- *Corresponding Author:
- S. A. Muhammad
Institute of Molecular Biology and Biotechnology, Bahauddin Zakariya University, Multan, Pakistan
E-mail: draun@bzu.edu.pk
| Date of Submission | 30 Jun 2015 |
| Date of Revision | 12 Mar 2016 |
| Date of Acceptance | 23 Apr 2016 |
| Indian J Pharm Sci, 2016;78(2):259-266 |
Abstract
Leishmaniasis is a protozoal infection which presents a broad spectrum drug resistance and causing diseases ranging from asymptomatic infections to significant mortality. The enzyme, trypanothione reductase from Leishmania infantumis is essential for the parasite survival. It is an attractive target to design new less toxic drugs against Leishmaniasis as found absent in mammalian cells. The antileishmanial activity of epicoccamide derivatives was studied by in silico approaches. The crystallographic structure of trypanothione reductase was obtained from the protein data bank (ID: 2W0H) database. Binding mode of the epicoccamide inhibitors showed that they are stabilized in the active site of enzyme through hydrogen bond and hydrophobic interactions. These derivatives (A-D) showed significant binding affinities with trypanothione reductase with binding energies: -13.21, -13.44, -13.31, and -13.52 kcal/mol, respectively.The present study showed new and effective antileishmanial compounds that can get pharmaceutical importance.
Keywords
Leishmaniasis, trypanothione reductase, epicoccamide derivatives, in silico analysis
Leishmaniasis is a protozoal disease that is closely related to stress factors like poverty, unhygienic conditions and poor living standards. This disease is most prominent in poor developing countries although WHO estimates that the disease threatens approximately 350 million individuals in 88 countries worldwide [1].
There are many species of the Leishmania that can infect humans and are responsible for the following different forms of leishmaniasis: mucocutaneous, cutaneous, visceral and diffuse cutaneous leishmaniasis [2-4]. Mucocutaneous leishmaniasis (ML) is caused by Leishmania braziliensis and its prognosis is sores on the skin while cutaneous leishmaniasis (CL) is caused by L. major. Visceral leishmaniasis (VL) is may be caused by any species like L. donovani, L. infantum or L. chagasi. This is the most severe form of the disease, which can be fatal if not treated. Diffuse cutaneous leishmaniasis (DCL) is caused by L.amazonensis and generates chronic ulcerations on the skin that are difficult to treat [5].
Currently pentavalent antimonials, sodium stibogluconate and meglumine antimoniate are the first-line drugs for leishmaniasis. However, they are highly toxic and require high doses and patient hospitalization for three to four weeks for parenteral administration [6]. Furthermore cases of increased resistance to antimonials have been reported and drugs used in case of resistance are also very toxic and needed to be administered parenterally [3,6,7]. Due to the increasing incidence of drug resistance and toxicity there is an urgent need to search for alternative therapies against new targets.
Several metabolic pathways are currently under study in order to identify critical enzymes for inhibition purposes. The aim of this study is identification of potential targets for the treatment of parasitic diseases [8]. The enzymes involved in metabolism of glucose, sterols, unsaturated fats and nucleotides as well as in protein biosynthesis and for the support of trypanothione and polyamine levels are considered targets for drug development. Recent studies showed that more than 29 enzymes from L. major, L. donovani, L. Mexicana and L. infantum could be used as a therapeutic targets [8]. Thiol-redox metabolism is present in leishmaniasis which is found absent in mammals and it is regulated/controlled by trypanothione reductase. Thiol-reductase plays a key role in survival of Leishmania sp. Trypanothione reductase is a validated target for rational drug design against visceral leishmaniasis, most lethal form of leishmaniasis, caused by Leishmania infantum [9-11].
Natural products are the most productive source for the discovery of new and novel compounds. Among natural sources, microorganisms especially fungi have been exemplary with the production of novel compounds that have proved to be effective antimicrobial, anticancer, antiviral and immunosuppressive agents [12]. Recently compounds with antileishmanial activity have been reported from isolated fungi [13-15]. Studies indicated that fungal metabolites might be potential lead molecules for antileishmanial drug discovery.
The current interest in the development of new antileishmanial lead molecules, combined with lack of adequate computer-aided drug discovery efforts in this area, prompted us to explore the fungal compounds for their trypanothione reductase inhibitory potential by using ligand based structure activity relationship model.
