- *Corresponding Author:
- Ke wang
Department of Clinical Laboratory, Yongchuan Hospital Affiliated to Chongqing Medical University, Yongchuan, Chongqing 402160, China
E-mail: wk125@126.com
| Date of Received | 05 January 2023 |
| Date of Revision | 11 July 2023 |
| Date of Acceptance | 10 November 2023 |
| Indian J Pharm Sci 2023;85(6):1677-1684 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Sepsis shock was associated with worse mortality than common sepsis caused by disseminated infection. The present study had shown that repression of adaptive immunity was a characteristic of sepsis shock rather than sepsis, but the mechanism in which it exerted the adverse reaction was still need to be studied. In this study, we first employed weighted gene co-expression network analysis for acquiring the key modules with sepsis shock. Then, the differentially expressed genes and transcription factors were screened in the module for further function analysis, protein-protein interaction network building and long noncoding ribonucleic acid-micro ribonucleic acid-transcription factor network study. The study strengthened the concept that adaptive immune repression typically featured the sepsis shock concerning antigen presentation via major histocompatibility complex class II, T-cell activation and T-cell depletion. Among which T-cell depletion ranked as an overwhelming contributor to sepsis shock outcome. The transcription factor tumor protein P53, forkhead box protein O1 and GATA binding protein 3 served as the master regulator for the T-cell depletion process mainly through driving hematopoietic stem cells out of quiescence to unrestrained hematopoietic stem cell expansion with detrimental deoxyribonucleic acid damage, which greatly decreased hematopoietic stem cells long-term differentiation potential into T lymphocyte hierarchy. The study enlightened that manipulating the expression level of tumor protein P53, forkhead box protein O1 and GATA binding protein 3 to an optimized state will guarantee the T cell replenishment for fighting sepsis shock, which might support a potential efficient therapeutic way for sepsis shock treatment.
Keywords
Sepsis, differentially expressed genes, protein-protein interaction, T-cell depletion, mortality
Sepsis would cause severe organ dysfunction, a devastating consequence of an infection that exceeds local tissue containment and disseminates to the whole body[1]. The sepsis clinical syndrome might range from severe sepsis to sepsis shock. Sepsis shock progresses and leads to the development of cardiovascular and associated with greater mortality compared to the severe sepsis[1,2]. Although the treatment of sepsis has significantly improved over the past 20 y, the mortality rate of sepsis is still higher in Intensive Care Units (ICUs) [3,4].
Traditionally, sepsis organ dysfunction was perceived as a result of an exaggerated hyperimmune response of the host. However, as all clinical trials of drugs targeting the inflammation proved as a failure to cure sepsis, the understanding of sepsis had been extended to the renewed paradigm that inflammation and immunosuppression concurrently existed within sepsis patients. The latter might serve as the overriding factor for sepsis death[3,5], because unrestrained inflammation accounted for about 15 % of mortality while the immunosuppression accounted for 85 % of deaths, even after nosocomial recovery with a secondary infection[5].
The manifestations of immunosuppression during the sepsis process might involve defective antigen presentation, adaptive immunity (T and B cell abnormalities), abnormalities in Natural Killer (NK) cell activity, unbalanced pro- and anti- inflammatory cytokines, increased production of Programmed Death 1 (PD-1), increased regulatory T cells and a shift in the Type I and II helper T cell balance[1,3,5,6]. A study of differentially expressed genes of sepsis shock relative to sepsis displayed that persistent repression of adaptive immunity genes and pathways might serve as a particular characteristic of septic shock but not sepsis[2].
Even though immunosuppression is the main characteristic of sepsis shock rather than sepsis but the mechanism in which it exerted the adverse reaction was still need to be studied. Since genes might function via networks of co-expressed genes with similar biological functions, we first planned to employ Weighted Gene Co-expression Network Analysis (WGCNA) for acquiring the key module. After that, a further step was taken to screen the differentially expressed genes and Transcription Factors (TFs) in the module and put all these genes for function analysis, Protein-Protein Interaction (PPI) network building and long noncoding Ribonucleic Acid (lncRNA)-micro Ribonucleic Acid (miRNA)-TF network study. The study aimed to provide more accurate insights into sepsis shock associated biological pathways and key regulators.
Materials and Methods
WGCNA:
For acquiring the key module, we used the R package “WGCNA”. Module detection was then performed with the soft-thresholding power 6. Correlation between module eigengenes and sample types (sepsis shock patients vs. healthy controls) was used to identify modules significantly associated with sepsis shock.
Consensus eigengene networks:
A pairwise comparison of eigengene networks was built and the difference between adjacencies was used to detect network preservation. The smaller the difference in the adjacency matrix between the two sets indicates that the modules are well preserved.
