- *Corresponding Author:
- V. K. Nathan
School of Chemical and Biotechnology,
SASTRA Deemed University,
Thanjavur,
Tamil Nadu 613401,
India
E-mail: vinodkumar@scbt.sastra.edu
| Date of Received | 23 November 2020 |
| Date of Revision | 31 July 2021 |
| Date of Acceptance | 05 December 2021 |
| Indian J Pharm Sci 2021;83(6):1273-1279 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Natural chemical compounds from medicinal plants used for the healing of diseases and disorders with fewer side effects, easy accessibility and economically cheap. The current study was aimed at finding novel drug like molecules as anti-diabetic compounds using in silico approach. Intermolecular interactions between target proteins and different antidiabetic compounds were observed. Five phytocompounds were selected from Plumbago zeylanica, Neolitsea cassia and Wrightia tinctoria and taken for molecular docking against human pancreatic alpha-amylase and human dipeptidyl peptidase IV using Autodock 4.2. Among the five phyto compounds, 6 urs-12-en-24-oic acid Plumbago zeylanica is the best compound for both the human pancreatic alpha-amylase and human dipeptidyl peptidase IV inhibition, as it possessed higher value in molecular dockings.
Keywords
Molecular docking, in silico study, Autodock, alpha-amylase, dipeptidyl peptidase IV
Type 2 Diabetes Mellitus (T2DM) is a critical metabolic failure characterized by less insulin action and high blood glucose level [1]. T2DM is signified as the quickest worldwide threat to people’s health [2] and it causes blindness, lower limb amputation and kidney disease [3]. As stated by WHO, 350 million people may be affected in 2030 globally, if the action has not been taken against T2DM [4]. Alpha (α)-Amylase and Dipeptidyl Peptidase IV (DPP IV) inhibitors used to protect the digestion of starch and other carbohydrates with T2DM patients [5].
α-Amylase universally distributed throughout the animal, plant and microbial kingdoms. Many mammals secrete two types of α-amylase, one is salivary α-amylase by the parotid gland and another one is pancreatic α-amylase secreted by the pancreas [6]. α-Amylase inhibitors extensively used in the therapy of T2DM. DPP IV inhibitor has the potential to be a novel, efficient and considerable agent to treat T2DM [7]. The usage of DPP IV inhibitors has fewer side effects like hypoglycemia, increasing body weight and gastrointestinal tract disorders [8]. The research of oral glucose tolerance tests on animals revealed that genetic deletion of DPP IV has increased the glucose tolerance and the secretion of insulin [9]. It encourages the study of α-amylase and DPP IV inhibitor bioactive compounds from medicinal plants which have fewer side effects, low cost and are easy to obtain.
Numerous drugs such as meglitinides, biguanides, sulfonylureas, thiazolidinediones, α-amylase inhibitors, DPP IV inhibitors used to treat the T2DM [10]. The side effects of these enzymatic inhibitors are flatulence, low blood sugar and hepatitis [11,12]. In recent years, a variety of research has done on plants [13-19]. Besides, several herbs and fruits have the property of inhibitors [16,17]. The known natural inhibitors of digestive enzymes include phytocompounds which operate through various mechanisms [17]. Today, researchers emphasis primarily on the finding of practical and low side effect therapeutic drugs to treat T2DM [20]. Medicinal plants produce secondary metabolites that have effective antidiabetic potentials to regulate blood glucose level. In conventional treatment, several medicinal plants were used such as Plumbago zeylanica (P. zeylanica), Neolitsea cassia (N. cassia) and Wrightia tinctoria (W. tinctoria) that avoid difficulties in the control of T2DM.
Molecular docking is an essential method in the improvement of new medications. Docking strategy permits describing the conduct of a test molecule in the coupling site of the receptor target of interest. A docking method must have the ability to predict the useful binding strength between the ligand and the receptor complex [21]. In computational chemistry, in silico analysis has led to the discovery of new active drugs with fewer side effects [22]. Molecular docking analysis has directed to predict the binding attractions of different bioactive compounds and to explain specific sites of interaction between the bioactive compounds and the target proteins [23]. The aim of this in silico study was to explore the antidiabetic properties of phytocompounds derivatives namely 6 urs-12-en- 24-oic acid, Beta (β)-Amyrin, 10,12-Docasadiyndioic acid, 1-(Ethynyltrimethylsilyl-2)-1-Chloro-2-Ethyl- 2-methyl cyclopropane and androstane via molecular docking using Autodock software.