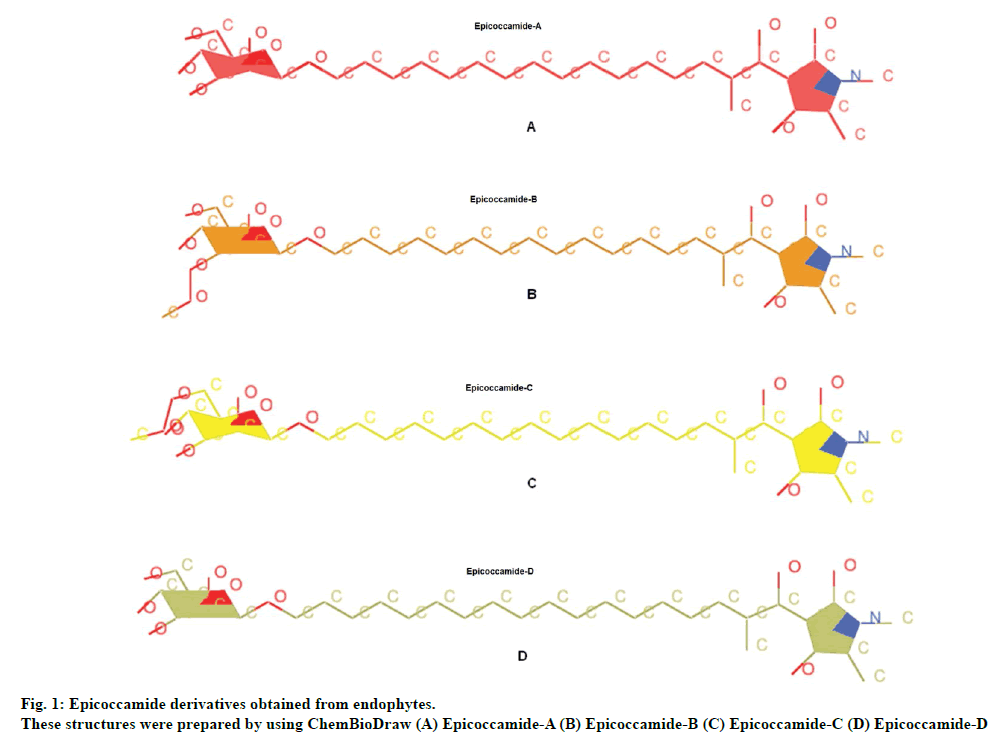
For current study tetramic acid containing natural fungal compounds (epicoccamides) were selected to prioritize them for in vitro evaluation. Naturally occurring tetramic acids constitute an important class of bioactive products with remarkably varied biological activity, spanning from antibiotic and antiviral, cytotoxicity, as well as inhibition of cell cycle [16,17]. Tetramic acid nuclei of epicoccamides are therefore found an interesting scaffold for drug designing. Epicoccamides are naturally occurring 3-acyltetramic acids bearing β-mannosylated fatty acid chains. Epicoccamide A (1) was isolated from the jellyfish derived fungus Epicoccum purpuraccerns (Epicoccum nigrum) by Wright et al., in 2003 [18], whereas epicoccamides B-D (2-4) were isolated from an Epicoccum sp. associated with fruiting body of tree fungus Pholiota squarrosa [19]. In current study, the potential and effective antileishmanial activity of these derivatives was determined by computational approaches.
Materials and Methods
The crystal structure of Leishmania trypanothione reductase (PDB ID: 2W0H) was obtained from the Protein Data Bank. Trypanothione reductase is essential for the parasite survival and virulence and has a potential role in specific trypanothione reductase system. Since this enzyme is not available in mammalian cells, it could be a potential drug target.
Antileishmanial ligands
Epicoccamide derivatives previously reported from endophytes were used as ligands molecules (fig. 1). These structures were prepared by using ChemBioDraw and MOL2 format of these ligands were converted to PDB file using OpenBabel tool, prior to upload onto ArgusLab drug design software [20].
Target and ligand optimization
For docking analysis, PDB coordinates of the target protein and ligand molecules were optimized by UCSF Chimera tool. These coordinates had minimum energy and stable conformation.
Analysis of drugable properties
The drugable properties (partition coefficient, molecular weight, toxicity) were evaluated using Mol inspiration server [21].
Analysis of target active binding sites
Proteins carry out their functions through interactions with other molecules and therefore accurately identifying the protein ligand binding site plays an important role in protein functional annotation and rational drug discovery. The active sites are the coordinates of the ligand in the original target protein grids and these active binding sites of target protein were analysed using the DoGSite Scorer: active site prediction and analysis server [22].
