- *Corresponding Author:
- W. Q. Song
Department of Rehabilitation, Xuanwu Hospital, Capital Medical University, Beijing 100053, China
E-mail: songwq66@126.com
| This article was originally published in a special issue, “Evolutionary Strategies in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2020:83(3) Spl issue;6-13 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
Osteoporosis is a common bone disease; however, its pathophysiology is yet unclear. This study aimed to investigate candidate genes in osteoporosis and its pathomechanism. Microarray datasets Genomic Spatial Event-7429, 13850, 56815, and 7158, comprising data obtained from blood samples of osteoporosis patients and healthy controls, were downloaded from the Gene Expression Omnibus database. Differentially expressed genes were identified via intersection of the four datasets, using Affy and Limma software packages. Functional and pathway enrichment analysis of differentially expressed genes were conducted using the database for annotation, visualization, and integrated discovery database. Thereafter, protein-protein interactions between the products of differentially expressed genes and key modules were analyzed using the search tool for the retrieval of interacting genes/proteins database and Cytoscape software. Furthermore, a transcriptional regulatory network was established with the differentially expressed genes, using the Web-based gene set analysis toolkit database. In total, 702 differentially expressed genes were filtered from the intersection of the four datasets, which were primarily enriched in functions and pathways associated with glutamate secretion, Adenosine triphosphate binding, extracellular region, and phosphatidylinositol 3-kinase-protein kinase B signaling pathway. Furthermore, 361 nodes and 846 edges were present in the protein-protein interaction network. Two significant modules were obtained, each containing 22 key genes. Transcriptional factor-target gene regulatory network analysis revealed two vital transcription factors, Forkhead Box O4 and LIM Homeobox 3. Genes including endothelin receptor type A, XC chemokine receptor 1, 5-hydroxytryptamine receptor 2a, phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta-4, glutamate metabotropic receptor 5, kiss-1 metastasis suppressor, 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta-2, cholinergic receptor muscarinic 3, g protein subunit gamma 4, neurotensin receptor 1, neuromedin U, neuromedin B, breast cancer gene 1 associated RING domain 1, exonuclease 1, replication factor C subunit 2, restriction site associated DNA 52 homolog DNA repair protein, DNA topoisomerase III alpha, Hemolytic uremic syndrome 1 checkpoint clamp component, timeless circadian regulator, breast cancer gene 1 DNA repair associated, tumor protein p53 and claspin are potential key genes for osteoporosis and may help elucidate the underlying pathomechanism, and transcription factors Forkhead Box O4 and LIM Homeobox 3 are potential therapeutic targets for osteoporosis.
Keywords
Osteoporosis, differentially expressed genes, bioinformatics, pathway, protein-protein interaction network
Osteoporosis is a common bone disease, characterized by low bone mass, damaged bone microstructure, increased bone brittleness, and increased susceptibility to fracture. Osteoporosis has no obvious early-stage manifestations; however, it presents with severe symptoms upon detection [1] Therefore, osteoporosis is also known as a “silent disease.” In recent years, osteoporosis and its complications including deformity, disability, and depression have been a major public health concern among ageing populations worldwide. Race, age, sex, other bone metabolic diseases (endocrine system diseases, rheumatic immune diseases, etc.), drugs, and lifestyle contribute to osteoporosis pathogenesis [2]. The cost of nursing and medical care for osteoporosis individuals is enormous, imposing a heavy burden on the family and society [3].
The potential mechanism underlying osteoporosis pathogenesis involves an imbalance between bone resorption and formation [2]. Branden reported that tumor necrosis factor (TNF) receptor-associated factor 3 (TRAF3) down regulation in bone tissue potentially increases osteoclastogenesis and reduces osteoblastogenesis through the nuclear factor kappa light chain enhancer of activated B cells (NF-Κb) signaling pathway [4]. H3 lysine 36 trimethylation (H3K36me3) mediated by SET-domain-containing 2 (SETD2) potentially regulates the cell fate of bone marrow stromal cells (BMSCs) [5]. Family with sequence similarity 210 member A (FAM210A) reportedly regulates bone structure and function and may impact osteoporosis [6]. Uehara reported that protein kinase N3 promotes osteoclast-mediated bone resorption in response to Wingless-related integration site (Wnt) family member 5A-receptor tyrosine kinase like orphan receptor 2 (Wnt5a-Ror2) signaling [7].
