- *Corresponding Author:
- S. Rathod
Department of Pharmaceutical Chemistry, Bharati Vidyapeeth College of Pharmacy, Kolhapur, Maharashtra 416013, Sangli, Maharashtra 416305, India
E-mail: sanket.rathod-copk@bvp.edu.in
| Date of Received | 14 July 2022 |
| Date of Revision | 16 March 2023 |
| Date of Acceptance | 12 February 2024 |
| Indian J Pharm Sci 2024;86(1):322-329 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Cancer is one of the leading causes of mortality worldwide and researchers are working to find new ways to cure it. Cancer is a disease characterized by abnormal cell development with the ability to invade and spread to other sections of the body. Breast cancers, colon cancers, lung cancers, liver cancers, rectum cancers, and stomach cancers are the most common cancers over the globe. Cyclin-dependent kinases are one of the causes of cancer and they are protein kinases. Cyclin-dependent kinases play important roles in controlling cell division in cancer cells. This study aims to find potential cyclin-dependent kinases 1 and cyclin-dependent kinases 2 inhibitors. Several phytochemicals were subjected to molecular docking against cyclin-dependent kinases 1 and cyclin-dependent kinases 2 and phytochemicals having good binding affinity were subjected to molecular dynamic simulation to determine the stability of protein-ligand complexes.
Keywords
Cancer, cyclin-dependent kinases 1, cyclin-dependent kinases 2, computational, molecular docking, molecular dynamics
Cancer is the second leading cause of mortality in the world, in 2019 with approximately 10.1 million deaths (17.83 % of total deaths)[1]. In 2020, 19.3 million new cases of cancer were estimated worldwide, according to predictions from the Global Cancer Observatory (GLOBOCAN). After China and United States of America, India came in third. According to GLOBOCAN, 2.08 million more cases of cancer would be diagnosed in India between 2020 and 2040, an increase of 57.5 %[2]. In 2020, the most prominent malignancies diagnosed globally were female breast cancer (2.26 million cases), lung (2.21) and prostate cancer (1.41) and the most common causes of cancer death were lung (1.79 million fatalities), liver (830000) and stomach cancer (769000)[3]. Worldwide, 6.9 million new cases of cancer will be detected among persons 80 y of age or older in 2050 (20.5 % of all cancer cases)[4-6].
Cyclin-Dependent Kinase (CDK) activation abnormalities, which are extremely common in human cancers, offered a rationale for developing synthetic CDK inhibitors as anticancer drugs[7].
Pharmaceutical companies have behaved focused on the proteins that regulate cellular proliferation, differentiation, transformation and metastasis. In the human genome, there are approximately 518 distinct protein kinases[8]. At present, 13 members of the CDK family have been reported, which are named from CDK1 to CDK13. Among these members, CDK1, CDK2, CDK4 and CDK6 are directly involved in cell cycle regulation, while CDK7 controls the cycle by activating other CDKs. In addition, CDK8, CDK9 and CDK7 are transcription regulatory factors. Moreover, CDK2, CDK4, and CDK6 mediate the G1 phase of the cell cycle, whereas CDK2 and CDK1 regulate S and G2 phases, and the M phase, respectively. Among them, CDK is a key class of cell cycle proteins. CDK activity requires the binding of the regulatory subunits known as cyclins. Cyclins are generated and eliminated at precise intervals during the cell cycle, allowing for timely control of kinase activity. Each stage in the cell cycle is regulated by CDKs belonging to a family of threonine/serine protein kinases. The CDKs are master regulators of the cell division cycle in association with the cyclin regulatory subunits[9]. The CDK1/cyclin B complex is responsible for completing the mitotic process. Despite the highly regulated nature of the process, CDK1 is the only CDK required for cell cycle modulation[10]. CDK1 controls the start of the cell cycle and its progression through mitosis. Recent research has linked loss of CDK1 function or abnormal CDK1 expression to G2 phase arrest and a variety of tumor types, validating CDK1 as a therapeutic target[11]. CDK2 is the family of serine/ threonine protein kinases that have developed as a potent, selective, and low-toxic cancer therapy target[12]. CDK2 binds to cyclins A, B, and E and plays an important role in the G1/S transition, the start of the synthesis of DNA, and the regulation of the exit from the S phase. Furthermore, altered expression of cyclins, whose interaction with CDKs is required for their catalytic activity, can promote cancer proliferation[13]. Highly selective CDK2 inhibitors will aid in the continuous discovery of cancer subtypes that are responsive to CDK2 inhibition by defining the additional unique role of CDK2 in cancer development[14].
Phytochemicals and their derivatives have the potential to improve cancer therapy efficiency while reducing side effects[15]. Several of these phytochemicals are physiologically active substances found in nature with high anticancer potential. Phytochemicals often work as anticancer agents by regulating molecular pathways that have been related to cancer growth and progression[15]. Because of their structural variety, natural products have remained one of the most important frameworks in future anticancer agent development[16]. More than 100 novel compounds derived from natural products such as Etoposide, Paclitaxel and Vinblastine have been introduced into the market[17].