Molecular docking analysis
A computational ligand target docking approach was used to analyze structural complexes of the trypanothione reductase target with epicoccamide derivatives. Molecular docking was carried out by ArgusLab drug design software. At each step of the simulation, the energy of interaction of ligand and protein was evaluated using atomic affinity potentials computed on a grid. The remaining parameters were set as default.
Results and Discussion
The genus Leishmania is trypanosomatid protozoa and parasitic in nature which causes the disease known as leishmaniasis, which infects some 15 million people around the world in three clinical forms: cutaneous, mucocutaneous and visceral [23-25]. Current therapies for the disease are associated with a risk of high toxicity and development of drug resistant strains. Thiol-redox metabolism involving amino acid trypanothione and enzyme trypanothione reductase, essential for survival of Leishmania sp., is considered important target for drug design [11]. Computer aided drug design is a broadly used tool for developing relationships between the effects (e.g. activities and properties of interest) of a series of molecules with their structural properties. It is a dynamic area that integrates new technologies at a staggering rate [25,26]. In the present study, we address the performance of a computer aided drug design protocol involving homology modeling of trypanothione reductase and structure based virtual screening as a tool for identifying novel classes of potent trypanothione reductase inhibitors. Epicoccamides, naturally occurring 3-acyltetramic acids were used for computer aided docking studies. There are many reports about their diverse pharmacological activities spanning from antibiotic and antiviral, cytotoxicity, as well as inhibition of cell cycle [18,19].
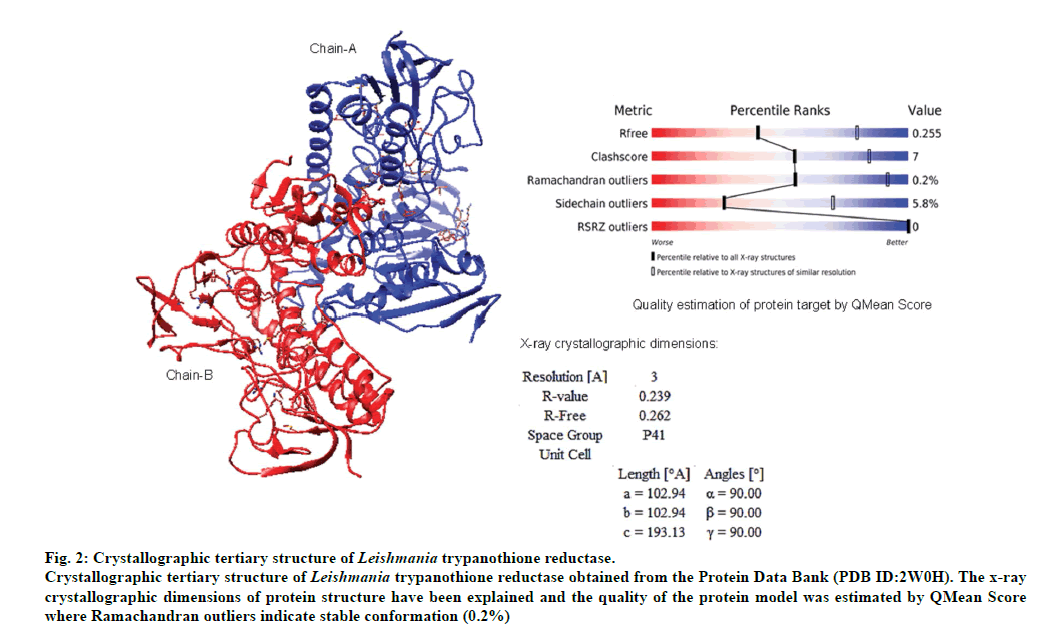
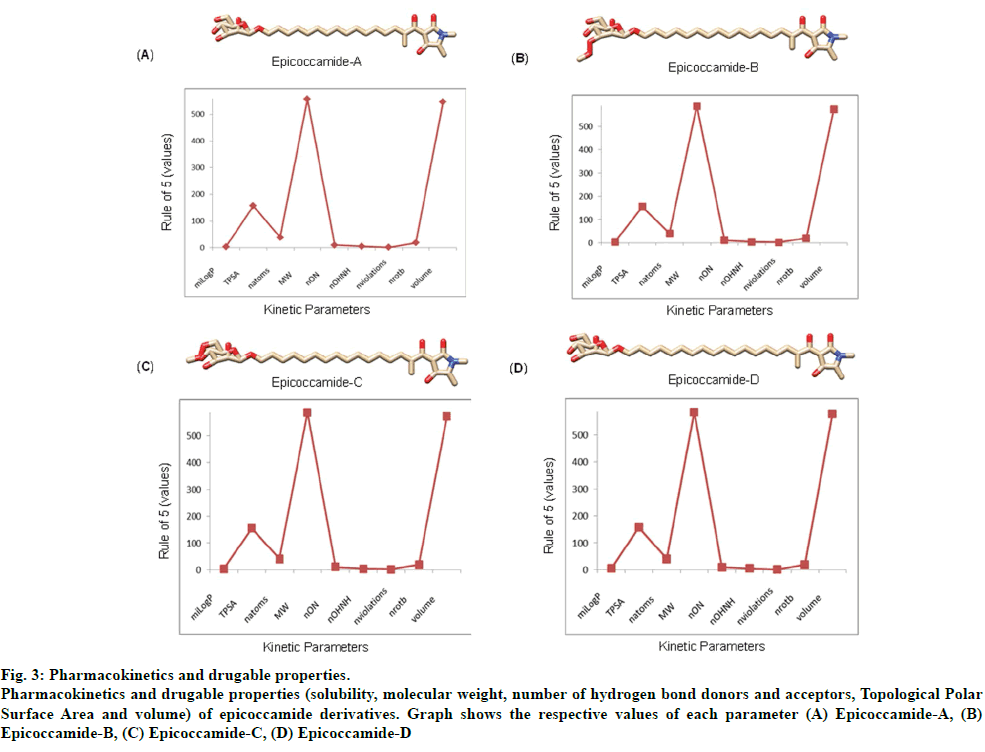
The validated tertiary crystallographic structure of Leishmania trypanothione reductase was used as target for molecular docking analysis (fig. 2). The minimum binding energy indicated that the target enzyme was successfully docked with epicoccamide derivatives (A-D). The pharmacokinetics and drugable properties of epicoccamide derivatives (AD) were calculated (fig. 3). These properties indicated that each derivative has drugable properties and can be effectively used to treat leishmaniasis.
Fig 2: Crystallographic tertiary structure of Leishmania trypanothione reductase.
Crystallographic tertiary structure of Leishmania trypanothione reductase obtained from the Protein Data Bank (PDB ID:2W0H). The x-ray
crystallographic dimensions of protein structure have been explained and the quality of the protein model was estimated by QMean Score
where Ramachandran outliers indicate stable conformation (0.2%)
Fig 3: Pharmacokinetics and drugable properties.