Furthermore, numerous studies have verified that the NF-κB signaling pathway, Notch signaling pathway, Wnt/β-catenin pathway, and phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) (PI3K/AKT) pathway are associated with the regulation of BMSC differentiation during osteoporosis. However, the mechanisms underlying BMSC differentiation remain unclear.
Microarray technology has contributed significantly to advancements in the identification of differentially expressed genes (DEGs), functional genes, and gene regulatory networks associated with disease [8]. It has advantages including small size, rapidity, accuracy, high sensitivity, and parallel detection of large data on the same chip. However, these results are generated via a single queue and are always inconsistent among different studies because of organizational or sample heterogeneity among individual experiments, different technological testing platforms, and various data processing methods [9]. In this study, 4 Messenger Ribonucleic acid (mRNA) microarray datasets from Gene Expression Omnibus (GEO) were downloaded and analyzed to identify DEGs in blood samples obtained from osteoporosis patients and healthy controls. Further, we carried out Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and constructed a protein-protein interaction (PPI) network and interaction network between transcription factors (TFs) and DEGs. This study aimed to identify more reliable potential modulators and pathophysiological mechanisms of osteoporosis.
Materials and Methods
Gene datasets:
We obtained four gene expression datasets, (Genomic spatial event (GSE) (GSE7429, GSE13850, GSE56815, and GSE7158) from the GEO database [10]. Thereafter, we screened the four original microarray datasets and analyzed differential gene expression profiles between blood samples from osteoporosis patients and healthy controls among premenopausal or postmenopausal women. According to the diagnostic criteria recommended by World Health Organization (WHO), bone mineral density (BMD) should be measured via dual energy X-ray absorptiometry (DXA). Patients with a hip or lumber T-score ≤-2.5 were identified to have osteoporosis [2]. All women were divided into low and high BMD groups.
Data preprocessing and analysis of DEGs:
The Affy package with the R package (version 3.5.1) was used to preprocess the original data from the four datasets, including quantile normalization, log2 transformation, and the K-nearest neighbors algorithm to supplement missing values [11]. The Limma package in R was used to identify DEGs in the high BMD and low BMD groups [12]. A corrected p-value <0.01 was selected as the threshold for DEG screening. Consequently, data from different datasets were intersected and analyzed to obtain 702 DEGs.
Gene ontology (GO) and signaling pathway enrichment analysis of DEGs:
GO and KEGG pathway enrichment analyses were performed using database for annotation, visualization and integrated discovery (DAVID) database to identify functional annotations (molecular function, biological process, and cellular component) and pathways involving DEGs. A p-value of <0.05 was considered the threshold [13].
Construction of the PPI network and module analysis:
Search tool for the retrieval of interacting genes/ proteins (STRING) database (version 11.0), a PPI network, helped to identify core regulatory genes [14]. We used a confidence score of >0.7 as the cut-off criterion. Furthermore, we constructed a visualized PPI network, using Cytoscape software (version 3.7.0), an open source software platform for visualizing complex networks and integrating them with any type of attribute data [15]. Molecular Complex Detection (MCODE), a plug-in for Cytoscape, was used to construct functional modules by clustering in a huge protein network and information of each node was determined to obtain the most important visualized model data [16]. The screening requirements were as follows: MCODE scores >5, degree cut-off=2, node score cut-off=0.2, Max depth=100, and k-score=2.
Construction of a TF-target gene regulatory network:
Regulation of target genes by TFs plays an important role in diseases [17]. We analyzed the WEB based gene set analysis toolkit (WebGestalt) database via the overrepresentation enrichment analysis (ORA) method to construct a TF-target gene regulatory network [18]. Cytoscape software (version 3.7.0) was used to construct a TF-target gene regulatory network. A false discovery rate of <0.05 was considered the cut-off criterion.
Results and Discussion
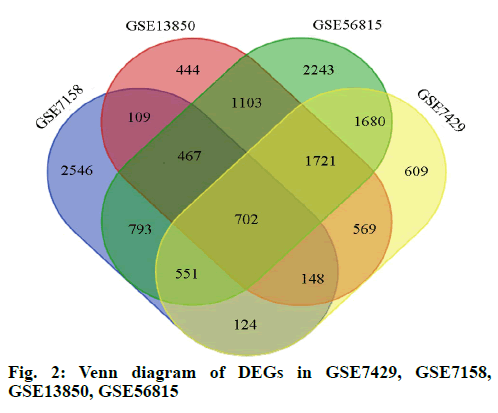
In this study, we performed four microarray analyses. After processing the data, we identified 5440 DEGs in GSE7158, 6104 DEGs in GSE7429, 5263 DEGs in GSE13850, and 9260 DEGs in GSE56815 (fig. 1). The intersection of the four datasets yielded 702 genes, as obtained through a Venn diagram (fig. 2) between low BDM and high BDM women.
