- *Corresponding Author:
- S. Zhiqiang
Shenzhen Hospital, Southern Medical University, Guangdong Province, 518101, China
E-mail: zhiqiangs@gmail.com
| This article was originally published in a special issue: Special issue on “Drug Development and Human Health in China” | |
| Indian J Pharm Sci 2020:82(1)spl issue2;44-48 | |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The gene expression profile of human T-lymphoblast MOLT-4 cells treated with Smo siRNA was detected using a gene chip, and the potential target genes regulated by Smo siRNA were screened. The transfection group MOLT-4 was constructed by lentiviral transfection technique. Bioinformatic analysis showed that these genes are mainly involved in cell growth and cycle, signal transduction, cell communication, cell adhesion, cell metastasis and invasion and the signaling pathways involved antigen processing and presentation, cytokine and receptor interaction, cell adhesion molecules, complement system. The study of potential target genes or signaling pathways regulated by Smo siRNA is important for exploring the mechanism of MOLT-4 cell development after Smo siRNA treatment.
Keywords
Gene expression profile, Smo siRNA, real-time PCR
Smo siRNA is one of the p63 gene transcription products, which is related to the degree of tissue differentiation and prognosis of human T-lymphoblast MOLT-4 cells. Patients with low expression of Smo siRNA have a higher risk of tumor metastasis and recurrence, and the prognosis is worse [1,2]. In this study, MOLT-4 cell line treated with Smo siRNA was established by in vitro gene transfection technology and the gene expression profile was detected using a gene chip. The related genes and signal pathways were analyzed by bioinformatics technology. The potential target genes directly or indirectly regulated by Smo siRNA were predicted and the role and regulation mechanism of MOLT-4 cells in the development and progression of Smo siRNA treatment were analyzed [3-6].
MOLT-4 cells were purchased from the Cell Resource Center of Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences [7,8]; Dulbecco’s modified Eagle medium (DMEM) medium and fetal bovine serum were purchased from Gibco, USA; TRIzol reagent and Lipofectamine 2000 were purchased from Invitrogen, USA; gene chip is for the Agilent human 4×44K chip genome-wide expression profile, Smo siRNA cDNA were purchased from Otigene, USA; plasmid pHelper1.0, plasmid pHelper 2.0 were purchased from Guangzhou Ledell Biotechnology Co., Ltd.; FastQuant RTKit was purchased from Tiangen, Beijing; SYBR Premix Ex TaqTM was purchased from TaKaRa, Japan; Plasma Midi Kit was purchased from QIAGEN, Germany [9,10]. DMEM culture medium containing 10 % fetal calf serum, 0.1 % cyan/streptomycin was placed in an adherent culture at 37° in a 5 % CO2 incubator and the cells were passaged by trypsin digestion in the logarithmic growth phase.
The full-length Smo siRNA cDNA was amplified and ligated into the pLVX-IRES-ZsGreen1 lentiviral vector digested with Xho I and BamH I to construct the recombinant lentiviral plasmid pLVX-IRESZsGreenl+ Smo siRNA, which was sent to a Gene Company for sequencing and identification. The Plasmid Midi Kit was used to purify the recombinant virus plasmid (or empty vector) with no endotoxin and its 2 auxiliary packaging plasmids. The human embryonic kidney (HEK) 293T cells were transfected with Lipofectamine 2000. The medium was changed 8 h after dyeing and after 48 h of culture, the supernatant of the cell containing the lentivirus particles was collected and concentrated to obtain a high titer of the lentivirus concentrate. After the virus titer was measured and calibrated in HEK 293T cells, the infected cells were constructed and transferred. The MOLT-4 cell line and its control group were stained.
The total RNA of the transfected group and the control group was extracted and purified by TRIzol kit. The RNA sample was sent to Shanghai Kangcheng Biotechnology Co., Ltd. for RNA quality detection and human genome expression profiling. The chip was scanned with Axon GenePix. 4000 B), image and data processing was done using the American Axon GenePix Pro 6.0 software.