Pharmacokinetics and drugable properties (solubility, molecular weight, number of hydrogen bond donors and acceptors, Topological Polar
Surface Area and volume) of epicoccamide derivatives. Graph shows the respective values of each parameter (A) Epicoccamide-A, (B)
Epicoccamide-B, (C) Epicoccamide-C, (D) Epicoccamide-D
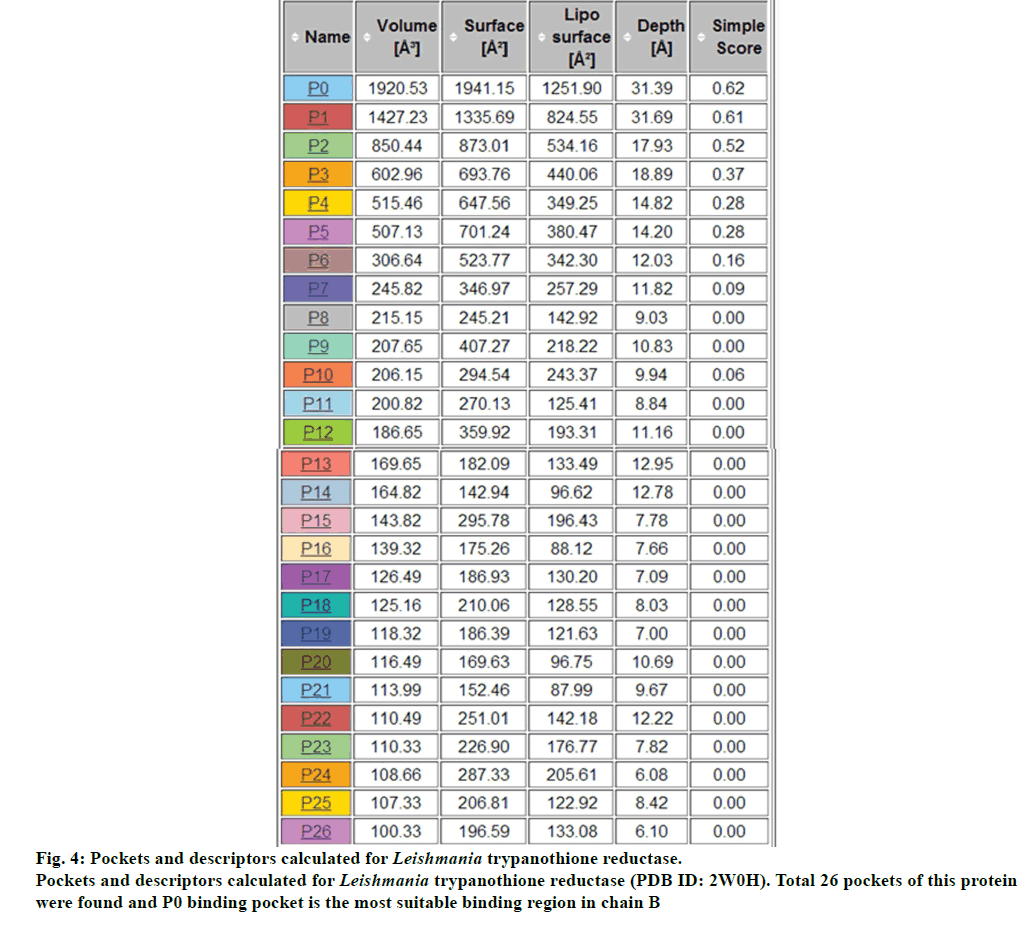
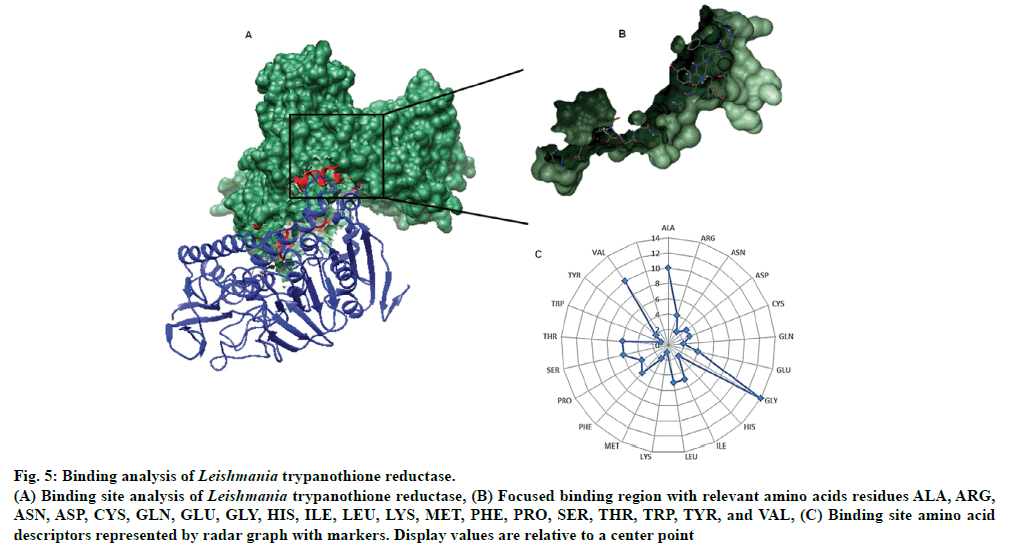
There were total 26-pockets of this protein and among them, P0 is the most suitable binding pocket with maximum simple score (0.62, fig. 4). In fig.5, amino acids descriptors of trypanothione reductase are shown: ALA, ARG, ASN, ASP, CYS, GLN, GLU, GLY, HIS, ILE, LEU, LYS, MET, PHE, PRO, SER, THR, TRP, TYR, and VAL. The various features of descriptors (size, shape, and element properties) for pocket P0 of trypanothione reductase was calculated and has been recorded in Tables 1 and 2.
| Descriptor | Value |
|---|---|
| volume [Å3] | 1920.53 |
| surface [Å2] | 1941.15 |
| lipophilic surface [Å2] | 1251.9 |
| depth [Å] | 31.39 |
| ellipsoid main axis ratio c/a | 0.16 |
| ellipsoid main axis ratio b/a | 0.45 |
| enclosure | 0.06 |
Table 1: Size And Shape Descriptors For Pocket P0 of Leishmania Trypanothione Reductase
| Descriptor | Value |
|---|---|
| # pocket atoms | 419 |
| # carbons (C) | 280 |
| # nitrogens (N) | 70 |
| # oxygens | 65 |
| # sulfors (S) | 4 |
| # other elements | 0 |
Table 2: Element Descriptors For Pocket P0 of Leishmania Trypanothione Reductase
Fig 5: Binding analysis of Leishmania trypanothione reductase.