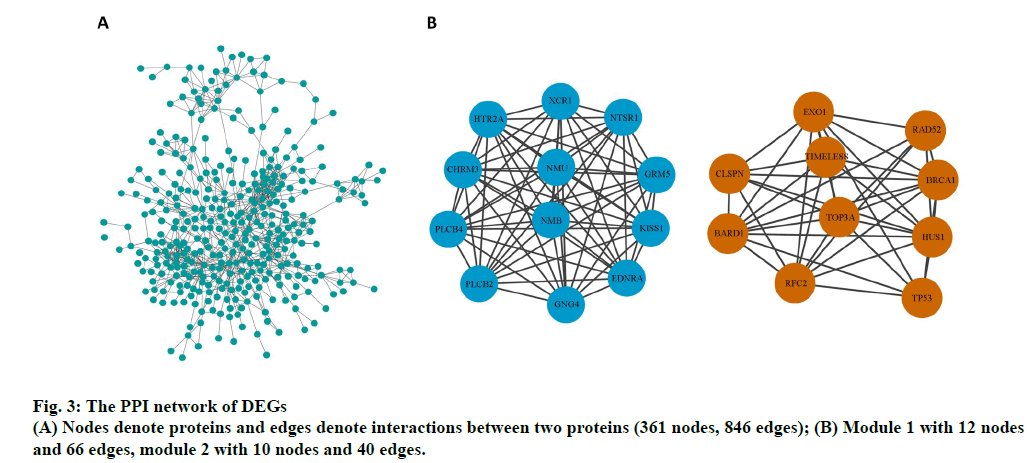
The PPI network of DEGs and the most significant module were established using Cytoscape software. The PPI network comprised 361 nodes and 846 edges (fig. 3A). Two significant modules (module 1and 2) were obtained using the MCODE plug-in. Genes in the two modules include endothelin receptor type A (EDNRA), XC chemokine receptor 1 (XCR1), 5-hydroxytryptamine receptor 2a (HTR2A), phosphatidylinositol-4,5- bisphosphate phosphodiesterase beta-4 (PLCB4), glutamate metabotropic receptor 5 (GRM5), kiss-1 metastasis suppressor (KISS1), 1-phosphatidylinositol- 4,5-bisphosphate phosphodiesterase beta-2 (PLCB2), cholinergic receptor muscarinic 3 (CHRM3), G protein subunit gamma 4 (GNG4), neurotensin receptor 1 (NTSR1), neuromedin U (NMU), neuromedin B (NMB), breast cancer gene 1 associated RING domain 1 (BARD1), exonuclease 1 (EXO1), replication factor C subunit 2 (RFC2), restriction site associated DNA 52 homolog DNA repair protein (RAD52), DNA topoisomerase III alpha (TOP3A), Hemolytic uremic syndrome 1 checkpoint clamp component (HUS1), timeless circadian regulator (TIMELESS), breast cancer gene 1 DNA repair associated (BRCA1), tumor protein p53 (TP53) and claspin (CLSPN) (fig. 3B).