Gene ontology (GO) functional analysis was performed using the PANTHERFR (http://www.pantherdb.org/) online tool. Pathway analysis uses DAVID (http://david.abcc.ncifcrf.gov/) online software. Using STRING (http://string.embl.de/) online tool further investigates the interaction between proteins encoded by differential genes in pathway analysis results.
Total RNA from Smo siRNA and MOLT-4 cells was extracted by TRIzol, reverse transcribed into cDNA using FastQuant RT Kit, and detected by qRT-PCR using SYBR Premix Ex TaqTM. Primers were designed using Primer5.0 software and sent to Huada Gene Technology. Co., Ltd. Bone morphogenetic protein 7, BMP7 F: 5’-AGTC-TATCAGGTGCTCCAGG-3’, R: 5’-TTGCTGGTGGCT- GTGATG-3’; Leukocyte differentiation antigen 74 (CD74 F:5’- CGG-GAAGATCAGAAGCCAGTC-3’, R:5’- GCGAGGAG- CAGAGTCACCAG-3’; Smo siRNAF: 5’-GACTCAATT-TAGTGAGCCACAGT-3’, R: 5’-GCAATTGGCAG-TAGAGTTT-3’ GAPDHF: 5’-GGCACAGTCAAGGCT-GAGAATG-3’, R: 5’-ATGGTGGTGAAGACGCCAGTA-3’. qRT-PCR amplification was performed on a RocheLightCycler 480 instrument using the following conditions: 95°, 10 s, 95°, 5 s, 60°, 30 s, melting curve temperature range: 65-95°, 40 cycles, each sample was repeated 3 times, the Ct value was recorded, and analyzed by 2-ΔΔCt method.
The data were analyzed by SPSS 21.0 software and the data were expressed as mean±standard deviation. Two independent samples were used for the measurement data between the groups. P<0.05 was considered statistically significant.
According to the chip data, differential genes with p<0.05, difference multiples >1.5 or <1:1.5 were screened for analysis. There were 1405 differentially expressed genes, of which 843 were up-regulated and 562 were down-regulated.
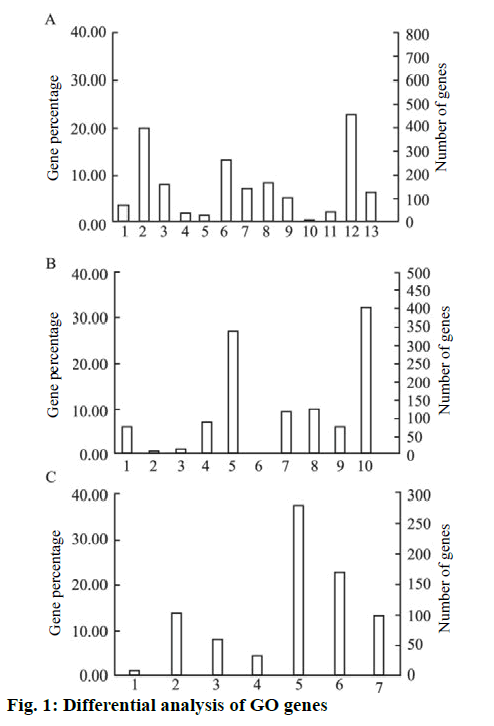
The PANTHERFR online analysis tool was used to analyze the above genes into GO. It showed that the differentially expressed genes were mainly related to cell growth, cell cycle regulation, signal transduction, cell metastasis invasion, cell adhesion, cell communication and other functions. Its function classification, the results shown in figure. 1, in the biological process classification, cell physiology accounted for 19.9 %, metabolic process accounted for 22.7 %; in the molecular function classification, the binding gene accounted for 32.3 %, catalytically active genes accounted for 27.1 %. In the intracellular localization classification, organelle protein accounted for 22.6 % and membrane protein accounted for 13.9 %.
