- *Corresponding Author:
- J. Zhang
Department of Pharmacy, Shandong Provincial Hospital Affiliated to Shandong University, Jinan 250016
E-mail: ttdoccn@126.com
| Date of Submission | 14 December 2015 |
| Date of Revision | 22 November 2016 |
| Date of Acceptance | 26 March 2017 |
| Indian J Pharm Sci 2017;79(2):307-311 |
Abstract
This study is aimed to develop and validate a high-performance liquid chromatography-tandem mass spectrometry method for the determination of bile acids in mouse serum. The serum samples were analysed after solid-phase extraction. The analytes were separated on a Diamonsil C18 column with a mobile phase of methanol and water containing 10 mM ammonium acetate and 0.005% formic acid (70:30) at a flow rate of 0.5 ml/min. The analytes were detected by tandem mass spectrometry in negative ion mode. The results demonstrated that the calibration curve was linear for all bile acids over a range of 10~10 000 μg/l. Specificity, matrix effect, recovery, linearity, accuracy and precision were validated for bile acids in mouse serum. The high-performance liquid chromatography-tandem mass spectrometry method was found to be selective, sensitive, simple and was successfully applied to determine the bile acids in more than 200 mouse serum samples. In conclusion, this method is suitable for the quantitative detection of bile acids.
Keywords
Liquid chromatography-tandem mass spectrometry, HPLC-MS/MS, Mice serum, Bile acid
Bile acids account for the majority of lipids in the biliary system, which are derived from cholesterol in liver microsomes and actively transported into bile by a secretory pathway [1]; 95-98% of the bile acids will be reabsorbed at the terminal ileum after reaching the small intestine and be transported back to the liver through the portal vein, resulting in the enterohepatic circulation of bile acids [2]. Hepatobiliary and gastrointestinal diseases have been reported to cause changes in bile acid metabolism, while many other diseases might cause changes in bile acid metabolism to different degrees [3-10]. Therefore, it is necessary to establish a simple, rapid and effective bioanalytical method for detecting bile acids in different biological matrices. However, this detection is difficult owing to the complex nature and low concentrations of bile acids in biological fluids.
Currently, the detection and quantification methods for bile acids mainly focus on high- performance liquid chromatography (HPLC), gas chromatography/mass spectrometry (GC/MS), and high-performance liquid chromatography-tandem mass spectrometry (HPLCMS/ MS), among other methods [11]. The GC/MS method required bile acids to be treated by hydrolysis and derivatization according to their category [12,13]. The HPLC method also required fluorescence derivatization [14], whereas taurine-conjugated bile acids require derivatization after hydrolysis, but the HPLC-MS/MS method did not require hydrolysis and derivatization, making it a relatively simple operating method for detecting various types of bile acids in serum at the same time with the advantages of being fast, reproducible and relatively sensitive with high separation efficiency.
In this investigation, our aim was to develop a HPLCMS/ MS method for the determination of bile acids with an advantage of being rapid, involving easy sample preparation and clean-up processes without requiring additional complex derivatization techniques. The developed method was successfully applied to quantifying bile acids in more than 200 mice sera samples.
Cholic acid (CA), taurocholic acid (TCA), deoxycholic acid (DCA), ursodeoxycholic acid (UDCA), lithocholic acid (LCA) and activated charcoal were purchased from Sigma-Aldrich, USA. β-Mouse cholic acid (β-MCA) was purchased from Steraloids Inc., USA. The internal standard, cholic-2,2,4,4-d4 acid (CA-d4) was purchased from CDN Isotopes, Quebec, Canada. Methanol and acetonitrile were purchased from J.T. Baker Solusorb Co., USA. Ethanol was purchased from Tianjin Kemiou Chemical Reagents Ltd. Co., China and all chemicals were of chromatographic purity. Analytical grade ammonium acetate and formic acid was purchased from Chemical Technology Academy of Shandong Province, China. Wahaha purified water was obtained from the local market.
Accurately weighed quantities (0.010 g) of CA, TCA, DCA, UDCA, LCA, and the β-MCA standard were dissolved in anhydrous methanol to prepare solutions of 10 ml. Then, for each standard sample, 1 mg/ml working standard solutions were prepared. Appropriate amounts of the working standards were accurately weighed out and diluted with anhydrous methanol to prepare three different concentrations (100 ng/ ml, 1 μg/ml and 10 μg/ml) of the six mixed standard solutions. All solutions were kept in a refrigerator at 4°. Accurately weighed amount of the CA-d4 standard was dissolved in anhydrous methanol to prepare a 1 μg/ ml internal standard solution.