Materials and Methods
Protein and ligand preparation:
The previously reported Three Dimensional (3D) crystal structure of CDK1 (PDB: 6GU7)[18-21] having 2.75 Å resolution and CDK2 (PDB: 6GUE) [22-24] having 1.99 Å resolution was retrieved from the RCSB Protein data bank[7]. The downloaded structures of the protein were prepared for docking as per the previously described protocol[25]. Thereafter prepared structures were evaluated for stereochemical adaptability and the quality of the protein structure using the PROCHECK and ProSAweb server[26,27]. The structures of phytochemicals were downloaded in mol2 file format from Indian Medicinal Plants, Phytochemistry, and Therapeutics (IMPPAT) (https://cb.imsc.res.in/imppat/home)[28-32]. IMPPAT is an Indian medicinal phytochemical compound database and it helps to find the accessible compounds of the required plant. This database has various filters and can help to find the compounds based on biological activity[33-35]. All downloaded mol2 files of phytochemicals were imported into BIOVIA Discovery Studio 2020 to optimize by adding hydrogen atoms and convert into a PDB file format. Further, all ligand groups were energy minimized using the OpenBabel module of the PyRx with MMFF94 force field and steepest descent algorithm and used for further molecular docking studies[36,37]. Dinaciclib was used as a standard to compare the docking results[38].
Molecular docking studies:
Molecular docking was carried out to investigate the binding affinity and interactions between the subjected phytochemicals and target protein (PDB: 6GU7 and PDB: 6GUE). AutoDock Vina of PyRx 0.8 was used to perform molecular docking[37-40]. The prepared structures of proteins and phytochemicals were imported in PyRx 0.8. A maximized grid box with center X: 22.0372, Y: 12.6379, Z: 4.7538 and dimensions X: 60.73 Å, Y: 41.37 Å, Z: 66.32 Å for PDB: 6GU7 and with center X: -10.889, Y: -7.5226, Z: 9.3097 and dimensions X: 94.9893Å, Y: 108.3853Å, Z: 98.3694Å for PDB: 6GUE was selected in the Vina workspace to cover the entire protein structure. The exhaustiveness was kept to default at eight and nine different conformations was predicted for each phytochemical with the selected target protein (PDB: 6GUE and 6GU7) [25,37]. Further, the best pose with the highest negative binding affinity was selected for each molecule[41]. Docking interaction, visualization, and analysis of saved conformations were carried out with the help of BIOVIA Discovery Studio.
Molecular dynamic simulation:
Protein-ligand complexes of phytochemicals having good binding affinity against both targeted proteins are subjected to Molecular Dynamic (MD) simulation. The selected 6GUE-107876 and 6GU7-4871complex files were subjected to MD simulation studies with the help of the GROMACS simulation package through the WebGRO and the GROMOS96 43a1 force field was chosen to perform this simulation[42]. Topology files of phytochemicals were built with the help of the PRODRG 2.5 server[43]. The entire protein-ligand complex system was solvated using the Simple Point Charge (SPC) water model with a triclinic box[44]. The 5000 steps of steepest descent were performed for Energy Minimization (EM) of the subjected complex[41]. The complex system was neutralized and the simulation was performed in the presence of 0.15 M NaCl[45]. Equilibration of the simulated systems was done using canonical (NVT) and isothermal-isobaric (NPT) ensembles after each step of EM[46-48]. The temperature on the system kept maintained using the Berendsen thermostat approach and pressure was kept maintained at 1.0 bar using the Parrinello-Rahman barostat approach to control the simulated proteinligand complex systems[49-51]. The simulation was performed under the 300 K temperature for 100 ns, respectively. The obtained trajectories of MD simulation of 100 ns were subjected to determine the conformational changes and stability of the protein-ligand complex. Obtained MD trajectories were subjected to analysis of the Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), Radius of gyration (Rg) and Hydrogen Bonds (HBs), of the subjected complex system of hit molecule[52,53].