(A) Binding site analysis of Leishmania trypanothione reductase, (B) Focused binding region with relevant amino acids residues ALA, ARG,
ASN, ASP, CYS, GLN, GLU, GLY, HIS, ILE, LEU, LYS, MET, PHE, PRO, SER, THR, TRP, TYR, and VAL, (C) Binding site amino acid
descriptors represented by radar graph with markers. Display values are relative to a center point
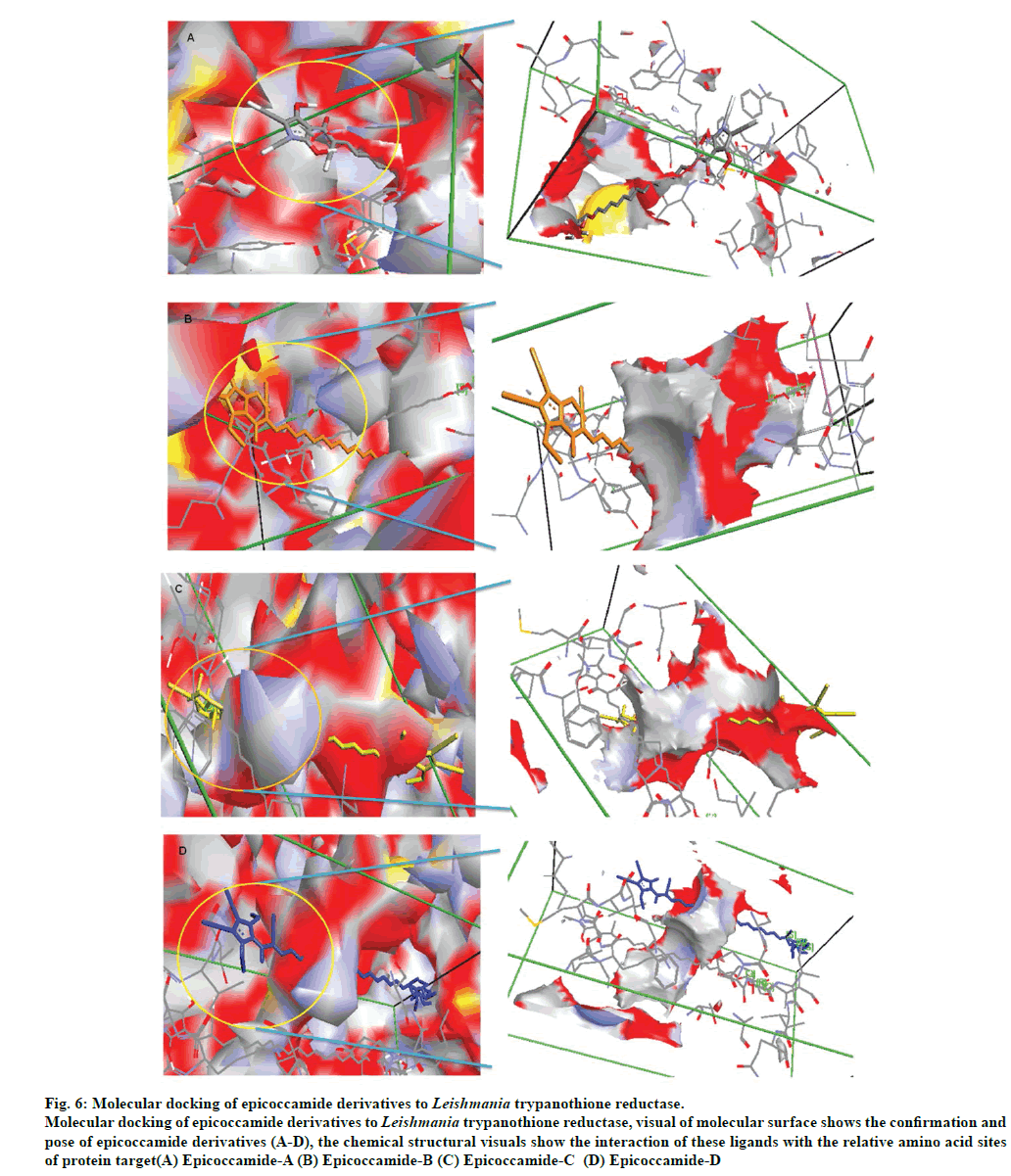
The molecular docking of trypanothione reductase with the epicoccamide derivatives revealed that all the computationally predicted lowest energy complexes of target protein are stabilized by intermolecular hydrogen bonds and stacking interactions. The molecular interaction presented the most energetically favorable binding mode of epicoccamide derivatives (A-D) to trypanothione reductase. The ligand showed the best interaction with target proteins based on the RMSD values [22]. Beside RMSD calculation; binding free energies of these interactive molecules were also calculated to find the best binding mode (Table 3). The calculated final docked energies for epicoccamide A-D are: -13.21, -13.44, -13.31, and -13.52 kcal/mol, respectively (fig.6). Docking results showed that these derivatives have potential to enter the substrate binding region of the active site and effective for leishmaniasis. However epicoccamide D was found most effective inhibitor of Try R due to lowest binding energy as compared to other ligands.