Materials and Methods
Selection of receptor:
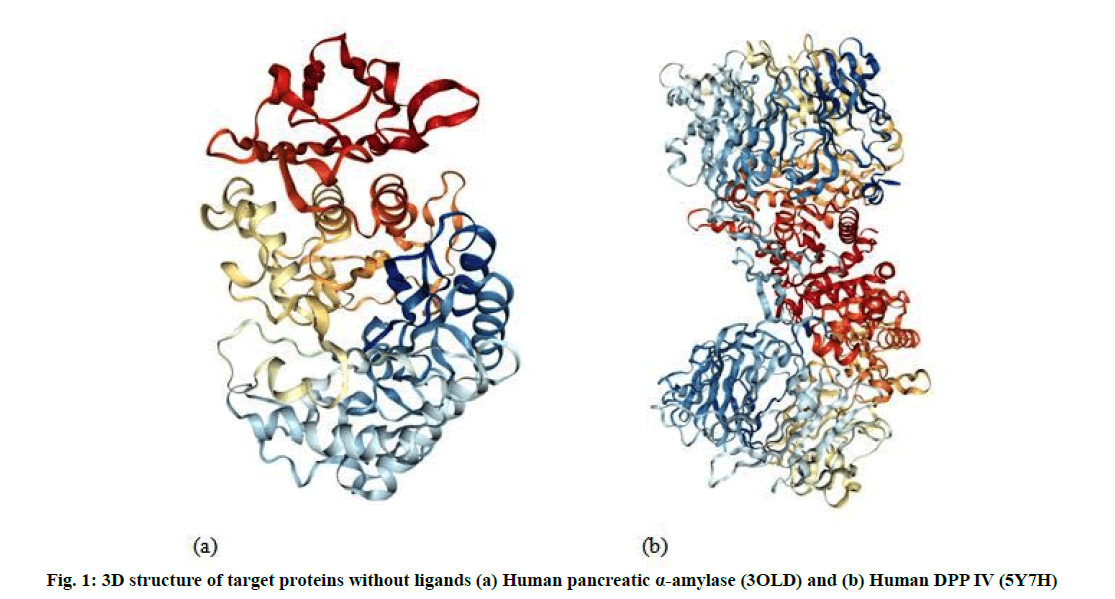
The Three Dimensional (3D) structure of the receptor binding sites of human Pancreatic α amylase (3OLD) (fig. 1a) and human DPP IV (5Y7H) (fig. 1b) retrieved from the Protein Data Bank (PDB) (http://www.rcsb.org).
Selection of ligand:
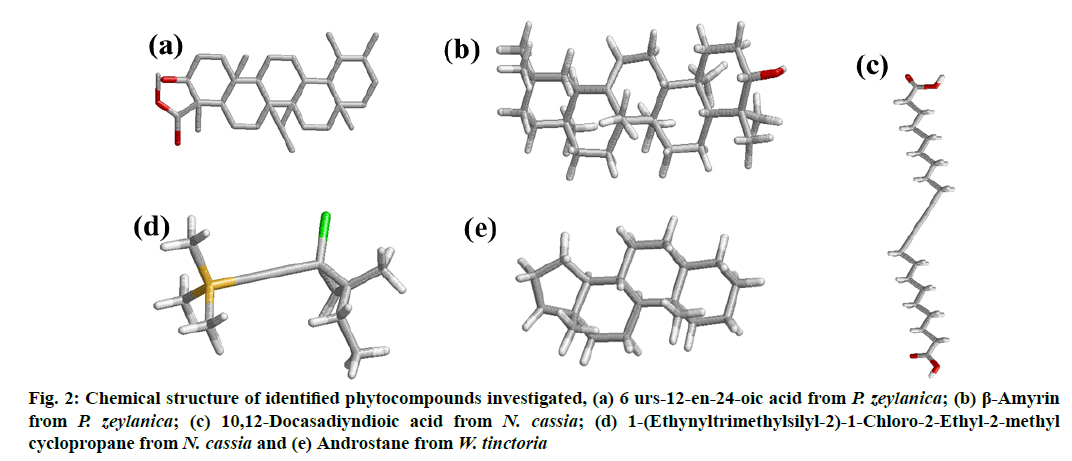
The 3D structures of bioactive compounds namely 6 urs-12-en-24-oic acid (fig. 2a) and β-amyrin (fig. 2b) from P. zeylanica, 10,12-Docasadiyndioic acid (fig. 2c) and 1-(Ethynyltrimethylsilyl-2)-1-Chloro-2- Ethyl-2-methyl cyclopropane (fig. 2d) from N. cassia and androstane (fig. 2e) from W. tinctoria collected from Pubchem, a compound database. The collected Structure Data File (SDF) files of identified bioactive compounds from the PubChem database were converted into PDB format using the EduPymol version 1.7.4.4 [24].