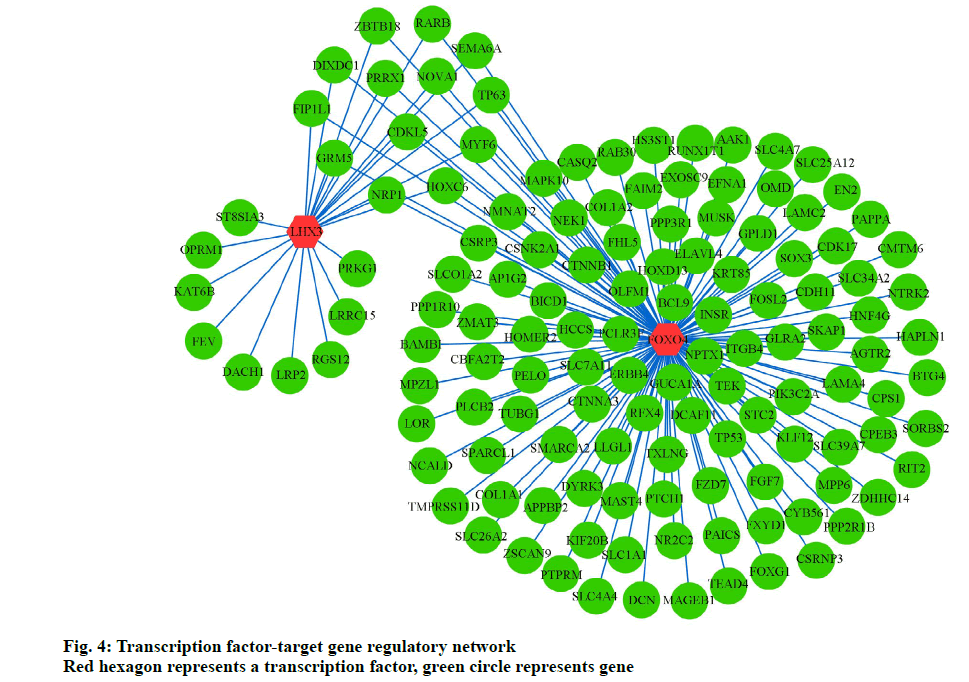
To further elucidate the association between TFs and target genes, we analyzed the WebGestalt database to construct a TF-target gene network, using Cytoscape software. Two most vital TFs, Forkhead Box O4 (FoxO4) and LIM Homeobox 3 (LHX3), and 145 interactions pairs were obtained (fig. 4).
Osteoporosis is a systemic skeletal disease characterized by low BMD, occurring in the aged population and represents increasing burdens on society. In summary, 702 DEGs among the 4 databases, 22 hub genes and 2 TFs were identified, which are potential key genes in osteoporosis.
To identify interactions among DEGs, GO and KEGG enrichment analyses were carried out. Molecular function upon GO enrichment analysis was primarily glutamate binding, protein kinase activity, and protein tyrosine kinase activity. Previous studies have reported that these functions play important roles in osteoporosis progression. The expression of glutamate occurs in both osteoblasts and osteoclasts. Glutamate is vital for bone formation and resorption. Glutamate transporters are potentially applicable for enhancing the osteogenic ability of osteoblasts [19]. BMSC proliferation during osteoporosis was primarily determined on the basis of protein kinase B (Akt)/mammalian target of rapamycin (mTOR) activity [19]. Protein tyrosine kinase activity is essential for the regulation of bone resorption in osteoclasts [20].
In this study, KEGG analysis revealed that the important pathways are the PI3K-AKT and Wnt signaling pathways. Recent studies reported that these pathways may regulate osteoclast and osteoblast activity during osteoporosis. Osteoblast differentiation is reportedly regulated by MicroRNA 216a (miR-216a) in the PI3K-AKT signaling pathway [1]. Similarly, Zhao reported that the Phosphatase and tensin homolog (PTEN)/PI3K-AKT signaling pathway promotes osteoclastogenesis during osteoporosis [21]. Ge reported that the Wnt/β‑catenin signaling pathway plays a vital role in promoting osteogenesis [22]. Boyden reported that Wnt signaling is involved in bone metabolism through Low-density lipoprotein receptor-related protein 5 (LRP5) [23]. Furthermore, some studies strongly suggest that BMD is associated with Wnt signaling.
On construction of the PPI network using Cytoscape software, we obtained two significant modules, including 22 key genes (EDNRA, XCR1, HTR2A, PLCB4, GRM5, KISS1, PLCB2, CHRM3, GNG4, NTSR1, NMU, NMB, BARD1, EXO1, RFC2, RAD52, TOP3A, HUS1, TIMELESS, BRCA1, TP53, and CLSPN) potentially significant in osteoporosis. BRCA1 inhibits postnatal osteoblastic osteogenesis through Mitogen-activated protein kinase (MAPK) signaling pathways [24]. A bioinformatics analysis reported that BRCA1 is a potential key gene for bone regeneration and repair during osteopenia in osteoporosis [25]. BRCA1 is a tumor suppressor in osteosarcomas and regulates tumorigenesis [26,27]. Johnson reported that EDNRA inhibits the marked endothelin-1-mediated increase in mineralization. NMU, a small peptide, influences bone rebuilding in conditions of low bone mass, such as osteoporosis, owing to leptin-dependent regulation [28]. Dimitri reported that neuromedin U is potentially involved in the regulation of the bone remodeling [29]. Knockdown of neuromedin B could expedite apoptosis in osteoclast lineage cells and reduce proliferation, thus decreasing bone mass [30]. A previous study suggested that TP53 regulates osteogenic differentiation of dental stem cells; however, the pathomechanism of osteogenesis has yet been unclear [31]. In this study, significant differences were observed in the expression of the aforementioned genes between blood samples of osteoporosis patients and healthy controls. This study shows that these genes are essential markers in bone metabolism. Further studies are required to further elucidate the mechanism underlying osteoporosis.