Results and Discussion
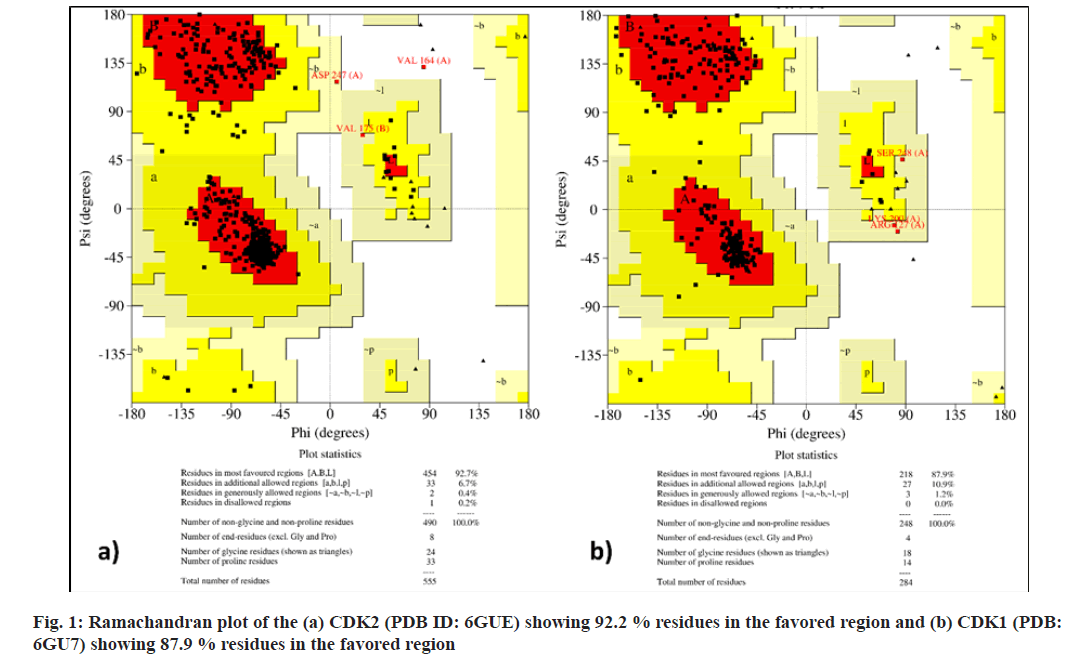
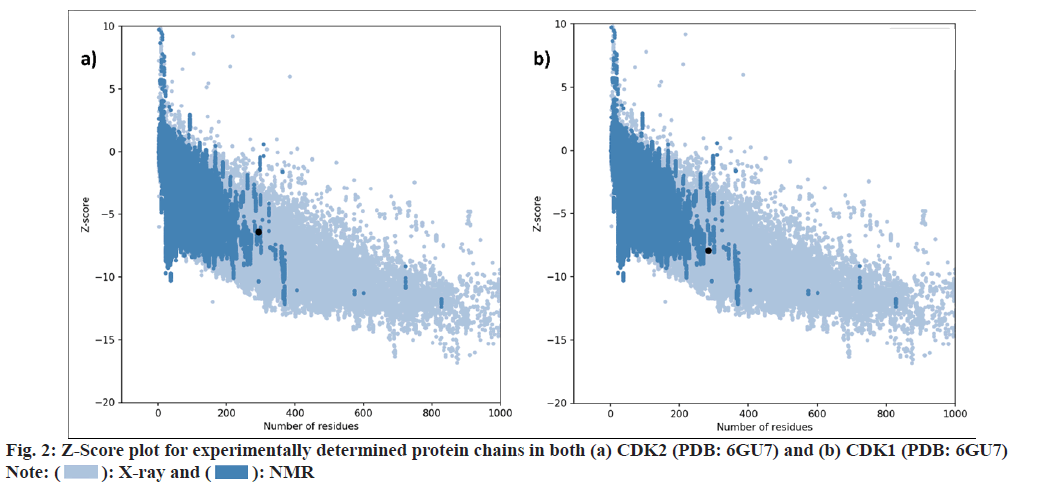
Virtual screening techniques help to increase the efficiency of the drug discovery process. Target proteins were selected on the basis of previously reported literatures. Selected phytochemicals were prepared for molecular docking using the free version of BIOVA Discovery Studio 2020. The selected protein structure was subjected to quality evaluation and binding site analysis by using PROCHECK and ProSA-web servers. The Ramachandran plot revealed that 92.7 % of PDB: 6GUE and 87.9 % of 6GU7 residues are present in favored regions as shown in fig. 1. The z-score indicates overall model quality for both the prepared protein structures was found to be -7.93 for PDB: 6GUE and -6.42 for PDB: 6GU7. Fig. 2 represents a plot that contains the z-scores of all experimentally determined protein chains in both PDB: 6GU7 and 6GUE.