Consensus modules relating to clinical traits:
Correlation analysis was performed to relate module eigengene with external traits, including disease group (sepsis shock patients vs. healthy controls), age and gender. Gene Significance (GS) of the disease group was correlated to the Module Membership (MM) to investigate whether genes significantly associated with the both disease group and MM. Each gene's MM (eigengene-based connectivity) was calculated by correlating its gene expression profile with the module eigengene of a given module. For a given module, a MM value of 0 indicates that a gene is not part of the module, whereas a MM of -1 or 1 indicates that it is highly connected to the module.
Enrichment analysis for biological function:
We obtained GSE57065 from Gene Expression Omnibus (GEO). Differential expression detection was performed using the R linear models for microarray data (limma) package. We took log Fold Change (FC) >1 and False Discovery Rate (FDR) <0.05 as the filter, the acquired genes are namely dgene. Using the count data of Ribonucleic Acid sequencing (RNAseq) of GSE57065, extracted the TFs from a dataset of JASPAR (http://jaspar.genereg.net/) database. Using R limma package for difference analysis, acquired 74 significantly different TFs including 55 up-regulated and 19 down-regulated (logFC>1 and FDR<0.05), namely dTF. Both dgene and dTF were intersected with MEblue and further analyzed for their enrichment using Gene Ontology (GO) (http://geneontology.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (www.kegg.jp). R package clusterProfiler was used to draw the bubble diagram.
PPI study:
The PPI network was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://www.string-db.org/), and a confidence score of ≥0.9 was set as the threshold. Protein nodes that did not interact with other proteins were removed. Furthermore, the PPI network was analyzed to screen the significant modules and hub genes using Cytoscape (version: 3.8.0). Subsequently, the cytoHubba (version: 0.1) plug-in was used to screen the PPI network and genes with degree >10 were identified as hub genes.
lncRNA-miRNA-TF network:
lncRNA-miRNA-TF network was drawn through Cytoscape network visualization and Hub lncRNA- miRNA-TF pathway was screened by cytoHubba (version: 0.1) plug-in.
Results and Discussion
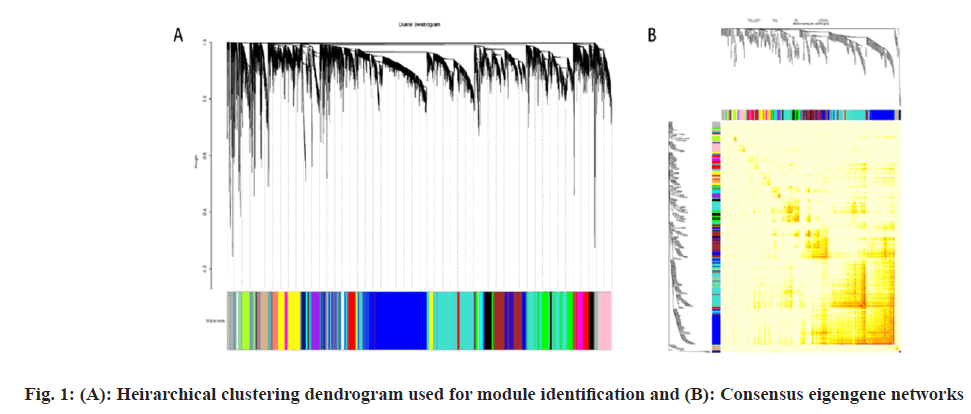
Dataset (GSE57065) was extracted from National Center for Biotechnology Information (NCBI), which contained genes of 82 sepsis shock sample and 25 normal controls. According to the soft- thresholding power 6, the hierarchical clustering dendrogram (fig. 1) was used for the module identification and a total 25 modules are detected (fig. 1A). The consensus eigengene network was shown in fig. 1B. The results showed that the eigengene networks of the datasets were well expressed in consensus with hierarchical clustering.
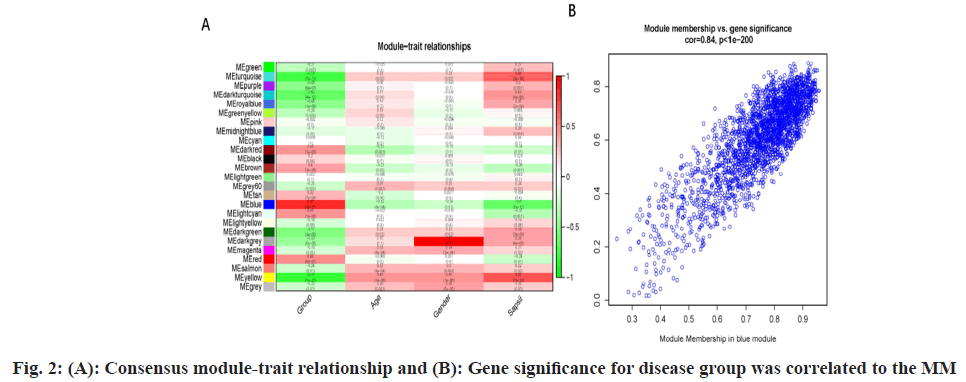
Pearson’s correlation analysis was used to explore the correlation between module eigengene and clinical traits (fig. 2A). The MEblue module was significantly associated with sepsis shock (coefficient=0.8 and p<0.001). The correlations between GS and MM of sepsis shock were explored, and the results showed that there was a highly significant correlation between GS and MM (coefficient=0.84 and p<0.001) (fig. 2B).