HUS1 protects against the toxic effects of formaldehyde on mouse bone marrow mesenchymal stem cells [32]. GRM5 expression is reportedly associated with bone cancer pain in a mouse model [33]. TOP3A, a vital microsatellite marker, plays a potentially significant role in high-grade osteosarcoma and influences tumorigenesis [34]. XCR1 is essential in cell proliferation and migration and has been correlated with bone metastasis [35]. XCR1 is a potential diagnostic marker for large B-cell lymphoma demonstrating bone marrow involvement [36]. These four genes are associated with bone tumor and bone mass; however, no direct evidence is available to determine the effect of these genes on osteoblast/osteoclast differentiation and the pathophysiology of osteoporosis.
HTR2A, BARD1, TIMELESS, PLCB2, RFC2, CHRM3, GNG4, PLCB4, CLSPN, EXO1, RAD52, NTSR1, and BRCA1 are key molecular players in osteoporosis in this study. HTR2A, a neuronal marker, is involved in differentiation of bone marrow mesenchymal stem cells into neuronal lineage cells [37]. BARD1 and TIMELESS are potential biomarkers for breast cancer [38,39]. PLCB2 and RFC2 play a potentially important role in osteosarcoma [40,41]. CHRM3, GNG4, and PLCB4 regulate colon cancer, glioblastoma, and gastrointestinal stromal tumors, respectively [42-44]. CLSPN, EXO1, and RAD52 are associated with DNA replication checkpoints, regulation of DNA, and DNA repair, respectively [45-47]. NTSR1 interacts with lipids [48]. No study has reported the association between these genes and osteoporosis. However, our results suggest that they are potential key factors involving in osteoporosis regulation and warrant further clinical and in vitro analyses.
In the present study, FoxO4 and LHX3 were identified as key TFs in the TF‑target gene network, playing an important role in osteoporosis. The FoxO TF family, comprising FoxO1, FoxO3a, FoxO4, and FoxO6, is involved in cellular survival, the oxidative stress response, and carcinogenesis [49]. FoxO4 mediates osteoblast differentiation by deacetylating FoxO4 in progenitors [50]. Iyer reported that FoxO4 attenuates osteogenesis during osteoporosis through the Wnt pathway [51]. Furthermore, Ambrogini reported that FoxO4 helps defend against oxidative stress in osteoblasts and regulates bone mass homeostasis in mice [52]. Moreover, FoxO4 participates in the tumorigenic regulation in different cancers including gastric cancer, colorectal cancer, hepatocellular carcinoma, prostate cancer, breast cancer, and lung cancer [49]. Roupe reported that FoxO4 is involved in wound healing and tissue regeneration [53]. LHX3, a typical LIM family TF, is reportedly associated with regulation of pituitary hormones, thyroid function, and the β-subunit of follicle-stimulating hormone (FSHB) [54-56]. This study predicts that LHX3 is a potentially important TF in monitoring osteoporosis. These results warrant verification through further studies.
This study has two primary limitations. First, hub genes were identified using bioinformatics approaches without experimental verification. Further studies are required to verify these results using in vitro analyses including quantitative real-time reverse transcription polymerase chain reaction (RT-PCR) and western blot analyses. Second, all data were standardized; however, there may be heterogeneity among individual microarray studies using different platforms and clinical samples, such as assets, sex, or geographical regions. In this study, we intersected 4 datasets, with differences among different algorithms.
In conclusion, this study revealed 702 DEGs via bioinformatics analysis. Thereafter, we 22 hub genes and 2 TFs were identified to be involved in osteoporosis pathophysiology and serve as potential therapeutic targets for osteoporosis. However, further studies are required to verify the function of these genes in osteoporosis.
Author’s contributions:
Huan Xin Xie and Lei Cao carried out the studies, participated in collecting data, and drafted the manuscript. Lin Lin Ye and Jie Hu performed the statistical analysis and participated in its design. Gui Xiang Shan and Wei Qun Song helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgements:
We would like to thank all the lab tutors for participating in this study.