Normal mouse serum was extracted and mixed with 100 mg/ml activated carbon with mild agitation. The mixture was stored at room temperature overnight (around 17 h) and centrifuged at 19 500 rpm for 1 h. The supernatant was filtered through a filter membrane (MCM0.22 μm, Agela Technologies Co.) and transferred to a clean tube to obtain mouse blank serum, which was either used for testing or stored at –70° [12].
The chromatographic system consisted of an Agilent 1200/6410 series HPLC system (Agilent Technologies, Waldbronn, Germany) with an on-line degasser. Chromatographic separation was carried out on a Diamonsil C18 chromatographic column (150×4.6 mm, 5 μm). The mobile phase consisted of methanol:water (containing 10 mM ammonium acetate and 0.005% formic acid, 70:30, v/v). The injection volume was 10 μl and the mobile phase flow rate was 0.5 ml/min. The column oven temperature was set to 25°.
The AP-electrospray ionization (ESI) ion source was operated in the negative ion mode, using the following settings: dry gas engine (nitrogen) flow velocity 9.0 l/min, dry gas pressure 45 psi, dry gas temperature 350° and capillary voltage 4000 V. Analytes were monitored using multiple reaction monitoring (MRM) and the selected ion pairs were, 514.3→514.3 (TCA, β-MCA), 407.4→407.4 (CA), 391.4→391.4 (UDCA, DCA), 375.4→375.4 (LCA), and 411.4→411.4 (CAd4) at debris voltages of 130, 120, 140, 130, and 120 V, respectively and the collision energy was 0 eV.
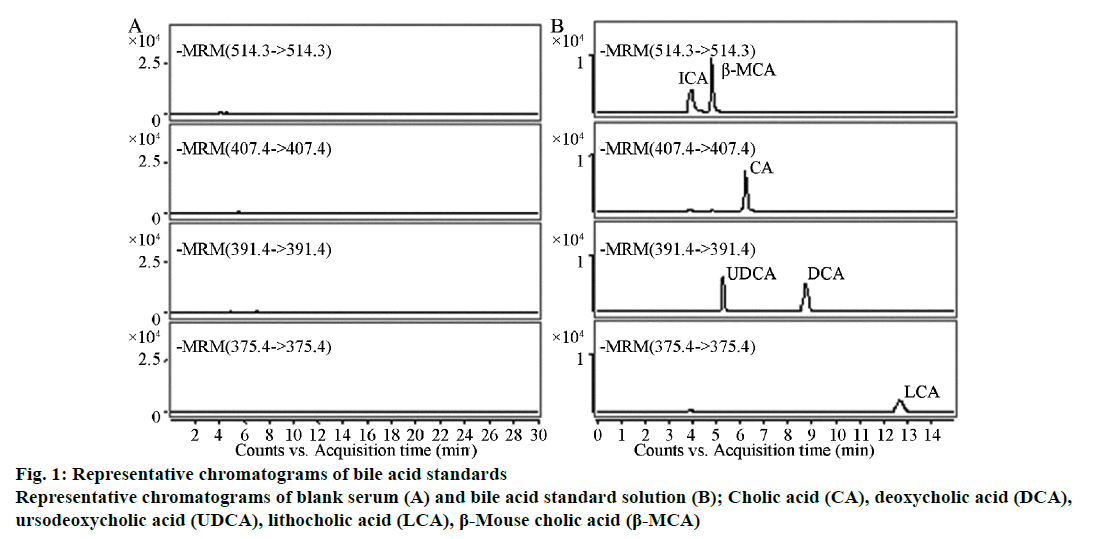
A 200-μl sample of mouse serum was mixed with 50 μl of a 1 μg/ml internal standard solution and 1 ml of 0.05% formic acid. After mixing thoroughly by vortexing for 2 min, the mixture was loaded onto the HLB Cartridge extraction column (Waters Co.), which had been pre-conditioned with 1 ml of methanol and 1 ml of 0.05% formic acid. The cartridge was subsequently washed with 1 ml of water and 1 ml of 5% methanol. Bile acids were eluted out with 1 ml of methanol and 2 ml of acetonitrile. The eluent was dried at 60° under a nitrogen stream, and the residue was dissolved in 100 μl of 90% methanol. After vortexing for 1 min until thoroughly mixed and ultrasonicating for 1 min, 10 μl of the sample was injected into the HPLC-MS/MS system. The blank serum (a) and bile acid standard solution (b) were separately injected into the HPLC-MS/MS system for analysis.