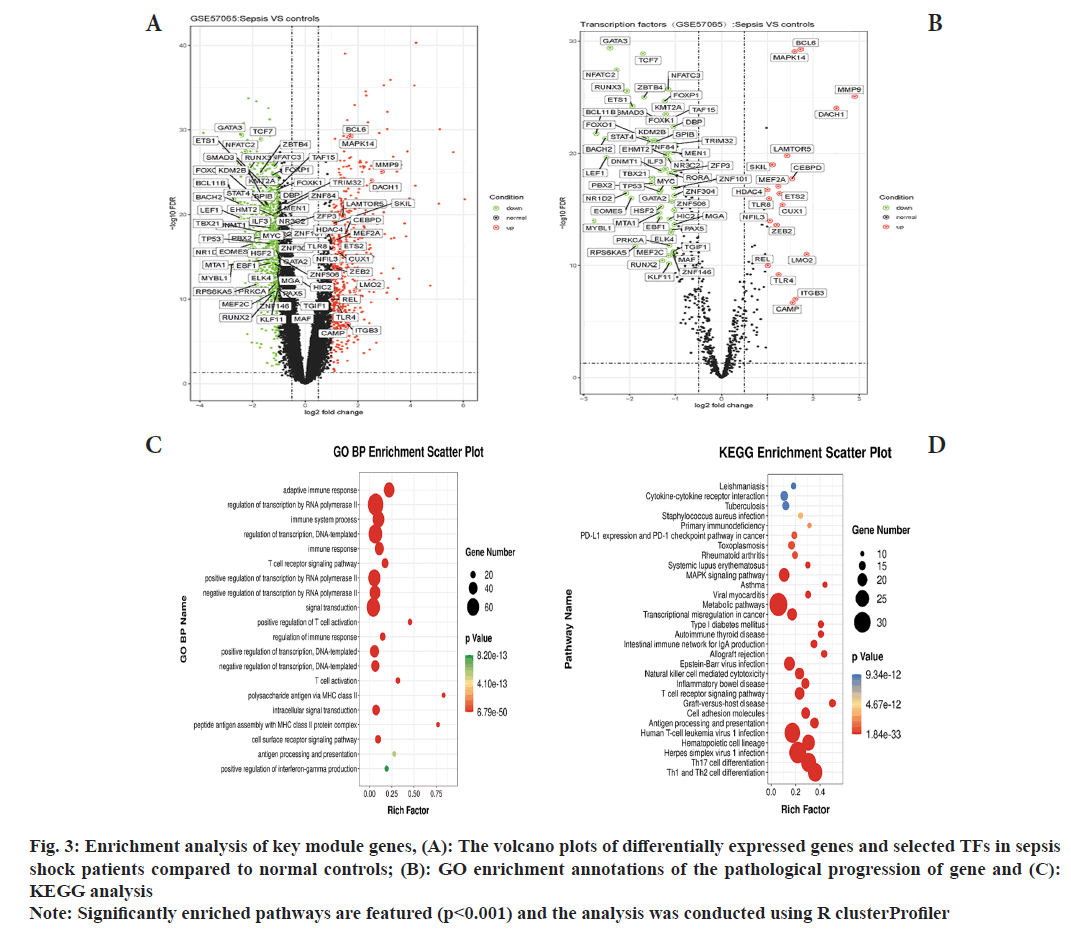
To analyze the biological function of the MEblue module, we first screened differential genes within sepsis shock patients relative to control (fig. 3A). Then we got an intersection between the differential genes and MEblue module genes. Enrichment analysis of the intersection genes was conducted using KEGG and GO analysis. The most enriched GO biological processes of the MEblue module included adaptive immune response, T-cell receptor signaling pathway, T-cell activation and immune system process (fig. 3B). KEGG pathway were enriched in type I and type II helper T cell differentiation, T helper 17 (Th17) cell differentiation, hematopoietic cell lineage, antigen processing and presentation, cell adhesion molecules and T cell receptor signaling pathway (fig. 3C).