Figure 2: Chemical structure of identified phytocompounds investigated, (a) 6 urs-12-en-24-oic acid from P. zeylanica; (b) β-Amyrin from P. zeylanica; (c) 10,12-Docasadiyndioic acid from N. cassia; (d) 1-(Ethynyltrimethylsilyl-2)-1-Chloro-2-Ethyl-2-methyl cyclopropane from N. cassia and (e) Androstane from W. tinctoria
Protein ligand docking:
Molecular docking analysis performed to evaluate the most preferred geometry of the protein ligand complex. Docking phase is meaningless without its two components such as target protein and ligand. Human pancreatic α amylase (3OLD) and human DPP IV (5Y7H) used for performing docking study as target proteins (fig. 1). Docking results identify native or native-like configurations of the protein-ligand complex. The selected proteins complex used after removal of already bonded ligand and water molecules. Docking was performed by AutoDock 4.2 software. The complete docking steps could be stated as follows: first of all, the water molecules were eliminated from the protein. After the removal of water molecules, the PDB file of 3OLD and 5Y7H which provided as an input to the software. Kollman charges were computed for the macromolecule by AutoDock. Then the macromolecule was checked for the missing atoms and repaired. After repairing missing atoms, the hydrogens were added by keeping all the parameters at default settings. The macromolecule after all these modifications was saved as .PDB in the same directory. Then the ligand preparation was carried out. Like macromolecule, Kollman and Gasteiger charges were computed for the ligand [25]. Some of the torsions of the ligands were defined. The root was detected; the rotatable bonds were converted into non-rotatable bonds and vice versa. Also the number of active torsions was to most atoms rather than fewest. A Protein Data Bank, Partial Charge (Q) and Atom Type (T) (PDBQT) file was then created for the modified ligand with extension .PDBQT. After the preparation of a macromolecule and ligand, the rigid residue was prepared using the grid module provided in the AutoDock 4.0 Grid module employed .PDB file. This program was run using a searching grid extended over ligand molecules with box spacing 70×70×60; spacing was 0.608, x, y, z coordinates 8.485, -5.766, 15.737 respectively for 3OLD; 80×80×80, spacing was 0.508, x, y, z coordinates 52.271, 58.538, 45.819 respectively for 5Y7H while other parameters were default. The flexible macromolecule was saved with the .PDBQT extension. For molecular docking AutoDock 4.2 software was used.
It employed a configuartion file referring to PDBQT files of macromolecules and compounds prepared using AutoDock 4.2 and grid properties. As an output AutoDock 4.2 generated log files and PDBQT files of energy models for the selected data set. The output file contained different energy models. Among these models, the lowest energy model against each ligand was selected and appended at the end of the original protein file. As a result of this step, docked files for the selected set were generated. For the interpretation of docking results; target protein and protein docked with the data set of compounds, the interactions between the active pocket of protein and compounds were found. The best docking poses were predicted to be the most stable conformation of each compound for binding to the protein active site. Consequently, the output of the docking process was analyzed utilizing EduPymol version 1.7.4.4.