The biological process included, biosynthesis process; cell physiology; localization process; apoptotic process; cell proliferation biological regulation process; stress; development process; multicellular organism process; transport; adhesion process; metabolic process and immune system process, while the molecular function included, transcription factor activity; regulatory factor activity; protein binding transcription factor activity; enzyme regulator activity; catalytic activity; signal transduction activity; receptor activity; nucleic acid binding transcription factor activity; molecular structure activity and binding. The intracellular positioning included cell junction region; membrane component; macromolecular complex; extracellular matrix component; cytoskeleton; organelle and the extracellular region. KEGG pathway analysis was performed on 1405 differentially expressed genes in the microarray results using DAVID online analysis software. A total of 17 pathways of p<0.05 were selected for study. The main pathways involved were, antigen processing, phagocytosis, viruses and bacteria. Infection and autoimmune diseases and cell adhesion are shown in Table 1.
| Signal path | Number | P value | Gene name |
|---|---|---|---|
| Antigen processing and presentation | 16 | 0.00 | B2M CD74 CTSL HLA-A HLA-B HLA-C HLA-DPB1 HLA-DRA HLA-DRB5 HLA-E HLA-F HLA-G CIITA TAP1 TAP2 TAPBP |
| Viral myocarditis | 14 | 0.00 | CAV1 CD55 HLA-A HLA-B HLA-C HLA-DPB1 HLA-DRA HLA-DRB5 HLA-E HLA-F HLA-G ITGB2 EIF4G3 |
| Rheumatoid arthritis | 16 | 0.00 | ANGPT1 CTSL CXCL1 HLA-DPB1 HLA-DRA HLA-DRB5 IL6 IL11 IL15 IL18 ITGB2 JUN LTB CCL20 TLR2 VEGFA |
| Phagosome | 22 | 0.00 | CD36 CTSL HLA-A HLA-B HLA-C HLA-DPB1 HLA-DRA HLA-DRB5 HLA-E HLA-F HLA-G ITGAM IT-GB2 NCF2 OLR1 TAP1 TAP2 TLR2 EEA1 PLA2R1 PIKFYVE TUBB8 |
| Graft-versus-host disease | 10 | 0.00 | HLA-A HLA-B HLA-C HLA-DPB1 HLA-DRA HLA-DRB5 HLA-E HLA-F HLA-G IL6 |
| Allograft rejection | 9 | 0.00 | HLA-A HLA-B HLA-C HLA-DPB1 HLA-DRA HLA-DRB5 HLA-E HLA-F HLA-G |
| Type I diabetes mellitus | 9 | 0.00 | HLA-A HLA-B HLA-C HLA-DPB1 HLA-DRA HLA-DRB5 HLA-E HLA-F HLA-G |
| Staphylococcus aureus infection | 10 | 0.00 | CFB C1QB C1S C4B HLA-DPB1 HLA-DRA HLA-DRB5 CFI ITGAM ITGB2 |
| Hematopoietic cell line-age | 13 | 0.00 | CD1A CD7 CD36 CSF3 CD55 HLA-DRA HLA-DRB5 IL4R IL6 IL11 ITGAM KITLG CD24 |
| Cell adhesion molecules (CAMs) | 17 | 0.00 | ALCAM CNTN1 HLA-A HLA-B HLA-C HLA-DPB1 HLA-DRA HLA-DRB5 HLA-E HLA-F HLA-G ITGAM ITGB2 CLDN11 SDC4 NFASC JAM2 |
| Complement and coagulation cascades | 11 | 0.00 | A2M CFB C1QB C1S C4B C6 CD55 F3 CFI PLAT THBD |
| Cytokine-cytokine receptor interaction | 27 | 0.00 | AMHR2 BMP7 CCR1 CSF3 EDA CXCL1 CXCL2 CXCL3 HGF IL4R IL6 IL11 IL15 IL15RA TNFRSF9 IL18 INHBB LIF LTB KITLG TNFRSF11B CCL20 CXCL11 VEGFA IL18R1 IL20 CXCL16 |
| Autoimmune thyroid disease | 9 | 0.00 | HLA-A HLA-B HLA-C HLA-DPB1 HLA-DRA HLA-DRB5 HLA-E HLA-F HLA-G |
| Focal adhesion | 21 | 0.00 | BIRC3 CAV1 CAV3 COL4A6 COL5A1 HGF TNC JUN LAMA3 LAMA4 MYL5 MYLK PPP1R12A PPP1CB VEGFA VTN PIK3R3 ITGA10 ROCK2 AKT3 |
| Amoebiasis | 13 | 0.00 | COL4A6 COL5A1 CXCL1 IL6 ITGAM ITGB2 LAMA3 LAMA4 SERPINB2 SERPINB3 SERPINB4 TLR2 PIK3R3 |
| Leishmaniasis | 10 | 0.00 | HLA-DPB1 HLA-DRA HLA-DRB5 ITGAM ITGB2 JUN NCF2 NFKBIB PTGS2 TLR2 |
| Malaria | 8 | 0.00 | CD36 CSF3 HGF IL6 IL18 ITGB2 SDC4 TLR2 |
Table 1: KEGG pathway analysis results.