Fig. 3: Enrichment analysis of key module genes, (A): The volcano plots of differentially expressed genes and selected TFs in sepsis shock patients compared to normal controls; (B): GO enrichment annotations of the pathological progression of gene and (C): KEGG analysis
Note: Significantly enriched pathways are featured (p<0.001) and the analysis was conducted using R clusterProfiler
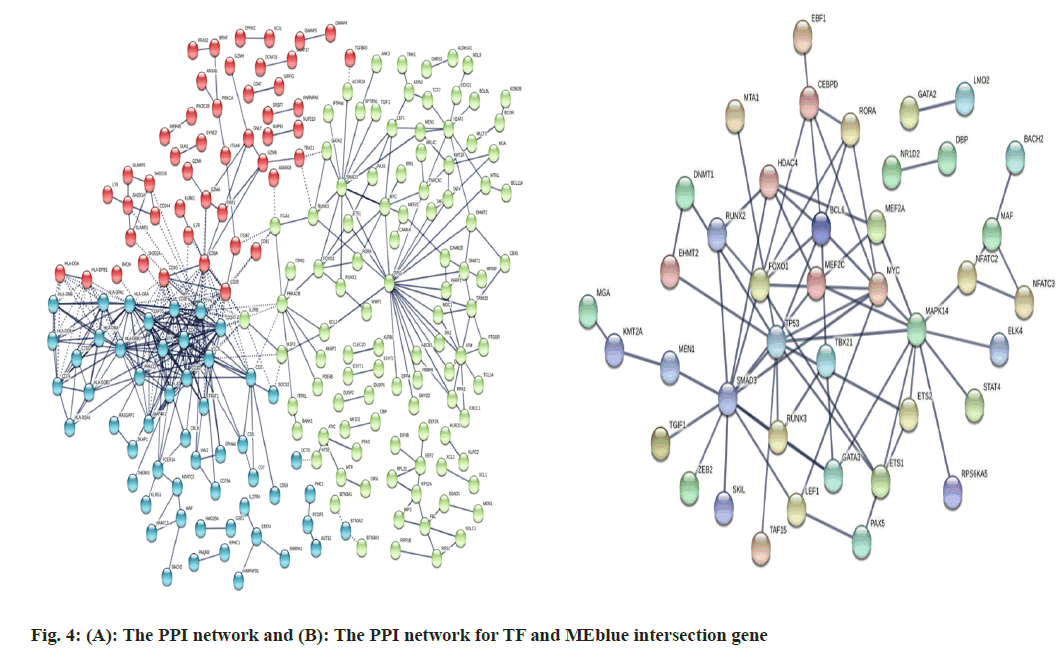
Modules were made up of co-expressed genes, indicating that they might be regulated by common TFs. Thus, we performed enrichment analysis for TFs. Similarly, differentially expressed TFs were first screened out for sepsis shock and compared with the control. The volcano map displayed that GATA binding protein 3 (GATA3) and Transcript Elongation Factor 7 (TEF7) were outstandingly downregulated within sepsis shock patients. PPI network analyzed for the intersection genes and showed that adaptive immunity genes formed a prominent cluster, mainly composing Cluster of Differentiation 247 (CD247) (CD3 Zeta chain), CD3 delta chain, CD3 gamma chain, Interleukin- 2-inducible T-cell Kinase (ITK)/tyrosine protein kinase ITK/TSK, Linker for Activation of T cells (LAT), Leukocyte C-terminal Src Kinase (LCK), FYN proto-oncogene, Src family tyrosine kinase (FYN), Zeta chain of T cell receptor Associated Protein (ZAP70) and other genes (fig. 4A). After the differential expressed TFs intersected with MEblue module genes, the result of PPI network for the intersection genes showed a module enriched with Tumor Protein P53 (TP53), Forkhead box protein O1 (FOXO1), Mothers against decapentaplegic homolog 3 (SMAD3), Myocyte Enhancer Factor 2C (MEF2C), T-Box transcription factor (TBX21) and other key regulators (fig. 4B).
lncRNAs (>200 nts) and messenger RNAs (mRNAs) bind to miRNAs via competitively and act as competing endogenous RNAs (ceRNAs). In this mechanism, lncRNAs act as a sponge for miRNAs and indirectly up-regulated the level of miRNAs targeting specific mRNAs. According to this, the lncRNA-miRNA-mRNA network was built to find the potential lncRNAs and miRNAs key regulators in sepsis shock.