Results and Discussion
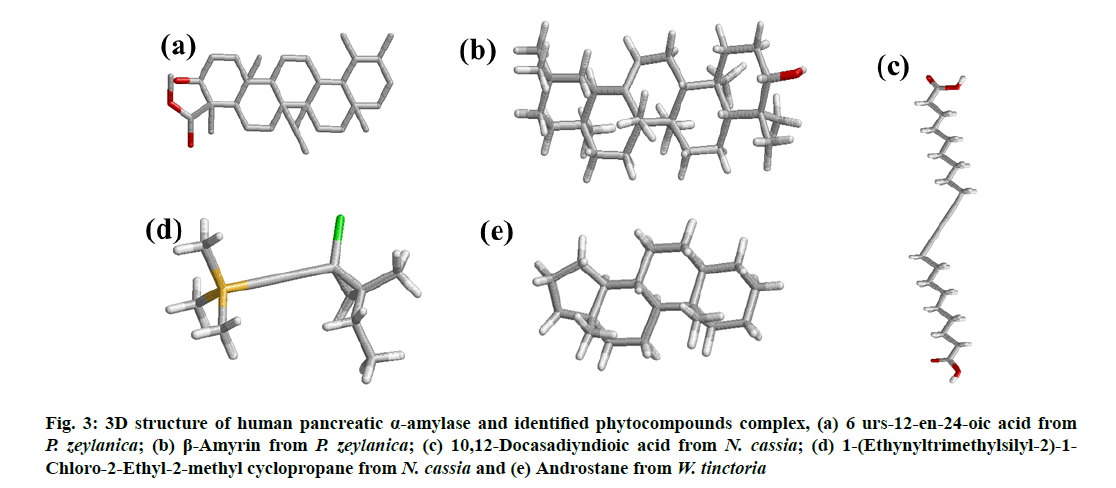
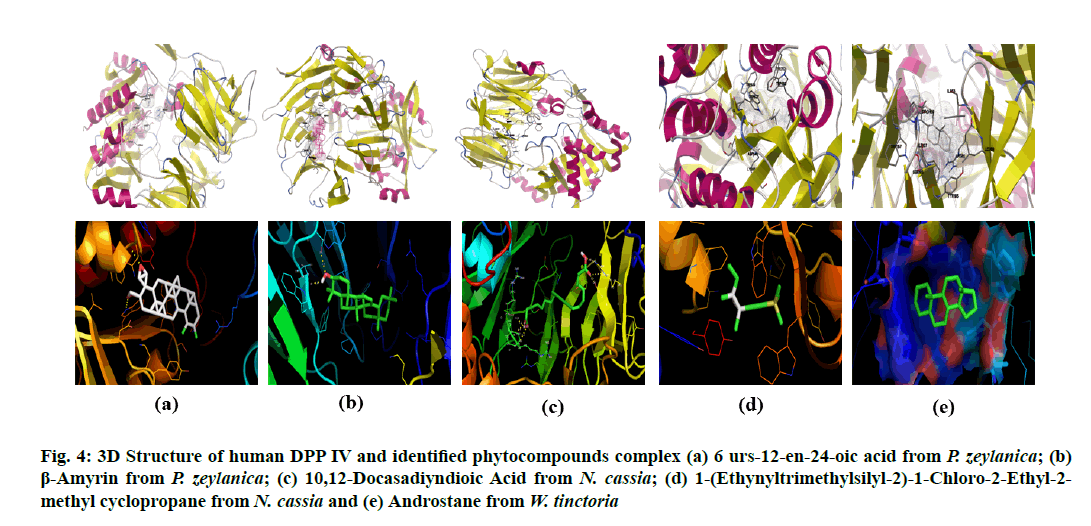
All the identified compounds (6 urs-12-en-24- oic acid (fig. 2a) and β-amyrin (fig. 2b) from P. zeylanica, 10,12-Docasadiyndioic acid (fig. 2c) and 1-(Ethynyltrimethylsilyl-2)-1-Chloro-2-Ethyl- 2-methyl cyclopropane (fig. 2d) from N. cassia and androstane (fig. 2e) from W. tinctoria) were docked into that active site of human pancreatic α amylase (fig. 1a) and DPP IV (fig. 1b) by using AutoDock 4.2. The binding affinities of these compounds to target proteins are shown in Table 1 and Table 2 and fig. 3 and fig. 4. The identified five compounds have higher binding interaction compared to the co-crystallized ligand of 3OLD and 5Y7H were found lying deep into the binding cavity of target proteins showing all the significant interaction and favorable energy of interaction ranging from -3.55 to -10.58 kcal/mol and -4.33 to -9.45 kcal/mol respectively (Table 1 and Table 2). The bonded and non-bonded interactions of thesecompoundsare shown in Table 1 and Table 2.Among all the tested compounds, 6 urs-12-en-24-oic acid of P. zeylanica exhibited the lowest binding energy -10.58 kcal/mol and -9.45 kcal/mol and highest affinity for 3OLD and 5Y7H respectively.