| Docking Pose | Complex | Binding Energy (kcal/mole) | RMSD/UBa | RMSD/LBa |
|---|---|---|---|---|
| 1 | 2W0H_Epicoccamide-A | -13.21 | 0 | 0 |
| 2 | 2W0H_Epicoccamide-A | -13.10 | 2.83 | 1.00 |
| 3 | 2W0H_Epicoccamide-A | -12.29 | 3.4 | 1.61 |
| 4 | 2W0H_Epicoccamide-A | -11.19 | 3.9 | 1.89 |
| 1 | 2W0H_Epicoccamide-B | -13.44 | 0 | 0 |
| 2 | 2W0H_Epicoccamide-B | -13.26 | 2.43 | 1.01 |
| 3 | 2W0H_Epicoccamide-B | -13.04 | 2.09 | 1.81 |
| 4 | 2W0H_Epicoccamide-B | -12.54 | 3.26 | 1.62 |
| 1 | 2W0H_Epicoccamide-C | -13.31 | 0 | 0 |
| 2 | 2W0H_Epicoccamide-C | -13.01 | 2.21 | 1.72 |
| 3 | 2W0H_Epicoccamide-C | -12.19 | 2.00 | 1.33 |
| 4 | 2W0H_Epicoccamide-C | -11.21 | 3.19 | 1.21 |
| 1 | 2W0H_Epicoccamide-D | -13.52 | 0 | 0 |
| 2 | 2W0H_Epicoccamide-D | -13.12 | 2.00 | 1.91 |
| 3 | 2W0H_Epicoccamide-D | -12.52 | 1.99 | 1.71 |
| 4 | 2W0H_Epicoccamide-D | -12.13 | 3.14 | 1.52 |
Table 3: Energy And Rmsd Values Obtained During Docking Analysis Of Epicoccamide Derivatives (A-D) With Leishmania Trypanothione Reductase (2WOH)
Fig 6: Molecular docking of epicoccamide derivatives to Leishmania trypanothione reductase.
Molecular docking of epicoccamide derivatives to Leishmania trypanothione reductase, visual of molecular surface shows the confirmation and
pose of epicoccamide derivatives (A-D), the chemical structural visuals show the interaction of these ligands with the relative amino acid sites
of protein target(A) Epicoccamide-A (B) Epicoccamide-B (C) Epicoccamide-C (D) Epicoccamide-D
In conclusion, we have identified molecular details of the binding modes of natural fungal metabolites epicoccamides for antileishmanial activity against trypanothione reductase. The present study is an excellent starting point for future studies of structural optimization of these compounds, whose observed details at the molecular level opened a route towards developing therapeutic epicoccamide based prototypes against Leishmania.
Financial Assistance
None.
Conflict of Interests
None declared.
References
- World Health Organization, Control of leishmaniases, Tech Report Series 2010;949:22-26.
- Ashford RW. The leishmaniases as emerging and reemergingzoonoses. Int J Parasitol 2000;30:1269-81.
- Avlonitis MR, Lekka E, Detsi A, Koufaki M, Calogeropoulou T, Scoulica E, et al. Antileishmanial ring-substituted ether phospholipids. J Med Chem 2003;46:755-67.
- Croft SL, Coombs GH. Leishmaniasis-current chemotherapy and recent advances in the search for novel drugs. Trends Parasitol 2003;19:502-8.
- Shukla AK, Singh BK, Patra S, Debuy VK. Rational approaches for drug designing against leishmaniasis. ApplBiochemBiotechnol 2010;160:2208-18.