Over the past few decades molecular docking technique established as an outstanding tool to determine the binding energies and best binding confirmations in the interaction of target protein and potential small molecules and nowadays, it is a widely applied computational tool. In the present work, molecular docking was done by using the AutoDock Vina package of PyRx 0.8 software[41]. Downloaded phytochemicals were subjected to molecular docking against CDK1 and CDK2 (PDB ID: 6GU7 and 6GUE). The binding affinity of phytochemicals against CDK1 (PDB: 6GU7) ranged from -4.1 to -10.4 kcal/mol and for phytochemicals against CDK2 (PDB: 6GUE) ranged from -4.5 to -9.9 kcal/mol. Among all the phytochemicals, PubChem CID 4871 showed a good binding affinity of -10.4 kcal/mol with CDK1 (PDB: 6GU7) and for CDK2 (PDB: 6GUE), PubChem CID 107876 showed good binding affinity of -9.9 kcal/mol. The binding affinity of the docked control i.e. Dinaciclib showed a binding affinity of -8.6kcal/mol with PDB:6GU and -9.2kcal/mol with PDB: 6GUE. Both the selected hits i.e. PubChem CID 4871 against CDK1 (PDB: 6GU7) and PubChem CID 107876 against CDK2 (PDB: 6GUE) showed better binding affinity than compared standard (Dinaciclib). The BIOVA Discovery Studio Visualizer was used to determination of detailed Two Dimensional (2D) and 3D interactions of best conformation. The interaction of PubChem CID 4871 with residues of CDK1 (PDB: 6GU7) and PubChem CID 107876 with CDK2 (PDB: 6GUE) showed the highest affinity and interaction. GLN132, ASP146, ALA31, LEU135, PHE82, ILE10, LEU83, GLU130, THR14, GLY13, LYS130, PHE80, VAL64, ALA145, VAL18 are the interacting amino acid residues between PubChem CID 4871 and CDK1 (PDB: 6GU7) as shown in fig. 3a and fig. 3b and they formed van der Waals, conventional hydrogen bond, carbon hydrogen bond, π-Cation, π-Sigma, Alkyl, π-Alkyl interactions. While in the case of PubChem CID 107876 and CDK2 (PDB: 6GUE), LYS178, PRO155, ARG157, VAL154, GLY153, THR316, GLN313, GLU230, GLN228, ASN229, TYR418, MET334, TYR413, SER331, HIS419, LYS417, GLU330, PHE319, TYR179, VAL156 are interacting amino acid residues as shown in fig. 3c and fig. 3d. van der Waals interaction was not indicated in the 2D and 3D visitation of interaction as it does not have a direct interaction between the protein-ligand complex. Both the phytochemicals were further subjected to a molecular dynamics simulation study to determine the stability of the complex over 100 ns of simulation time.
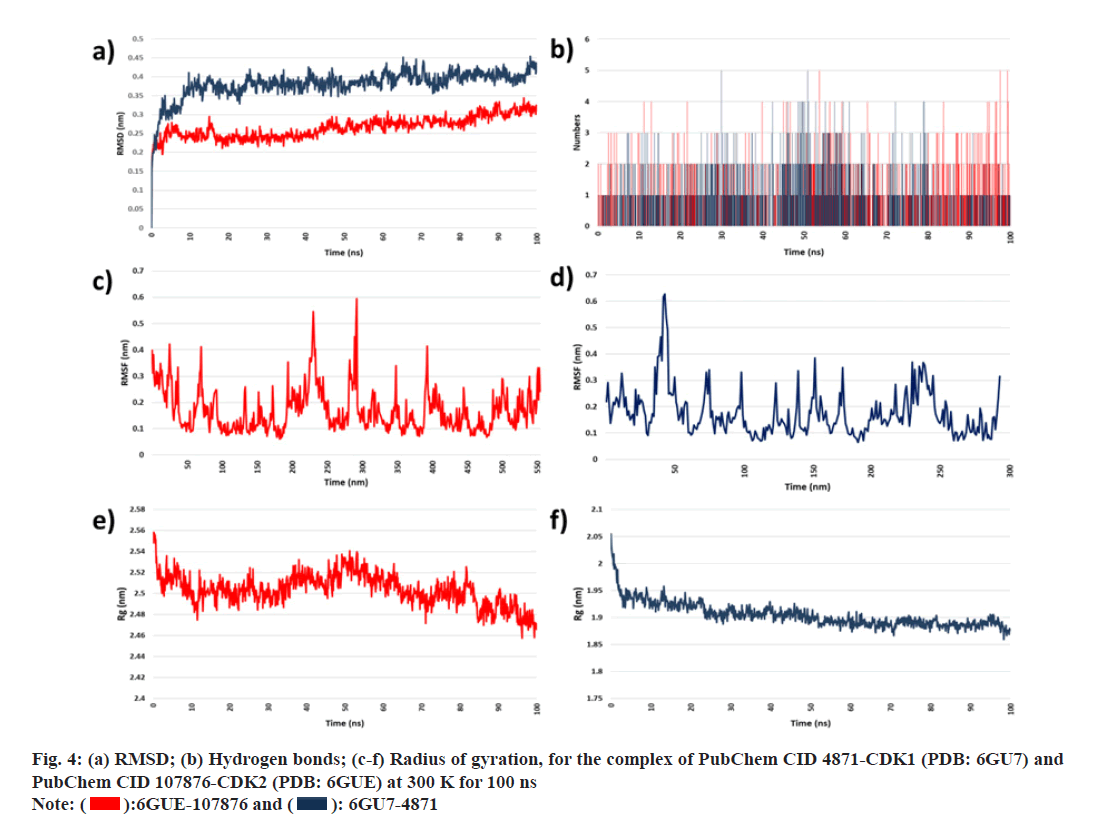
MD simulation studies were performed on proteinligand complexes having good binding affinity. Through the MD simulation technique, the dynamic behavior and configurational changes between 6GUE-107876 and 6GU7-4871 complexes were analyzed over 100 ns. Analysis of parameters like RMSD, RMSF, Rg and HBs were done over 100 ns using generated MD trajectories from WebGro. The conformational stability of the protein-ligand complex system was analyzed with the help of RMSD. RMSD of the protein-ligand complex was calculated to obtain the equilibrium time of the simulated complexes (6GUE-107876 and 6GU7- 4871). Low RMSD indicates more stability of the protein-ligand complex system. Fig. 4a represents the RMSD plot for the 6GUE-107876 and 6GU7- 4871. Through the results, it is observed that the RMSD of complexes showed minimum deviation over the simulation time of 100 ns and it indicates that both complexes have good conformational stability. The number of hydrogen bonds present in the complexes with its consistency throughout the 100 ns simulation was determined at 300 K, as shown in fig. 4b. There were not many considerable changes observed in the hydrogen bond