Funding:
The work was supported by the National Science Foundation of China (No. 303-01-002-0155).
Conflict of Interests:
The authors declared no conflict of interest.
References
- Li H, Li T, Fan J, Fan L, Wang S, Weng X, et al. miR-216a rescues dexamethasone suppression of osteogenesis, promotes osteoblast differentiation and enhances bone formation, by regulating c-Cbl-mediated PI3K/AKT pathway. Cell Death Differ 2015;22:1935-45.
- Compston J, Cooper A, Cooper C, Gittoes N, Gregson C, Harvey N, et al. UK clinical guideline for the prevention and treatment of osteoporosis. Arch Osteoporos 2017;12:43.
- Tarantino U, Iolascon G, Cianferotti L, Masi L, Marcucci G, Giusti F, et al. Clinical guidelines for the prevention and treatment of osteoporosis: summary statements and recommendations from the Italian Society for Orthopaedics and Traumatology. J Orthop Traumatol 2017;18:3-6.
- Boyce BF, Li J, Xing L, Yao Z. Bone remodeling and the role of TRAF3 in osteoclastic bone resorption. Front Immunol 2018;9:2263.
- Wang L, Niu N, Li L, Shao R, Ouyang H, Zou W. H3K36 trimethylation mediated by SETD2 regulates the fate of bone marrow mesenchymal stem cells. PLoS Biol 2018;16:e2006522.
- Tanaka KI, Xue Y, Nguyen-Yamamoto L, Morris JA, Kanazawa I, Sugimoto T, et al. FAM210A is a novel determinant of bone and muscle structure and strength. Proc Natl Acad Sci USA 2018;115:E3759-68.
- Uehara S, Udagawa N, Mukai H, Ishihara A, Maeda K, Yamashita T, et al. Protein kinase N3 promotes bone resorption by osteoclasts in response to Wnt5a-Ror2 signaling. Sci Signal 2017;10.
- Yang X, Zhu S, Li L, Zhang L, Xian S, Wang Y, et al. Identification of differentially expressed genes and signaling pathways in ovarian cancer by integrated bioinformatics analysis. Onco Targets Ther 2018;11:1457.
- Xia B, Li Y, Zhou J, Tian B, Feng L. Identification of potential pathogenic genes associated with osteoporosis. Bone Joint Res 2017;6:640-8.
- Foj L, Filella X. Identification of potential miRNAs biomarkers for high-grade prostate cancer by integrated bioinformatics analysis. Pathol Oncol Res 2019;25:1445-56.
- Li GL, Xu XH, Wang BA, Yao YM, Qin Y, Bai SR, et al. Analysis of protein-protein interaction network and functional modules on primary osteoporosis. Eur J Med Res 2014;19:1-9.
- Ma M, Luo S, Zhou W, Lu L, Cai J, Yuan F, et al. Bioinformatics analysis of gene expression profiles in B cells of postmenopausal osteoporosis patients. Taiwan J Obstet Gynecol 2017;56:165-70.
- Cao Y, Li R, Li Y, Zhang T, Wu N, Zhang J, et al. Identification of transcription factor-gene regulatory network in acute myocardial infarction. Heart Lung Circ 2017;26:343-53.
- Zhang Y, Wang N, Ma J, Chen XC, Li Z, Zhao W. Expression profile analysis of new candidate genes for the therapy of primary osteoporosis. Eur Rev Med Pharmacol Sci 2016;20:433-40.
- Zhou Y, Layton O, Hong L. Identification of genes and pathways involved in ovarian epithelial cancer by bioinformatics analysis. J Cancer 2018;9:3016.
- Li L, Lei Q, Zhang S, Kong L, Qin B. Screening and identification of key biomarkers in hepatocellular carcinoma: evidence from bioinformatic analysis. Oncol Rep 2017;38:2607-18.
- Zhou Y, Zhu W, Zhang L, Zeng Y, Xu C, Tian Q, et al. Transcriptomic data identified key transcription factors for osteoporosis in caucasian women. Calcif Tissue Int 2018;103:581-8.
- Zhu X, Wang Z, Zhao Y, Jiang C. Investigation of candidate genes and mechanisms underlying postmenopausal osteoporosis using bioinformatics analysis. Mol Med Rep 2018;17:1561-72.
- Yang B, Cai Z, Zhang W, Yin D, Zhao W, Yang M. Autophagy alleviates the decrease in proliferation of amyloid β1‑42‑treated bone marrow mesenchymal stem cells via the AKT/mTOR signaling pathway. Mol Med Rep 2019;19:4091-100.