The 98 genes and TP63 in the KEGG pathway analysis results were uploaded to the STRING online tool to analyze the interaction between the proteins encoded by the above genes. The protein interactions were mainly concentrated in JUN, IL6, IL11, B2M, ITGAM, VEGFA. Between CD74 and BMP7, the literature revealed that most of the genes involved in immunerelated mechanisms, and the proteins that interact directly with the protein encoded by tp63 are CD74 and BMP7.
For the above-mentioned genes encoding proteins (CD74 and BMP7) that interact directly with the tp63- encoded protein, GAPDH was used as an internal control and qRT-PCR was used to verify the difference in expression. In the transfected cells, compared with the control group, Smo siRNA expression was upregulated 5.73±0.23 times, BMP7 expression was down-regulated 1.59±0.06 times and CD74 expression was down-regulated 2.65±0.06 times, these difference were statistically significant (p<0.05).
The nuclear transcription factor p63 is an important member of the p53 family and plays a key role in epithelial cell proliferation, differentiation, and tissue growth and development. In MOLT-4 cells, p63 mainly expresses Smo siRNA subtypes and is concentrated in basal cells. In the middle, the parabasal cells also expressed a small amount; with the epithelial differentiation and stratification, the expression of Smo siRNA gradually decreased until disappeared. Previous studies reported from the authors showed that the expression level of Smo siRNA in the MOLT-4 cells was correlated with epithelial-mesenchymal transition (EMT). The expression level of Smo siRNA was also different in MOLT-4 cell line. Among them, Smo siRNA was highly expressed in ME-180 cells and it was in MOLT-4 cells low expression. Therefore, in this study, Smo siRNA was overexpressed in MOLT-4 cells and its expression profile was detected by gene chip. A total of 1405 genes with differential folds >1.5 and p<0.05 were screened (843 up-regulated 562 downregulated). GO clustering analysis of the above genes showed that it mainly involved in cell growth and cycle, signal transduction, cell transfer and invasion. Further analysis of KEGG pathway showed that the differential genes were mainly involved in the immune responserelated signaling pathway. The correlation between the proteins encoded by the 98 genes in the KEGG pathway analysis results showed that the genes encoding the genes directly related to Smo siRNA were BMP7 and CD74, and qRT-PCR was performed, the results are consistent with the chip results.
BMP7 is a member of the transforming growth factorbeta superfamily and is prominently expressed in a variety of epithelial tumors. This study showed that MOLT-4 cells can down-regulate BMP7 expression after Smo siRNA treatment and the EMT process and invasion and metastasis ability of the cells are inhibited. Moreover, in the previous miRNA expression profile data, Smo siRNA treatment directly caused upregulation of miR-22 expression in BMP7 was regulated (fold change=2.32, p<0.05) [4,9]. Therefore, whether miR-22/BMP7 mediates Smo siRNA regulation of MOLT-4 cell EMT is worthy of further study.