interaction within the protein-ligand complexes. RMSF was plotted at 300 K as given in fig. 4c and fig. 4d. Through the RMSF values, it is observed that fluctuations occur in 6GUE-107876 complex and LEU25 (RMSF value: 2.3530 Å), THR97 (RMSF value: 1.9860 Å), GLU138 (RMSF value: 2.0070 Å), ARG157 (RMSF value: 1.9930 Å), TYR159 (RMSF value: 2.9340 Å), SER171 (RMSF value: 3.7090 Å), PRO272 (RMSF value: 1.9310 Å) amino acid residues showed fluctuations. And in case of 6GU7-4871, GLU40 (RMSF value: 3.8220 Å), GLU41 (RMSF value: 3.9000 Å), PRO95 (RMSF value: 2.4580 Å), PRO96 (RMSF value: 3.0990 Å), GLY97 (RMSF value: 3.0930 Å), ASN 224 (RMSF value: 4.4950 Å), ASN225 (RMSF value: 5.4240 Å), GLU226 (RMSF value: 3.4240 Å) amino acid residues showed fluctuations, in which ASN225 showed highest RMSD value. However, the RMSF values of both complexes remain in the acceptable range indicating the stability of particular amino acid residues. Rg values for both 6GUE-107876 and 6GU7-4871complexes have been studied over 100 ns at 300 K and it is observed that the Rg values for the 6GUE-107876 ranged between 2.46-2.58 nm and for 6GU7-4871 Rg value ranged between 1.85 to 1.95 nm with showing conformational stability of the complex with minimum fluctuation in the plot, as shown in fig. 4e and fig. 4f.
Phytochemicals having anticancer activity were virtually screened against CDK1 (PDB: 6GU7) and CDK2 (PDB: 6GUE) to determine the binding affinity and stability. Through the results of molecular docking, it is observed that out of all, PubChem CID 4871 and PubChem CID 107876 showed a good binding affinity with the targeted proteins and the visualization of interaction formed between both the complex reviled that VDW, Conventional H Bond, Carbon H Bond, π-Cation, π-Sigma, Alkyl, π-Alkyl played a major role in the binding of the ligand with targeted protein. MD simulation of protein-ligand complexes having good binding affinity was performed over 100 ns using the RMSD, RMSF, Rg and hydrogen bonds was analyzed. Through the results of MD simulation, it is observed that subjected 6GUE- 107876 and 6GU7-4871 complexes were stable throughout the performed 100 ns simulation time, although the CDK1/CDK2 inhibitory activity of subjected phytochemicals requires an in vivo or in vitro experimental confirmation.
Conflict of interest:
The authors confirm that this article's content has no conflict of interest.
References
- Wang B, He F, Hu Y, Wang Q, Wang D, Sha Y, et al. Cancer incidence and mortality and risk factors in member countries of the " Belt and Road " initiative. BMC Cancer 2022;22(1):582.
[Crossref] [Google Scholar] [PubMed]
- Sathishkumar K, Chaturvedi M, Das P, Stephen S, Mathur P. Cancer incidence estimates for 2022 and projection for 2025: Result from National Cancer Registry Programme, India. Indian J Med Res 2022;156(4-5):598-607.
[Crossref] [Google Scholar] [PubMed]
- Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, et al. Cancer statistics for the year 2020: An overview. Int J Cancer 2021;149(4):778-89.
[Crossref] [Google Scholar] [PubMed]
- Pilleron S, Soto-Perez-de-Celis E, Vignat J, Ferlay J, Soerjomataram I, Bray F, et al. Estimated global cancer incidence in the oldest adults in 2018 and projections to 2050. Int J Cancer 2021;148(3):601-8.
[Crossref] [Google Scholar] [PubMed]
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin 2022;72(1):7-33.
[Crossref] [Google Scholar] [PubMed]
- Kocarnik JM, Compton K, Dean FE, Fu W, Gaw BL, Harvey JD, et al. Cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life years for 29 cancer groups from 2010 to 2019 a systematic analysis for the global burden of disease study 2019. JAMA Oncol 2022;8(3):420-44.