Figure 3: 3D structure of human pancreatic α-amylase and identified phytocompounds complex, (a) 6 urs-12-en-24-oic acid from P. zeylanica; (b) β-Amyrin from P. zeylanica; (c) 10,12 Docasadiyndioic acid from N. cassia; (d) 1-(Ethynyltrimethylsilyl-2)-1- Chloro-2-Ethyl-2-methyl cyclopropane from N. cassia and (e) Androstane from W. tinctoria
Figure 4: 3D Structure of human DPP IV and identified phytocompounds complex (a) 6 urs-12-en-24-oic acid from P. zeylanica; (b) β-Amyrin from P. zeylanica; (c) 10,12-Docasadiyndioic Acid from N. cassia; (d) 1-(Ethynyltrimethylsilyl-2)-1-Chloro-2-Ethyl-2- methyl cyclopropane from N. cassia and (e) Androstane from W. tinctoria
| Parameters | 6 urs-12-en-24-oic acid from P. zeylanica | β-Amyrin from P. zeylanica | 10,12-Docasadiyndioic Acid from N. cassia | 1?(Ethynyltrimethylsilyl-2)-1-Chloro-2-Ethyl-2-methyl cyclopropane from N. cassia | Androstane from W. tinctoria |
|---|---|---|---|---|---|
| Binding energy (Kcal/mol) | -10.58 | -8.9 | -3.55 | -4.3 | -7.58 |
| Ligand Efficiency | -0.31 | -0.29 | -0.14 | -0.33 | -0.4 |
| Inhibition Constant | 17.49nM | 297.25nM | 2.52mM | 705.84µM | 2.77 µM |
| Intermolecular Energy (Kcal/mol) | -11.48 | -9.2 | -9.81 | -5.19 | -7.58 |
| vdW+Hbond+desolv Energy (Kcal/mol) | -11.28 | -9.05 | -8.7 | -5.16 | -7.58 |
| Electrostatic Energy | -0.19 | -0.15 | -1.11 | -0.03 | 0.0 |
| Total Internal Energy | 0.01 | 0.07 | -0.52 | -0.37 | 0.0 |
| Torsional Free Energy | 0.89 | 0.3 | 6.26 | 0.89 | 0.0 |
| RMSD | 30.14 | 31.87 | 27.89 | 31.01 | 30.29 |
| Number of Hydrogen bonds | 1 | 2 | 3 | 0 | 0 |
| Hydrogen bond Interaction | Lys200-NZ: O31-Ligand | Asp197-OD1: H32-Ligand and Arg195-NH2: O31-Ligand | Ala106-N: O27-Ligand, Ile235-N: O14-Ligand and Lys200-NZ:015-Ligand | NA | NA |
| Hydrogen Bond Length (A?) | 3.2 | 1.9 and 3.2 | 3.0, 3.0 and 2.6 | NA | NA |
Table 1: Molecular Docking Statistics of the Human Pancreatic Α-AMYLASE and Identified Phytocompounds Complex
| Parameters | 6 urs-12-en-24-oic acid from P. zeylanica | β-Amyrin from P. zeylanica | 10,12-Docasadiyndioic acid from N. cassia | 1?(Ethynyltrimethylsilyl-2)-1-Chloro-2-Ethyl-2-methyl cyclopropane from N. cassia | Androstane from W. tinctoria |
|---|---|---|---|---|---|
| Binding energy (Kcal/mol) | -9.45 | -8.81 | -4.41 | -4.33 | -8.06 |
| Ligand efficiency | -0.28 | -0.28 | -0.17 | -0.33 | -0.42 |
| Inhibition constant | 117.59 nm | 350.78 nm | 582.5 µm | 672.36 µm | 1.24 µm |
| Intermolecular energy (Kcal/mol) | -10.35 | -9.1 | -10.68 | -5.22 | -8.06 |
| vdW+Hbond+desolv energy (Kcal/mol) | -10.2 | -9.01 | -9.32 | -5.29 | -8.06 |
| Electrostatic energy | -0.15 | -0.09 | -1.35 | 0.05 | 0 |
| Total internal energy | 0.08 | 0.07 | -1.05 | -0.31 | 0 |
| Torsional free energy | 0.89 | 0.3 | 6.26 | 0.89 | 0 |
| RMSD | 86.09 | 89.38 | 83.73 | 84.55 | 86.88 |
| Number of hydrogen bonds | 2 | 2 | 6 | 0 | 0 |
| Hydrogen bond interaction | TRP627-NE1: O33Ligand and TRP563-CA: O31Ligand | SER212-OG: O31-Ligand and Leu214-O: H32-Ligand | Arg471NH1: O14Ligand, Arg471NE: O15 Ligand, Ser458OG: O14 Ligand, Arg358O: H28Ligand, Arg356N: O27Ligand and Phe357N: O26Ligand | NA | NA |