To prepare different concentrations (10, 25, 50, 100, 200, 500, 1000, 10 000 ng/ml) of bile acid serum samples, appropriate amounts of standard solution was dried with nitrogen and then dissolved in 200 μl of mouse blank serum. Following the sample processing operation, five samples were analysed for each concentration, and the chromatograms were recorded; the bile acid concentration (x) was set as the abscissa, the bile acid and internal standard peak area ratio (y) was set as the ordinate, and a weighted least squares method (W=1/x2) was used to establish the standard curve.
The low, medium, and high concentrations (10, 100, 10 000 ng/ml) of the bile acid standard solutions were analysed, and the corresponding peak area was designated A1; blank serum was used for sample processing (without the internal coordinate), using 100 μl of the 5, 50, or 5000 ng/ml bile acid standard solution to dissolve it, and the corresponding peak area was designated A2. Appropriate amounts of blank serum were prepared with the low, medium and high concentrations (10, 100, 10 000 ng/ml) of the bile acid serum samples following the sample processing operation (without the internal coordinate) and the corresponding peak area was designated A3. The A2/A1 ratio was used to calculate the influence of the matrix on bile acids at the low, medium and high concentrations. The A3/A2 ratio was used to calculate the absolute recovery of bile acids at the low, medium and high concentrations.
For determining the precision and accuracy, appropriate amount of blank serum was used to prepare serum samples of low, medium and high (10, 100, 10 000 ng/ ml) concentrations following the sample processing operation. The intra-day precision and accuracy were evaluated by analysing five duplicates of the low, medium and high concentrations in 1 d. The inter-day precision and accuracy were obtained by continuous detection of the low, medium, and high concentrations over 3 d.
Results indicated that each ingredient had no other peak impurities to interfere with the analysis and the reserving times were TCA, 4.18 min; β-MCA, 4.82 min; CA, 6.09 min; UDCA, 5.36 min; DCA, 8.76 min; and LCA, 12.96 min (Figure 1). The blank serum processed with activated carbon contained no significant endogenous impurities that interfered at the peak positions of the six types of bile acid standards, indicating that the method had good specificity.
All the bile acid standard curves and correlation coefficients were listed in Table 1. The results showed that the curves for all bile acid standards were linear between 10 and 10 000 ng/ml, which indicated a good range. The results presented in Table 2 indicated that the blank serum processed with activated carbon was appropriate for use as a standard curve for the blank matrix and had no significant matrix effect. The results of Table 3 showed that the RSD for all ingredients was below 12% with good precision. The average extraction recovery for all the ingredients was within 50~90% and the average relative recovery was within 90~115%.