[Crossref] [Google Scholar] [PubMed]
- Ikwu FA, Isyaku Y, Obadawo BS, Lawal HA, Ajibowu SA. In silico design and molecular docking study of CDK2 inhibitors with potent cytotoxic activity against HCT116 colorectal cancer cell line. J Genet Eng Biotechnol 2020;18(1):51.
[Crossref] [Google Scholar] [PubMed]
- Sapra Sharma P, Sharma R, Tyagi R. Inhibitors of cyclin dependent kinases: Useful targets for cancer treatment. Curr Cancer Drug Targets 2008;8(1):53-75.
[Crossref] [Google Scholar] [PubMed]
- Rajhans S, Pandya PN, Rawal RM, Pandya HA. CDK1 and CDK2 as potential targets for anticancer activity. Plant Arch 2020;20(1):895-901. [Crossref]
[Google Scholar] [PubMed]
- Sanchez-Martínez C, Lallena MJ, Sanfeliciano SG, de Dios A. Cyclin Dependent Kinase (CDK) inhibitors as anticancer drugs: Recent advances (2015-2019). Bioorg Med Chem Lett 2019;29(20):126637.
[Crossref] [Google Scholar] [PubMed]
- Saikat ASM. An in silico approach for potential natural compounds as inhibitors of protein CDK1/Cks2. Chem Proc 2022;8(1):5.
- Liang JW, Wang MY, Wang S, Li SL, Li WQ, Meng FH. Identification of novel CDK2 inhibitors by a multistage virtual screening method based on SVM, pharmacophore and docking model. J Enzyme Inhib Med Chem 2020;35(1):235-44.
[Crossref] [Google Scholar] [PubMed]
- Tadesse S, Anshabo AT, Portman N, Lim E, Tilley W, Caldon CE, et al. Targeting CDK2 in cancer: Challenges and opportunities for therapy. Drug Discov Today 2020;25(2):406-13.
[Crossref] [Google Scholar] [PubMed]
- Chohan T, Qian H, Pan Y, Chen JZ. Cyclin-dependent kinase-2 as a target for cancer therapy: Progress in the development of CDK2 inhibitors as anti-cancer agents. Curr Med Chem 2014;22(2):237-63.
[Crossref] [Google Scholar] [PubMed]
- Choudhari AS, Mandave PC, Deshpande M, Ranjekar P, Prakash O. Phytochemicals in cancer treatment: From preclinical studies to clinical practice. Front Pharmacol 2020;10:1614.
[Crossref] [Google Scholar] [PubMed]
- Naeem A, Hu P, Yang M, Zhang J, Liu Y, Zhu W, et al. Natural products as anticancer agents: Current status and future perspectives. Molecules 2022;27(23):8367.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Shi W, Wu C, Wan L, Zhao Y, Zhang C, et al. Pyrazole ring-containing isolongifolanone derivatives as potential CDK2 inhibitors: Evaluation of anticancer activity and investigation of action mechanism. Biomed Pharmacother 2021;139:111663.
[Crossref] [Google Scholar] [PubMed]
- Stepchenkova EI, Zhuk AS, Cui J, Tarakhovskaya ER, Barbari SR, Shcherbakova PV, et al. Compensation for the absence of the catalytically active half of DNA polymerase ε in yeast by positively selected mutations in CDC28. Genetics 2021;218(2):iyab060
[Crossref] [Google Scholar] [PubMed]
- Varun E, Bhakti K, Aishwarya K, Suraj RH, Jagadish MR, Mohana KP. Rohitukine content across the geographical distribution of Dysoxylum binectariferum Hook F. and its natural derivatives as potential sources of CDK inhibitors. Heliyon 2023;9(2):e13469.
[Crossref] [Google Scholar] [PubMed]
- Khalipha AB, Bagchi R, Hossain MS, Mondal M, Biswas S, Ray P, et al. A literature based review on rohitukine and molecular docking studies of flavopiridol (rohitukine derived) and its derivatives against Cyclin-Dependent Kinases (CDKs) for anticancer activity. Int J Sci Res 2019;1(1):15-28.
- Sofi S, Mehraj U, Qayoom H, Aisha S, Almilaibary A, Alkhanani M, et al. Targeting Cyclin-Dependent Kinase 1 (CDK1) in cancer: Molecular docking and dynamic simulations of potential CDK1 inhibitors. Med Oncol 2022;39(9):133.
[Crossref] [Google Scholar] [PubMed]
- Ahmad W, Ansari MA, Alsayari A, Almaghaslah D, Wahab S, Alomary MN, et al. In vitro, molecular docking and in silico ADME/Tox studies of emodin and chrysophanol against human colorectal and cervical carcinoma. Pharmaceuticals (Basel) 2022;15(11):1348.
[Crossref] [Google Scholar] [PubMed]
- Govindarasu M, Ganeshan S, Ansari MA, Alomary MN, AlYahya S, Alghamdi S, et al. In silico modeling and molecular docking insights of kaempferitrin for colon cancer-related molecular targets. J Saudi Chem Soc 2021;25(9):101319.