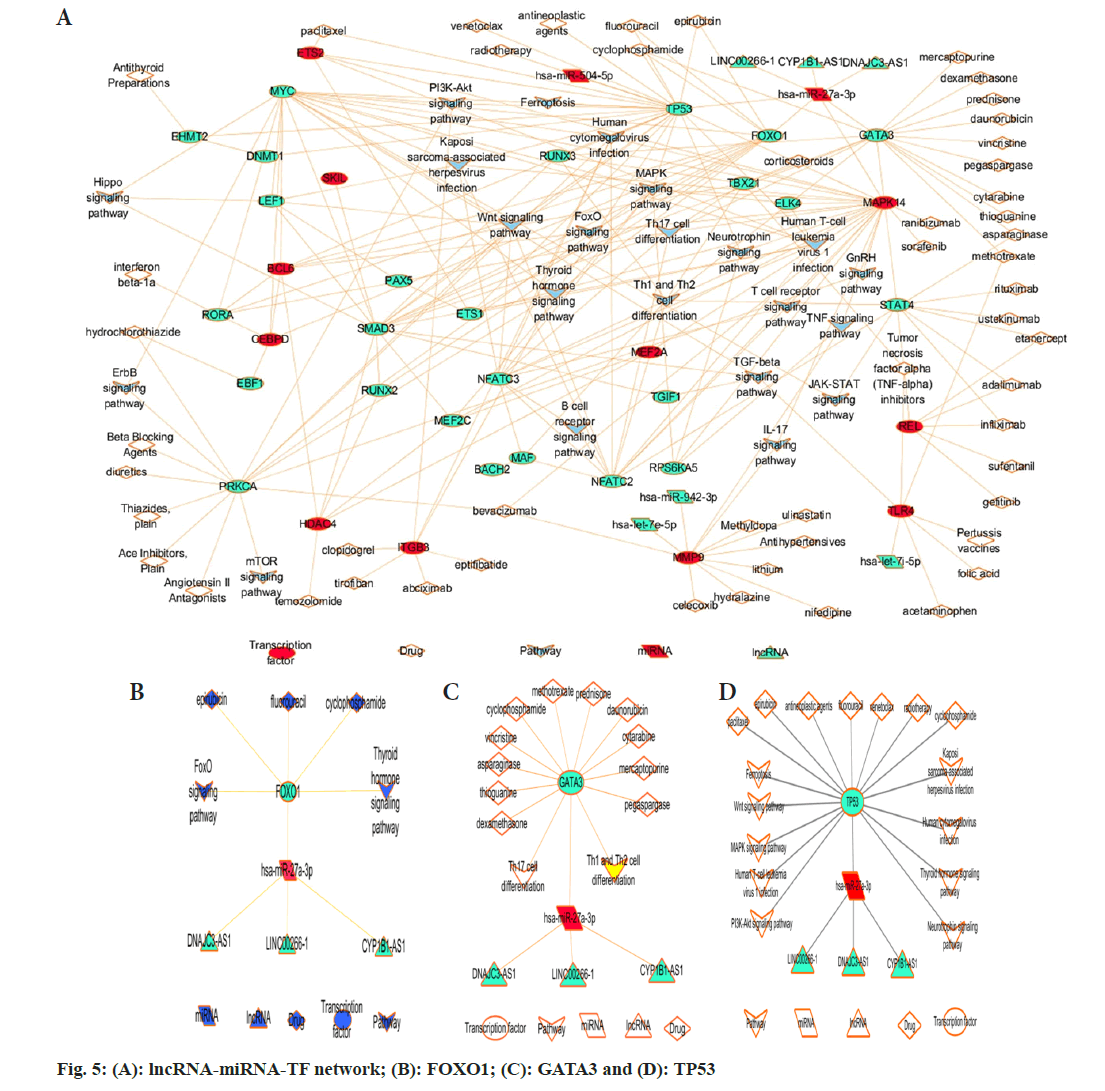
miRNAs that were potential regulators of the MEBlue module were predicted using the Gene annotations Co-occurrence discovery (GeneCodis) database (https://genecodis.genyo.es/), and miRcode was used to predict the lncRNA target of miRNA. Then, the integrated lncRNA-miRNA- TF-drug-pathway network was built as shown, and the hub transcriptional factors and miRNA were excavated by the Cytoscape and CytoHubba plugin. The result demonstrated that TP53, FOXO1 and GATA3 might serve as the master regulators for the sepsis shock process (fig. 5A), and Homo sapiens (has)-miR-26b-5p concurrently regulated the three genes negatively. At the same time, three lncRNA LINC00266-1, DNALC3-Antisense RNA 1 (AS1), and Cytochrome P450 family 1 subfamily B-AS1 (CYP1B-AS1) might positively regulate the three gene levels via the ceRNA mechanism (fig 5B-5D).