- Shalev M, Elson A. The roles of protein tyrosine phosphatases in bone-resorbing osteoclasts. Biochim Biophys Acta Mol Cell Res 2019;1866:114-23.
- Zhao C, Sun W, Zhang P, Ling S, Li Y, Zhao D, et al. miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway. RNA Biol 2015;12:343-53.
- Ge L, Cui Y, Liu B, Yin X, Pang J, Han J. ERα and Wnt/β‑catenin signaling pathways are involved in angelicin‑dependent promotion of osteogenesis. Mol Med Rep 2019;19:3469-76.
- Karner CM, Long F. Wnt signaling and cellular metabolism in osteoblasts. Cell Mol Life Sci 2017;74:1649-57.
- Whitehouse CA, Waters S, Marchbank K, Horner A, McGowan NW, Jovanovic JV, et al. Neighbor of Brca1 gene (Nbr1) functions as a negative regulator of postnatal osteoblastic bone formation and p38 MAPK activity. Proc Natl Acad Sci USA 2010;107:12913-8.
- Zhang W, Dong R, Diao S, Du J, Fan Z, Wang F. Differential long noncoding RNA/mRNA expression profiling and functional network analysis during osteogenic differentiation of human bone marrow mesenchymal stem cells. Stem Cell Res Ther 2017;8:1-3.
- Boulton SJ. BRCA1-mediated ubiquitylation. Cell Cycle 2006;5:1481-6.
- Engert F, Kovac M, Baumhoer D, Nathrath M, Fulda S. Osteosarcoma cells with genetic signatures of BRCAness are susceptible to the PARP inhibitor talazoparib alone or in combination with chemotherapeutics. Oncotarget 2017;8:48794.
- Sato S, Hanada R, Kimura A, Abe T, Matsumoto T, Iwasaki M, et al. Central control of bone remodeling by neuromedin U. Nat Med 2007;13:1234-40.
- Dimitri P, Rosen C. The central nervous system and bone metabolism: an evolving story. Calcif Tissue Int 2017;100:476-85.
- Yeo CE, Kang WY, Seong SJ, Cho S, Lee HW, Yoon YR, et al. Neuromedin B and its receptor silencing suppresses osteoclast generation by modulating precursor proliferation via M-CSF/c-Fms/D-type cyclins. Exp Cell Res 2017;359:112-9.
- Oliver F, Viale-Bouroncle S, Driemel O, Reichert TE, Schmalz G, Morsczeck C. Transcription factors TP53 and SP1 and the osteogenic differentiation of dental stem cells. Differentiation 2012;83:10-6.
- She Y, Li Y, Liu Y, Asai G, Sun S, He J, et al. Formaldehyde induces toxic effects and regulates the expression of damage response genes in BM-MSCs. Acta Biochim Biophys Sin 2013;45:1011-20.
- Ren BX, Gu XP, Zheng YG, Liu CL, Wang D, Sun YE, et al. Intrathecal injection of metabotropic glutamate receptor subtype 3 and 5 agonist/antagonist attenuates bone cancer pain by inhibition of spinal astrocyte activation in a mouse model. Pain Med 2012;116:122-32.
- Van Dartel M, Cornelissen PW, Redeker S, Tarkkanen M, Knuutila S, Hogendoorn PC, et al. Amplification of 17p11. 2∼ p12, including PMP22, TOP3A, and MAPK7, in high-grade osteosarcoma. Cancer Genet Cytogenet 2002;139:91-6.
- Wang T, Han S, Wu Z, Han Z, Yan W, Liu T, et al. XCR1 promotes cell growth and migration and is correlated with bone metastasis in non-small cell lung cancer. Biochem Biophys Res Commun 2015;464:635-41.
- Yamashita Y, Kajiura D, Tang L, Hasegawa Y, Kinoshita T, Nakamura S, et al. XCR1 expression and biased VH gene usage are distinct features of diffuse large B-cell lymphoma initially manifesting in the bone marrow. Am J Clin Pathol 2011;135:556-64.
- Fila-Danilow A, Borkowska P, Paul-Samojedny M, Kowalczyk M, Kowalski J. The influence of TSA and VPA on the in vitro differentiation of bone marrow mesenchymal stem cells into neuronal lineage cells: Gene expression studies. Postepy Hig Med Dosw 2017;71:236-42.