- Wood DJ, Korolchuk S, Tatum NJ, Wang LZ, Endicott JA, Noble MEM, et al. Differences in the conformational energy landscape of CDK1 and CDK2 suggest a mechanism for achieving selective CDK inhibition. Cell Chem Biol 2019;26(1):121-130.e5.
[Crossref] [Google Scholar] [PubMed]
- Bagal VK, Rathod SS, Mulla MM, Pawar SC, Choudhari PB, Pawar VT, et al. Exploration of bioactive molecules from Tinospora cordifolia and Actinidia deliciosa as an immunity modulator via molecular docking and molecular dynamics simulation study. Nat Prod Res 2023;37(23):4053-7.
[Crossref] [Google Scholar] [PubMed]
- Roman Laskowski BA, Macarthur MW, Thornton JM. Computer programs PROCHECK: A program to check the stereochemicai quality of protein structures. J Appl Crystallogr 1983;26(2):283-91.
- Wiederstein M, Sippl MJ. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007;35(Suppl.2):W407-10.
[Crossref] [Google Scholar] [PubMed]
- Mohanraj K, Shanmugam Karthikeyan B, Vivek-Ananth RP, Bharath Chand RP, Aparna SR, Mangalapandi P, et al. IMPPAT: A curated database of Indian medicinal plants, phytochemistry and therapeutics. Sci Rep 2018;8:4329.
[Crossref] [Google Scholar] [PubMed]
- Islam MR, Awal MA, Khames A, Abourehab MAS, Samad A, Hassan WMI, et al. Computational identification of druggable bioactive compounds from Catharanthus roseus and Avicennia marina against colorectal cancer by targeting thymidylate synthase. Molecules 2022;27(7):2089.
[Crossref] [Google Scholar] [PubMed]
- Palanichamy C, Pavadai P, Panneerselvam T, Arunachalam S, Babkiewicz E, Pandian SRK, et al. Aphrodisiac performance of bioactive compounds from Mimosa pudica linn.: In silico molecular docking and dynamics simulation approach. Molecules 2022;27(12):3799.
[Crossref] [Google Scholar] [PubMed]
- Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M, Lyu WY, et al. m6A modification: Recent advances, anticancer targeted drug discovery and beyond. Mol Cancer 2022;21(1):52.
[Crossref] [Google Scholar] [PubMed]
- Ahammad F, Alam R, Mahmud R, Akhter S, Talukder EK, Tonmoy AM, et al. Pharmacoinformatics and molecular dynamics simulation-based phytochemical screening of neem plant (Azadiractha indica) against human cancer by targeting MCM7 protein. Brief Bioinform 2021;22(5):bbab098.
[Crossref] [Google Scholar] [PubMed]
- Islam SI, Mou MJ, Sanjida S, Mahfuj S. An in-silico approach for identifying phytochemical inhibitors against Nervous Necrosis Virus (NNV) in asian sea bass by targeting capsid protein. Genet Aquat Org 2022;6(2):1-13. [Crossref]
[Google Scholar] [PubMed]
- Jha V, Thakur K, Kaur N, Dhamapurkar V, Bhosale O, Mhatre P, et al. Computational screening of medicinal plant phytochemicals to discover potent inhibitors against hepatitis B virus. J Adv Biol Biotechnol 2022;25(4):22-38.
- Nath A, Nair AS. Fingerprint-based similarity search identified p-anisidine as an anticancer agent in HeLa and a prospective phytochemical ETV1 transcription factor inhibitor. J Biomol Struct Dyn 2020;39(14):4973-80.
[Crossref] [Google Scholar] [PubMed]
- O’boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform 2011;3(33):1-14.
[Crossref] [Google Scholar] [PubMed]
- Gaikwad R, Rathod S, Shinde A. In-silico study of phytoconstituents from tribulus terrestris as potential anti-psoriatic agent. Asian J Pharm Res 2022;12(4):267-74.
- Kumar SK, Laplant B, Chng WJ, Zonder J, Callander N, Fonseca R, et al. Dinaciclib, a novel CDK inhibitor, demonstrates encouraging single-agent activity in patients with relapsed multiple myeloma. Blood 2015;125(3):443-8
[Crossref] [Google Scholar] [PubMed]
- Eberhardt J, Santos-Martins D, Tillack AF, Forli S. AutoDock vina 1.2.0: New docking methods, expanded force field, and python bindings. J Chem Inf Model 2021;61(8):3891-8.
[Crossref] [Google Scholar] [PubMed]
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol 2015;1263:243-50.
[Crossref] [Google Scholar] [PubMed]
- Rathod S, Shinde K, Porlekar J, Choudhari P, Dhavale R, Mahuli D, et al. Computational exploration of anti-cancer potential of flavonoids against cyclin-dependent kinase 8: An in silico molecular docking and dynamic approach. ACS Omega 2022;8(1):391-409.