Previous studies had suggested that sepsis shock was characterized by repression of adaptive immunity involving the pathway of antigen presentation via Major Histocompatibility Complex (MHC) class II was permanently down regulated within sepsis shock patients compared to sepsis[2,8]. Our study was quite in consensus with the suggestions as demonstrated by the fact that the MEblue module significantly associated with sepsis shock and was enriched with down- regulated genes for antigen processing and presentation, and type 1 and type 2 helper T cell differentiation[9-11], which exhibited inhibition of T cell activation and differentiation. Besides, the MEblue module was enriched with decreased hematopoietic cell lineage genes including CD2, CD5 and CD7 markers for T-cell progenitors, which meant competence of primitive Hematopoietic Stem Cell (HSC) to produce T lymphocytes might be severely compromised in sepsis shock patients. Furthermore, the MEblue module's PPI network focused on the hub genes composing CD3D, CD3G, CD247, ITK, LCK and ZAP70, all being indispensable for T-cell development, activation and differentiation. Among them, CD3 known as the marker for thymus immature T cells, which might also represent T-cell depletion due to the reduction in producing them. Our study might add evidence of adaptive immunity repression within sepsis shock, apart from MHC II antigen presentation, T-cell activation, and T-cell depletion originating from abnormal HSC, serving as another overwhelming contributor for disease progression.
HSCs also termed as Long-Term HSCs (LT- HSCs) residing in the bone marrow were required for replenishment of all mature blood and immune cells through a hierarchically organized lineage commitment program[12]. LT-HSCs first differentiate into Short-Term HSCs (ST-HSCs) that produce increasingly lineage-restricted progenitor cells, namely Common Myeloid Progenitors (CMP) and Common Lymphoid Progenitors (CLP), which would undergo terminal differentiation into the myeloid, lymphoid lineages[12,13]. In steady-state, HSC was kept in a relatively quiescent or dormant state, presumably a safeguard mechanism to maintain the integrity and long-term competence of an HSC pool[14-16]. However, when met with extensive demand for blood cells, like blood loss or infection, HSCs could re-enter the cell cycle for stress-induced self- renewing proliferation and lineage reconstitution. However, the two should be balanced without depletion of the stem cell pool[12,13,17]. However, it was reported that during prolonged systemic exposure to Lipopolysaccharide (LPS), mimicking that of sepsis, the potential of HSC to generate B and T lymphocytes was severely affected[18-20], just in agreement with the results mentioned in this study. It was suggested because excessive LPS exposure induce aberrant expansion of HSCs but permanently impeded the normal self-renew and differentiation capacity[20,21].
Previous studies showed that the HSCs expresses on Toll-Like Receptor 4 (TLR4) receptors for directly sensing LPS[22], which was mediated via TLR4-TIR domain-containing adaptor-inducing Interferon-beta (TRIF)-Reactive Oxygen Species (ROS)-p38[21]. The increase in ROS levels could decrease the HSCs' life span and induce Deoxyribonucleic Acid (DNA) damage[23-25]. Thus, prolonged LPS exposure in sepsis would inevitably provoke detrimental HSC DNA damage and attrition of normal HSCs, and this adversity could be partially blocked by ROS-p38 inactivation[21]. Also, it had been recommended that both p53 and FOXO subfamily able to protect from oxidative stress[26,27], helped to lower ROS levels, thereby reducing DNA damage within the hematopoietic compartment[15,27,28]. This cytoprotection was strengthened by p53 negatively regulating proliferation and enforcing HSC into quiescence, which avoided inheritable damage to achieve maintenance of stem cell integrity[15]. The same situation went to FOXOs, and its deficiency drove the HSCs out of quiescence into the cell cycle, leading to impaired HSCs' long-term regenerative potential[27-29]. The function of TP53 and FOXOs on regulating cell cycle, DNA damage and resistance to oxidative stress might greatly explain the down- regulation of both in sepsis shock, which might be due to over- interaction of LPS-TLR4, which made the HSCs as a supervisory gatekeeper of TP53 and FOXOs surrendered, leaving increased ROS level, permitted unrestrained proliferation and accumulated DNA damage. These three factors might make a vicious spiral, rendering HSCs too exhausted to produce their progeny, namely re- population capacity.
HSCs defective differentiation was subsequently featured with the decreased expression of GATA-3 and TEF7[30,31]; both TFs emerged as essential for development, differentiation and function throughout the T lymphocyte hierarchy, loss of which might cause relentlessly depleted T-cell lineage[31]. This study demonstrated about the CD3 downregulation, depression of type 1/2 helper T cell and Th17 differentiation. Our study suggested that HSCs defective derived T-cells might be the predominant reason for T-cell number reduction, despite sepsis shock it also induces apoptosis or any other unfavourable conditions as reported[10,32]. A clinical study had documented that lymphopenia and T-cell depletion were critically associated with increased mortality. At the same time, patients who survived sepsis showed restored higher lymphocyte numbers, especially T and NK cells[33]. It further supported that T-cell numbers might be overridden in combating a sepsis challenge.