| Bile acids | Linear regression equation y=ax+b | Correlation coefficient γ |
|---|---|---|
| CA | y=0.0049x+0.0054 | 0.9909 |
| TCA | y=0.0052x-0.0063 | 0.9903 |
| DCA | y=0.0052x-0.0056 | 0.9913 |
| UDCA | y=0.0036x-0.0086 | 0.9927 |
| LCA | y=0.0028x-0.0045 | 0.9919 |
| β-MCA | y=0.0036x+0.0313 | 0.9908 |
Table 1:The calibration curve of all the bile acids
| Bile acids | Low concentration (10 ng/ml) | Moderate concentration (100 ng/ml) | High concentration (10 000 ng/ml) |
|---|---|---|---|
| CA | 99.23±15.18 | 99.75±5.20 | 98.54±4.09 |
| TCA | 111.38±17.40 | 99.44±4.03 | 101.70±3.48 |
| DCA | 102.93±14.49 | 100.09±7.96 | 98.97±2.96 |
| UDCA | 109.51±16.29 | 89.88±3.09 | 108.36±5.53 |
| LCA | 90.17±15.99 | 100.40±7.67 | 96.12±7.42 |
| β-MCA | 97.34±11.47 | 101.11±2.36 | 100.53±5.15 |
Table 2:The matrix effects of blank serum
| Bile acids | Precision (RSD%) | Recovery (%) | |||
|---|---|---|---|---|---|
| Inter-day | Intra-day | Extraction | Relative | ||
| CA | Low concentration (10 ng/ml) | 4.3 | 3.0 | 78.25 | 108.52 |
| Moderate concentration (100 ng/ml) | 3.1 | 4.4 | 89.16 | 95.80 | |
| High concentration (10 000 ng/ml) | 1.4 | 1.4 | 82.65 | 104.86 | |
| TCA | Low concentration (10 ng/ml) | 3.6 | 11.3 | 80.73 | 105.22 |
| Moderate concentration (100 ng/ml) | 2.2 | 7.2 | 72.36 | 96.16 | |
| High concentration (10 000 ng/ml) | 7.9 | 1.1 | 72.44 | 93.44 | |
| DCA | Low concentration (10 ng/ml) | 9.0 | 6.9 | 79.15 | 107.56 |
| Moderate concentration (100 ng/ml) | 5.3 | 6.4 | 68.92 | 93.20 | |
| High concentration (10 000 ng/ml) | 0.7 | 4.4 | 65.03 | 112.70 | |
| UDCA | Low concentration (10 ng/ml) | 5.1 | 6.6 | 87.52 | 100.30 |
| Moderate concentration (100 ng/ml) | 1.7 | 4.8 | 87.17 | 90.82 | |
| High concentration (10 000 ng/ml) | 2.8 | 2.8 | 76.91 | 109.34 | |
| LCA | Low concentration (10 ng/ml) | 10.6 | 7.5 | 59.52 | 102.14 |
| Moderate concentration (100 ng/ml) | 4.4 | 5.7 | 56.76 | 93.36 | |
| High concentration (10 000 ng/ml) | 2.6 | 1.0 | 64.34 | 111.30 | |
| β-MCA | Low concentration (10 ng/ml) | 5.2 | 7.0 | 89.75 | 96.80 |
| Moderate concentration (100 ng/ml) | 2.3 | 5.0 | 83.05 | 111.28 | |
| High concentration (10000 ng/ml) | 3.5 | 0.7 | 82.95 | 91.50 | |
Table 3:The precision and recovery of all the bile acids
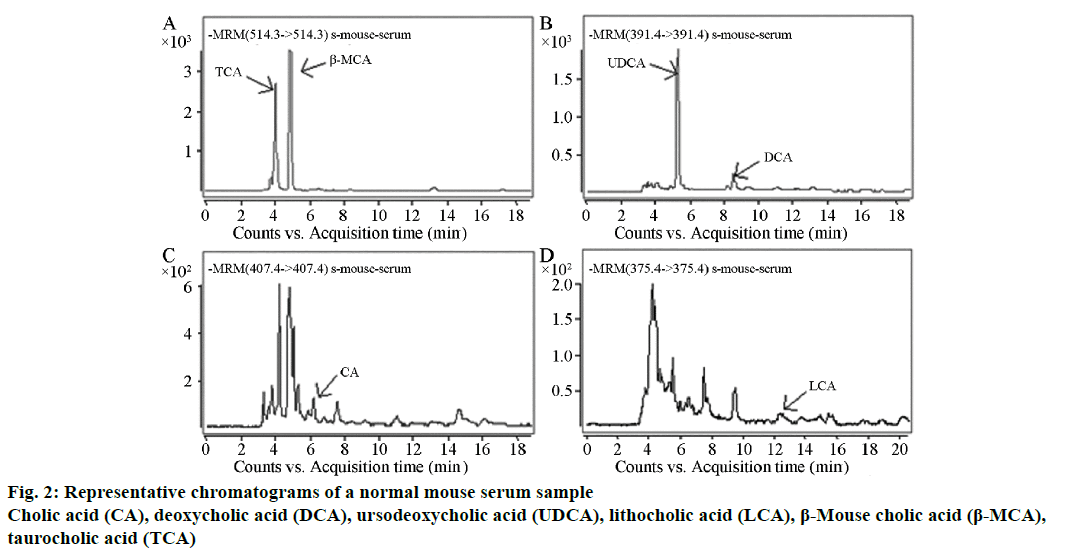
Forty samples of mouse serum were analysed by the established method. The bile acids that needed to be determined were completely separated within 20 min; this method performed well in terms of stability. One of the results from a normal mouse is shown in Figure 2, TCA, 96.25 ng/ml; β-MCA, 115.95 ng/ml; UDCA, 88.12 ng/ml; DCA, 9.02 ng/ml; CA, 3.31 ng/ml and LCA, 3.21 ng/ml.
