 34.5 μg/ml, whereas acarbose with 29.5 μg/ml (Table 2). The results of α-glucosidase inhibitory assay proved the antidiabetic potentiality of T. nepalense.
| COMPOUND | Inhibition (%)* | IC50 values (µg/ml) | |||
|---|---|---|---|---|---|
| 25 µg/ml | 50 µg/ml | 75 µg/ml | 100 µg/ml | ||
| TNEE | 44.94±6.40 | 58.02±5.06 | 65.27±4.65 | 72.43±5.38 | 34.5 |
| Acarbose | 46.11±4.74 | 68.25±6.47 | 76.06±5.78 | 85.79±5.24 | 29.5 |
TNEE is Taxithelium napalense ethanol extract *Mean±SD values (n=3)
Table 2: α-Glucosidase inhibitory activity of tnee
To conclude, this is the first report on chemical components from stressed moss T. nepalense. This study confirmed the presence of various compounds in the moss T. nepalense and also justified the use of this moss as a remedy to diabetes in traditional medicine. This research helped to predict the active metabolites present in the T. nepalense, in addition, it also helped in establishing the chemical composition of this moss as well. From these results, it could be concluded that T. nepalense contained various bioactive metabolites. Additionally, the ethanol extract has more total phenolic content than total flavonoid content. However, the isolation of individual secondary metabolites and investigating the biological activity possessed by these would give impetus for further research on the antidiabetic potential of this plant.
Acknowledgements
The authors thank the authorities of AU College of Pharmaceutical Sciences, Andhra University, Visakhapatnam, Andhra Pradesh, India for providing the necessary facilities to complete present work. The authors also thank Dr. Ankita Srivastava, CSIRNational Botanical Research Institute (NBRI), Lucknow for identifying the moss species collected.
Conflicts of interest:
The authors declare that there is no conflict of interest.
References
- Sastry GV, Bharadwaj VT. Occurrences of mosses in Indian mangrove forests. J Integr Sci 2018;1:1-6.
- Decker EL, Reski R. Moss bioreactors producing improved biopharmaceuticals. Curr Opin Biotechnol 2007;18:393-8.
- Kang HR, Lee D, Eom HJ, Lee SR, Lee KR, Kang KS, et al. Identification and mechanism of action of renoprotective constituents from peat moss Sphagnum palustre in cisplatin-induced nephrotoxicity. J Funct Foods 2016;20:358-68.
- Eom HJ, Park YJ, Kang HR, Kim HR, Bang IJ, Park HB, et al. Inhibitory effect of Sphagnum palustre extract and its bioactive compounds on aromatase activity. Bangladesh J Pharmacol 2016;11:661-5.
- Ramsay HP, Schofield WB, Tan BC. The genus Taxithelium (Bryopsida, Sematophyllaceae) in Australia. Aust Syst Bot 2002;15:583-96.
- Tatipamula VB, Killari KN, Ketha A, Vedula GS. Taxithelium napalense acts against free radicals and diabetes mellitus. Bangladesh J Pharmacol 2017;12:197-203.
- Tatipamula VB, Vedula GS, Rathod BB, Shetty PR, Sastry AVS. Study of phytochemical analysis, total flavonoid and phenolic content, antimicrobial properties and chemical constituents of two manglicolous lichens extracts. Planta Activa 2018;2018:129-34.
- Tatipamula VB, Kolli MK, Lagu SB, Paidi KR, Reddy RP, Yejella RP. Novel indolizine derivatives lowers blood glucose levels in Streptozotocin-induced diabetic rats: A histopathological approach. Pharmacol Rep 2019;71(2):233-42.