To sum up, using the consensus network, our study strengthened the concept that adaptive immune repression typically featured sepsis shock concerning antigen presentation via MHC class II, T-cell activation and T-cell depletion. PPI network building of the MEblue module suggested that T-cell depletion ranked as an overwhelming contributor for sepsis shock outcome, and the TFs TP53, FOXO1, GATA3 served as the master regulator for T-cell depletion process, largely through driving HSCs out of quiescence into the cell cycle, leading to unrestrained HSC expansion with detrimental DNA damage, which greatly decreased HSCs long-term differentiation potential into T lymphocyte hierarchy. The study enlightened that T cell number could be feasible and efficient predictive biomarkers and potential therapeutic targets for sepsis shock, such as manipulating the expression level of TP53, FOXO1, and GATA3 to an optimized state, to guarantee the T cell replenishment for fighting sepsis shock, significantly increase the survivor rate.
Acknowledgements:
This study was supported by Chongqing Natural Science Foundation (Grant No. cstc-2019jcyj- msxmX0371), Chongqing Health and the Health Committee and Chongqing Science and Technology Commission (Grant No. 2020FYYX124).
Author contributions:
Ke Wang and Ying Zhao contributed equally to this work.
Conflict of interests:
The authors declared no conflict of interests.
References
- Nedeva C. Inflammation and cell death of the innate and adaptive immune system during sepsis. Biomolecules 2021;11(7):1-15.
[Crossref] [Google Scholar] [PubMed]
- Kong C, Zhu Y, Xie X, Wu J, Qian M. Six potential biomarkers in septic shock: A deep bioinformatics and prospective observational study. Front Immunol 2023;14:1-17.
[Crossref] [Google Scholar] [PubMed]
- Liu D, Huang SY, Sun JH, Zhang HC, Cai QL, Gao C, et al. Sepsis-induced immunosuppression: Mechanisms, diagnosis and current treatment options. Mil Med Res 2022;9(1):1-19.
[Crossref] [Google Scholar] [PubMed]
- Angus DC, Poll T. Severe sepsis and septic shock. N Engl J Med 2013;369(9):840-51.
[Crossref] [Google Scholar] [PubMed]
- Torres LK, Pickkers P, van der Poll T. Sepsis-induced immunosuppression. Ann Rev Physiol 2022;84:157-81.
[Crossref] [Google Scholar] [PubMed]
- Bline KE, Muszynski JA, Guess AJ, Menocha S, Moore-Clingenpeel MD, Popelka JK, et al. Novel identification of myeloid-derived suppressor cells in children with septic shock. Pediatr Crit Care Med 2022;23(12):e555-63.
[Crossref] [Google Scholar] [PubMed]
- Arora S, Singh P, Dohare R, Jha R, Syed MA. Unravelling host-pathogen interactions: ceRNA network in SARS-CoV-2 infection (COVID-19). Gene 2020;762:1-13.
[Crossref] [Google Scholar] [PubMed]
- Whelan AJ, Ricci M, Harthan AA, Deshpande G. Calcium responsive pediatric septic shock refractory to isotonic crystalloids and inotropic agents. J Pediatr Pharmacol Ther 2022;27(8):765-9.
[Crossref] [Google Scholar] [PubMed]
- Xu J, Li J, Xiao K, Zou S, Yan P, Xie X, et al. Dynamic changes in human HLA-DRA gene expression and Th cell subsets in sepsis: Indications of immunosuppression and associated outcomes. Scand J Immunol 2020;91(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Ma Y, Cheng Z, Zheng Y, Wang W, He S, Zhou X, et al. Low dose of esmolol attenuates sepsis-induced immunosuppression via modulating T-lymphocyte apoptosis and differentiation. Shock 2023;59(5):771-8.
[Crossref] [Google Scholar] [PubMed]
- Heidarian M, Griffith TS, Badovinac VP. Sepsis-induced changes in differentiation, maintenance, and function of memory CD8 T cell subsets. Front Immunol 2023;14:1-11.
[Crossref] [Google Scholar] [PubMed]
- Sezaki M, Hayashi Y, Wang Y, Johansson A, Umemoto T, Takizawa H. Immuno-modulation of hematopoietic stem and progenitor cells in inflammation. Front Immunol 2020;11:1-16.
[Crossref] [Google Scholar] [PubMed]
- Kwok AJ, Allcock A, Ferreira RC, Cano-Gamez E, Smee M, Burnham KL, et al. Neutrophils and emergency granulopoiesis drive immune suppression and an extreme response endotype during sepsis. Nat Immunol 2023;24(5):767-79.
[Crossref] [Google Scholar] [PubMed]
- Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, et al. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell 2011;147(5):1146-58.
[Crossref] [Google Scholar] [PubMed]
- Kalafati L, Hatzioannou A, Hajishengallis G, Chavakis T. The role of neutrophils in trained immunity. Immunol Rev 2023;314(1):142-57.
[Crossref] [Google Scholar] [PubMed]
- Pattarabanjird T, Marshall M, Upadhye A, Srikakulapu P, Garmey JC, Haider A, et al. B-1b cells possess unique bHLH-driven P62-dependent self-renewal and atheroprotection. Circ Res 2022;130(7):981-93.