- Guerin PJ, Olliaro P, Sundar S, Boelaert M, Croft SL, Desjeux P, et al. Visceral leishmaniasis: current status of control, diagnosis, and treatment, and a proposed research and development agenda. Lancet Infect Dis 2002;2:494-501.
- Alavi-Naini R. Topical morphine for the treatment of cutaneous leishmaniasis. Med Hyp 2008;70:81-4.
- Bernal FA, Coy-Barrera E. In silico analyses of sesquiterpene-related compounds on selected leishmania enzyme-based targets. Molecules 2014;19:5550-69.
- Dumas C, Ouellette M, Tovar J, Cunningham ML, Fairlamb AH, Tamar S, et al. Disruption of the trypanothionereductase gene of Leishmania decreases its ability to survive oxidative stress in macrophages. EMBO J 1997;16:2590-8.
- Tovar J, Cunningham ML, Smith AC, Croft SL, Fairlamb AH. Down regulation of Leishmaniadonovanitrypanothionereductase by heterologous expression of a transdominant mutant homologue: effect on parasite intracellular survival. ProcNatlAcadSci USA 1998;95:5311-16.
- Venkatesan SK, Shukla AK, Dubey VK. Molecular docking studies of selected tricyclic and quinone derivatives on trypanothionereductase of Leishmaniainfantum. J Comp Chem 2010;31:2463-75.
- Pimentel MR, Molina G, Dionisio AP, Junior MRM, Pastore GM. The Use of endophytes to obtain bioactive compounds and their application in biotransformation process. Biotechnol Res Int 2011;1:11.
- Metwaly AM, Kadry HA, El-Hela AA, Mohammad AI, Ma G, Cutler SJ, et al.Antileukemic, antileishmanial and antifungal activities of secondary metabolites from the endophytic fungus Nigrosporasphaerica. Planta Med 2013;79:P52.
- Malak LG, Ibrahim MA, Bishay DW, Abdel-baky AM, Moharram AM, Tekwani B, et al. Antileishmanial metabolites from Geosmithia langdonii. J Nat Prod 2014;77:1987-91.
- Balunas MJ, Grosso MF, Villa FA, Engene N, McPhail KL, Tidgewell K, Pineda et al. Antileishmanial marine cyanobacterialpolyketides with intriguing biosynthetic origins. Org Lett 2012;14:3878-81.
- Royles BJL. Naturally occurring tetramic acids: Structure, isolation and synthesis. Chem Rev 1995;95:1981-2001.
- Ghisalberti EL. Bioactive Tetramic Acid Metabolites. Atta-ur-Rahman, editor. Bioactive Natural Products, Part I. Netherlands: Elsevier Science; 2003, p. 109-63.
- Wright AD, Osterhage C, König GM. Epicoccamide, a novel secondary metabolite from a jellyfish-derived culture of Epicoccumpurpurascens. Org BiomolChem 2003;1:507-10.
- Wangun HVK, Dahse HM, Hertweck C. Epicoccamides B-D, glycosylated tetramic acid derivatives from an Epicoccum sp. associated with the tree fungus Pholiotasquarrosa. J Nat Prod 2007;70:1800-03.
- Humkey RN, Purvis GD, Thompson MA, Nigel R. Enhancing the PMF scoring function for molecular docking and virtual screening. J Med Chem 2005.
- Chemoinformatic on the web, Slavak republic. http://www.molinspiration.com
- Volkamer A, Kuhn D, Grombacher T, Rippmann F, Rarey M. Combining global and local measures for structure-based drug ability predictions. J ChemInf Model 2012;52:360-72.
- Reithinger R, Dujardin JC, Louzir H, Pirmez C, Alexander B, Brooker S. Cutaneous leishmaniasis. Lancet Infect Dis 2007;7:581-96.
- Loría-Cervera EN, Andrade-Narváez FJ. Animal models for the study of leishmaniasis immunology. Rev Inst Med Trop Sao Paulo 2014;56:1-11.
- Abu-Khalaf R, Abu-Sheikha G, Bustanji Y, Taha MO. Discovery of new cholesteryl ester transfer protein inhibitors via ligand-based pharmacophoremodeling and QSAR analysis followed by synthetic exploration. Eur J Med Chem 2010;45:1598-617.
- Mbarki S, Dguigui K, Hallaoui ME. Construction of 3D-QSAR models to predict antiamoebic activities of pyrazoline and dioxazoles derivatives. J Mater Environ Sci 2011;2:61-70.