[Crossref] [Google Scholar] [PubMed]
- Pol-Fachin L, Fernandes CL, Verli H. GROMOS96 43a1 performance on the characterization of glycoprotein conformational ensembles through molecular dynamics simulations. Carbohydr Res 2009;344(4):491-500.
[Crossref] [Google Scholar] [PubMed]
- Schüttelkopf AW, van Aalten DMF. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 2004;60(8):1355-63.
[Crossref] [Google Scholar] [PubMed]
- Izadi S, Anandakrishnan R, Onufriev AV. Building water models: A different approach. J Phys Chem Lett 2014;5(21):3863-71.
[Crossref] [Google Scholar] [PubMed]
- Gorai S, Junghare V, Kundu K, Gharui S, Kumar M, Patro BS, et al. Synthesis of dihydrobenzofuro [3,2-b] chromenes as potential 3CLpro inhibitors of SARS-CoV-2: A molecular docking and molecular dynamics study. ChemMedChem 2022;17(8):e202100782.
[Crossref] [Google Scholar] [PubMed]
- Nose SU. A molecular dynamics method for simulations in the canonical ensemble. Mol Phys 2002;100(1):191-8.
- Huang C, Li C, Choi PYK, Nandakumar K, Kostiuk LW. A novel method for molecular dynamics simulation in the isothermal-isobaric ensemble. Mol Phys 2011;109(2):191-202.
- Bepari AK, Reza HM. Identification of a novel inhibitor of SARS-CoV-2 3CL-PRO through virtual screening and molecular dynamics simulation. PeerJ 2021;9:e11261.
[Crossref] [Google Scholar] [PubMed]
- Berendsen HJC, Postma JPM, van Gunsteren WF, Dinola A, Haak JR. Molecular dynamics with coupling to an external bath. J Chem Phys 1984;81(8):3684-90.
- Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys 1981;52(12):7182-90.
- Kushwaha PP, Singh AK, Bansal T, Yadav A, Prajapati KS, Shuaib M, et al. Identification of natural inhibitors against SARS-CoV-2 drugable targets using molecular docking, molecular dynamics simulation, and MM-PBSA approach. Front Cell Infect Microbiol 2021;11:730288.
[Crossref] [Google Scholar] [PubMed]
- Jiang Z, You L, Dou W, Sun T, Xu P. Effects of an electric field on the conformational transition of the protein: A molecular dynamics simulation study. Polymers (Basel) 2019;11(2):282.
[Crossref] [Google Scholar] [PubMed]
- Jain AS, Sushma P, Dharmashekar C, Beelagi MS, Prasad SK, Shivamallu C, et al. In silico evaluation of flavonoids as effective antiviral agents on the spike glycoprotein of SARS-CoV-2. Saudi J Biol Sci 2021;28(1):1040-51.
[Crossref] [Google Scholar] [PubMed]
 ): X-ray and (
): X-ray and ( ): NMR
): NMR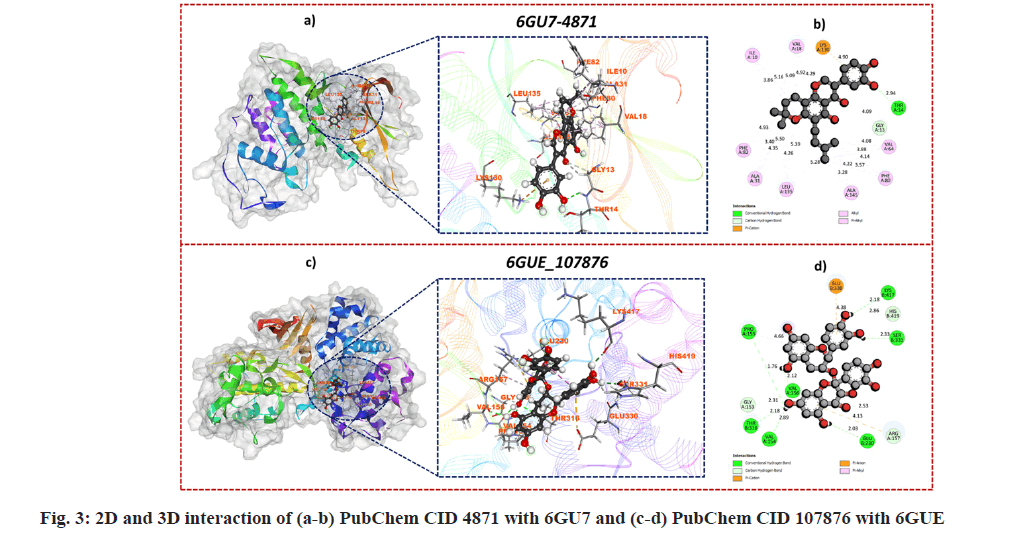
 ):6GUE-107876 and (
):6GUE-107876 and ( ): 6GU7-4871
): 6GU7-4871