[Crossref] [Google Scholar] [PubMed]
- Lu Y, Yang L, Shen M, Zhang Z, Wang S, Chen F, et al. Tespa1 facilitates hematopoietic and leukemic stem cell maintenance by restricting c-Myc degradation. Leukemia 2023;37(5):1039-47.
[Crossref] [Google Scholar] [PubMed]
- Chahi AK, Belew MS, Xu J, Chen HT, Rentas S, Voisin V, et al. PLAG1 dampens protein synthesis to promote human hematopoietic stem cell self-renewal. Blood 2022;140(9):992-1008.
[Crossref] [Google Scholar] [PubMed]
- Zhang H, Rodriguez S, Wang L, Wang S, Serezani H, Kapur R, et al. Sepsis induces hematopoietic stem cell exhaustion and myelosuppression through distinct contributions of TRIF and MYD88. Stem Cell Rep 2016;6(6):940-56.
[Crossref] [Google Scholar] [PubMed]
- Sallustio F, Picerno A, Tatullo M, Rampino A, Rengo C, Valletta A, et al. Toll-like receptors in stem/progenitor cells. Handb Exp Pharmacol 2021;276:175-212.
[Crossref] [Google Scholar] [PubMed]
- Takizawa H, Fritsch K, Kovtonyuk LV, Saito Y, Yakkala C, Jacobs K, et al. Pathogen-induced TLR4-TRIF innate immune signaling in hematopoietic stem cells promotes proliferation but reduces competitive fitness. Cell Stem Cell 2020;27(1):177.
[Crossref] [Google Scholar] [PubMed]
- Deravi N, Poudineh M, Pirzadeh M, Yavarpour-Bali H, Mehrabi H, Erabi G, et al. The Yin and Yang of toll-like receptors in endothelial dysfunction. Int Immunopharmacol 2022;108:108768.
[Crossref] [Google Scholar] [PubMed]
- Salimi U, Dummula K, Tucker MH, dela Cruz CS, Sampath V. Postnatal sepsis and bronchopulmonary dysplasia in premature infants: Mechanistic insights into “new BPD”. Am J Respir Cell Mol Biol 2022;66(2):137-45.
[Crossref] [Google Scholar] [PubMed]
- Walter D, Lier A, Geiselhart A, Thalheimer FB, Huntscha S, Sobotta MC, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature 2015;520(7548):549-52.
[Crossref] [Google Scholar] [PubMed]
- Leite-Avalca MC, Zampronio A, Lehmann C. Cannabinoid receptor 1 and 2 signaling pathways involved in sepsis. Shock 2021;56(5):673-81.
[Crossref] [Google Scholar] [PubMed]
- Mehta S, Campbell H, Drummond CJ, Li K, Murray K, Slatter T, et al. Adaptive homeostasis and the p53 isoform network. EMBO Rep 2021;22(12):1-24.
[Crossref] [Google Scholar] [PubMed]
- Leduc-Gaudet JP, Franco-Romero A, Cefis M, Moamer A, Broering FE, Milan G, et al. MYTHO is a novel regulator of skeletal muscle autophagy and integrity. Nat Commun 2023;14(1):1-20.
[Crossref] [Google Scholar] [PubMed]
- Bigarella CL, Li J, Rimmelé P, Liang R, Sobol RW, Ghaffari S. FOXO3 transcription factor is essential for protecting hematopoietic stem and progenitor cells from oxidative DNA damage. J Biol Chem 2017;292(7):3005-15.
[Crossref] [Google Scholar] [PubMed]
- Hajishengallis G, Li X, Chavakis T. Immunometabolic control of hematopoiesis. Mol Aspects Med 2021;77:1-11.
[Crossref] [Google Scholar] [PubMed]
- She H, Tan L, Yang R, Zheng J, Wang Y, Du Y, et al. Identification of featured necroptosis-related genes and imbalanced immune infiltration in sepsis via machine learning. Front Genet 2023;14:1-12.
[Crossref] [Google Scholar] [PubMed]
- Zhao X, Shan Q, Xue HH. TCF1 in T cell immunity: A broadened frontier. Nat Rev Immunol 2022;22(3):147-57.
- Martin MD, Badovinac VP, Griffith TS. CD4 T cell responses and the sepsis-induced immunoparalysis state. Front Immunol 2020;11:1-11.
[Crossref] [Google Scholar] [PubMed]
- Hohlstein P, Gussen H, Bartneck M, Warzecha KT, Roderburg C, Buendgens L, et al. Prognostic relevance of altered lymphocyte subpopulations in critical illness and sepsis. J Clin Med 2019;8(3):1-11.
[Crossref] [Google Scholar] [PubMed]