- Irminger-Finger I. BARD1, a possible biomarker for breast and ovarian cancer. Gynecol Oncol 2010;117:211-5.
- Chi L, Zou Y, Qin L, Ma W, Hao Y, Tang Y, et al. TIMELESS contributes to the progression of breast cancer through activation of MYC. Breast Cancer Res 2017;19:1-10.
- Hoberg M, Gratz HH, Noll M, Jones DB. Mechanosensitivity of human osteosarcoma cells and phospholipase C β2 expression. Biochem Biophys Res Commun 2005;333:142-9.
- Peng C, Yang Q, Wei B, Yuan B, Liu Y, Li Y, et al. Investigation of crucial genes and microRNAs in conventional osteosarcoma using gene expression profiling analysis. Mol Med Rep 2017;16:7617-24.
- Felton J, Hu S, Raufman JP. Targeting M3 muscarinic receptors for Colon Cancer therapy. Curr Mol Pharmacol 2018;11:184-90.
- Pal J, Patil V, Mondal B, Shukla S, Hegde AS, Arivazhagan A, et al. Epigenetically silenced GNG4 inhibits SDF1α/CXCR4 signaling in mesenchymal glioblastoma. Genes Cancer 2016;7:136-47.
- Li CF, Liu TT, Chuang IC, Chen YY, Fang FM, Chan TC, et al. PLCB4 copy gain and PLCß4 overexpression in primary gastrointestinal stromal tumors: Integrative characterization of a lipid-catabolizing enzyme associated with worse disease-free survival. Oncotarget 2017;8:19997-20010.
- Kim JM, Kakusho N, Yamada M, Kanoh Y, Takemoto N, Masai H. Cdc7 kinase mediates Claspin phosphorylation in DNA replication checkpoint. Oncogene 2008;27:3475-82.
- Desai A, Qing Y, Gerson SL. Exonuclease 1 is a critical mediator of survival during DNA double strand break repair in nonquiescent hematopoietic stem and progenitor cells. Stem Cells 2014;32:582-93.
- Strike P, Jones NJ. Mechanisms of genome maintenance and rearrangement: current research and recent advances in DNA repair and recombination. Mutat Res 1999;435:163-9.
- Bolivar JH, Munoz-Garcia JC, Castro-Dopico T, Dijkman PM, Stansfeld PJ, Watts A. Interaction of lipids with the neurotensin receptor 1. Biochim Biophys Acta Biomembr 2016;1858:1278-87.
- Jiang S, Yang Z, Di S, Hu W, Ma Z, Chen F, et al. Novel role of forkhead box O 4 transcription factor in cancer: bringing out the good or the bad. Semin Cancer Biol 2018;50:1-12.
- Kim HN, Han L, Iyer S, de Cabo R, Zhao H, O'Brien CA, et al. Sirtuin1 suppresses osteoclastogenesis by deacetylating FoxOs. Mol Endocrinol 2015;29:1498-509.
- Iyer S, Ambrogini E, Bartell SM, Han L, Roberson PK, de Cabo R, et al. FOXOs attenuate bone formation by suppressing Wnt signaling. J Clin Invest 2013;123:3409-19.
- Ambrogini E, Almeida M, Martin-Millan M, Paik JH, DePinho RA, Han L, et al. FoxO-mediated defense against oxidative stress in osteoblasts is indispensable for skeletal homeostasis in mice. Cell Metab 2010;11:136-46.
- Rajendran NK, Kumar SS, Houreld NN, Abrahamse H. Understanding the perspectives of forkhead transcription factors in delayed wound healing. J Cell Commun Signal 2019;13:151-62.
- Porcu E, Medici M, Pistis G, Volpato CB, Wilson SG, Cappola AR, et al. A meta-analysis of thyroid-related traits reveals novel loci and gender-specific differences in the regulation of thyroid function. PLoS Genet 2013;9:e1003266.
- Yoshida S, Kato T, Nishimura N, Kanno N, Chen M, Ueharu H, et al. Transcription of follicle-stimulating hormone subunit genes is modulated by porcine LIM homeobox transcription factors, LHX2 and LHX3. J Reprod Dev 2016;62:241-48.
- Ramzan K, Bin-Abbas B, Al-Jomaa L, Allam R, Al-Owain M, Imtiaz F. Two novel LHX3 mutations in patients with combined pituitary hormone deficiency including cervical rigidity and sensorineural hearing loss. BMC Endocr Disord 2017;17:1-10.