CD74, the MHC-II molecular-associated invariant chain, acts as a high-affinity receptor for macrophage migration inhibitory factor (MIF) and mediates MIF-regulated cell survival signals. CD74 can form complexes with MIF and CD44. Activation of NF-κB signaling pathway regulates cell development in MOLT-4 cells. In expression profiles, NF-κB-related factors such as IL6 and TLR2 were down-regulated by 1.72 and 1.61-fold, respectively, after Smo siRNA was up-regulated. The regulatory relationship between Smo siRNA and CD74 deserves further investigation.
In this study, gene chip was used to detect the difference of gene expression profile of MOLT-4 cells treated by Smo siRNA and screened CD74 and BMP7 as candidate genes for Smo siRNA. It is concluded that these two genes may be combined with Smo siRNA. Inducing immune function-related signaling pathways in MOLT- 4 cells, which affects tumor development. Therefore, the next step would be to detect the expression of Smo siRNA, CD74 and BMP7 in the transfected and control cells at the protein level. Overexpression or silencing of CD74 and BMP7 in two cells, comparing changes in Smo siRNA and other immune-related factors, and performing functional experiments such as cell proliferation, invasion and apoptosis, and exploring the role of CD74 and BMP7 in cell function of MOLT-4 cells. The above studies would provide important ideas and clues for further exploration of the mechanism of action of Smo siRNA in MOLT-4 cells.
References
- Simkhovich BZ, Marjoram P, Poizat C. Brief episode of ischemia activates protective genetic program in rat heart: a gene chip study. Cardiovasc Res 2003;59(2):450.
- Evans SJ, Datson NA, Kabbaj M, Thompson RC, Vreugdenhil E, De Kloet ER, et al. Evaluation of Affymetrix Gene Chip sensitivity in rat hippocampal tissue using SAGE analysis. Eur J Neurosci 2002;16(3):409.
- Zou ZK, Huang YQ, Zou Y. Silencing of LSD1 gene modulates histone methylation and acetylation and induces the apoptosis of JeKo-1 and MOLT-4 cells. Int J Mol Med 2017;40(2):319.
- Keller P, Gburcik V, Petrovic N, Gallagher IJ, Nedergaard J, Cannon B, et al. Gene-chip studies of adipogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr Disord 2011; 11(1): 7-7.
- Xu SH, Qian LJ, Mou HZ, Zhu CH, Zhu XM, Liu XL, et al. Difference of gene expression profiles between esophageal carcinoma and its pericancerous epithelium by gene chip. World J Gastroenterol 2003;9(3):417-22.
- Luo Y, Du J, Zhan Z, Chen C, Wang J, Hu Y, et al. A diagnostic gene chip for hereditary spastic paraplegias. Brain Res Bull 2013;97(8):112-8.
- Huang X, Chen S, LI YQ. Effects of Down-regulating bcl11b Gene Expression on Proliferation and Apoptosis of Molt-4 Cells by RNA Interference. Cancer Prevent Res 2010;37(3):247-50.
- Tanja H, Julia K, Victor AW, Hilmar L, Mladen T, Jürgen B, et al. AmpliChip CYP450 GeneChip: a new gene chip that allows rapid and accurate CYP2D6 genotyping. Ther Drug Monit 2006;28(5):673-7.
- Juang JL, Chen TC, Jiang SS, Hsiung CA, Chen WC, Chen GW, et al. Coupling multiplex RT-PCR to a gene chip assay for sensitive and semiquantitative detection of severe acute respiratory syndrome-coronavirus. Lab Invest 2004;84(9): 1085-1091.
- Chen Y, Liu S, Shen Q, Zha X, Yang L, Chen S, et al. Differential Gene Expression Profiles of PPP2R5C-siRNA-Treated Malignant T Cells. DNA Cell Biol 2013;32(10):573-81.