According to a previous report [15], most bile acid HPLC/ MS/MS determination methods applied the gradient elution method. In this study, the mobile phase was methanol-water containing 10 mM ammonium acetate and 0.005% formic acid (70:30), which also achieved good separation. The MS conditions used were an AP-ESI ion source, negative ion mode and MRM modes. The response enhancement was not obvious when attempting to use different collisions to select the detection of ion pairs, so the final choice for the collision could be located for 0 eV. Agilent and Waters column extractors were used in this study and owing to the lower recovery with the Agilent column, the HLB Cartridge Waters column was selected. After the extract was dried with a nitrogen stream, an attempt was made to dissolve it with the mobile phase, but the peak form of β-MCA was not good. Many different proportions of methanol-water solutions were evaluated, and a 90% methanol solution was found to be the best for dissolving β-MCA to obtain a good peak.
The method established in this study had good specificity, as each bile acid composition produced a good linear curve over 10~10 000 μg/l, the inter-day and intra-day precision RSD values were all below 12%, the average extraction recovery of each ingredient was 50~90%, and the average relative recovery was 90~115%. Therefore, this method is sensitive, specific, simple and reproducible with a short analysis time and a wider linear range, which makes it suitable for the quantitative detection of bile acids in mouse serum for scientific research.
Conflict of interests:
Authors report no conflict of interests.
Financial support and sponsorship:
Nil.
References
- Steiner C, von Eckardstein A, Rentsch KM. Quantification of the 15 major human bile acids and their precursor 7α-hydroxy-4-cholesten-3-one in serum by liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2010;878:2870-80.
- Roberts MS, Magnusson BM, Burczynski FJ, Weiss M. Enterohepatic circulation: physiological, pharmacokinetic and clinical implications. Clin Pharmacokinet 2002;41:751-90.
- Tazuma S, Kanno K, Sugiyama A, Kishikawa N. Nutritional factors (nutritional aspects) in biliary disorders: Bile acid and lipid metabolism in gallstone diseases and pancreaticobiliary maljunction. J Gastroenterol Hepatol 2013; 4:103-7.
- HofmannAF. Bile acids: trying to understand their chemistry and biology with the hope of helping patients. Hepatology 2009;49:1403-18.
- Wilcox C, Turner J, Green J.Systematic review: the management of chronic diarrhoea due to bile acid malabsorption.Aliment Pharmacol Ther 2014;39:923-39.
- Chiang GY. Bile acids: regulation of synthesis. J Lipid Res 2009;50:1955-66.
- Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 2009;89:147-91.
- Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res 2009;50:1509-20.
- Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011;141:1773-81.
- Tsuei J, Chau T, Mills D, Wan YJ. Bile acid dysregulation, gut dysbiosis, and gastrointestinal cancer. Exp Biol Med 2014;239:1489-504.
- Burkard I, von Eckardstein A, Rentsch KM. Differentiated quantification of human bile acids in serum by high-performance liquid chromatography-tandem mass spectrometry. J Chromatogr B Analyt Technol Biomed Life Sci 2005;826:147-59.
- Birk JJ, Dippold M, Wiesenberg GL, Glaser B. Combined quantification of faecal sterols, stanols, stanones and bile acids in soils and terrestrial sediments by gas chromatography-mass spectrometry. J Chromatogr A 2012;1242:1-10.
- Matysik S, Schmitz G. Application of gas chromatography-triple quadrupole mass spectrometry to the determination of sterol components in biological samples in consideration of the ionization mode. Biochimie 2013;95:489-95.
- You J, Fu Y, Sun Z, Suo Y. 2-(5-Benzoacridine)ethyl-p-toluenesulfonate as sensitive reagent for the determination of bile acids by HPLC with fluorescence detection and online atmospheric chemical ionization-mass spectrometric identification. Anal Bioanal Chem 2010;396:2657-66.
- Xiang X, Han Y, Neuvonen M, Laitila J, Neuvonen PJ, Niemi M. High performance liquid chromatography-tandem mass spectrometry for the determination of bile acid concentrations in human plasma. J Chromatogr B Analyt Technol Biomed Life Sci 2010;878:51-60.