| Hydrogen bond length (A?) | 2.9 and 2.7 | 2.5 and 1.8 | 2.9, 3.3, 2.7, 2.1, 3.1 and 2.8 | NA | NA |
Table 2: Molecular Docking Statistics of the Dpp IV and Identified Phytocompounds Complex
Molecular docking is now widely used to discover new ligands for the target of known structure. The free energy binding score denotes the interactions of ligand to the target protein; the lower free energy binding, the higher binding affinity [26]. Also, inhibition constant can be calculated using computational methods. The lowest inhibition constant denotes the most active compound. It signifies the molecular interaction between ligands and the target protein. The high value of surface interaction indicates the possibilities of compounds binding with the target protein [27].
Based on the in silico analysis, 6 urs-12-en-24-oic acid of P. zeylanica (fig. 3a and fig. 4a) has the lowest value (-10.58 Kcal/mol for α- amylase and -9.45 Kcal/mol for DPP IV) in binding free energy while β-amyrin of P. zeylanica (fig. 3b and fig. 4b) and androstane of W. tinctoria (fig. 3e and fig. 4e) were in the second (-8.9 Kcal/mol for α-amylase and -8.81 Kcal/mol for DPP IV) and third position (-7.58 Kcal/mol for α-amylase and -8.06 Kcal/mol for DPP IV) with two target proteins (Table 1 and Table 2). 1-(Ethynyltrimethylsilyl-2)-1- Chloro-2-Ethyl-2-methyl cyclopropane (fig. 3c and fig. 4c) and 10,12-Docasadiyndioic acid from N. cassia have shown the fourth (-4.3 Kcal/mol) and fifth (-3.55 Kcal/mol) positions for α-amylase respectively and fifth (-4.33 Kcal/mol) and fourth places (-4.41 Kcal/mol) for DPPIV respectively when compared with binding energies with other ligands (Table 1 and Table 2). Hydrogen bonds are a significant factor in defining the specificity of ligand binding [28]. 6 urs-12- en-24-oic acid of P. zeylanica ranked highest in binding affinity with 3OLD and 5Y7H using a hydrogen bond (Table 1 and Table 2). Lys200 of 3OLD formed one hydrogen bonds with 6 urs-12-en-24-oic acid of P. zeylanica between NZ of the residue and hydroxyl (O31) of the ligand played in binding affinity with a bond length of 3.2 A?. The TRP627 and TRP563 of 5Y7H active site residues formed two hydrogen bonds with O31and H32 of 6 urs-12-en-24-oic acid with the length of 2.9 and 2.7A?. β-amyrin from P. zeylanica formed two hydrogen bonds with both the target proteins.
β-amyrin and 3OLD complex formed hydrogen bonds by Asp197-OD1 and Arg195-NH2 of 3OLD bonded with H32 and O31 of Ligand respectively. Their hydrogen bond length was 1.9 and 3.2A?. β-amyrin and 5Y7H complex also formed through two hydrogen bond: SER212-OG and Leu214-O of 5Y7H with O31and H32 of ligand and the length were 2.5 and 1.8A? respectively. Similarly, 10,12-Docasadiyndioic acid from N. cassia formed three hydrogen bonds with 3OLD and 6 hydrogen bonds with 5Y7H (Table 1 and Table 2). Though, 1-(Ethynyltrimethylsilyl- 2)-1-Chloro-2-Ethyl-2-methyl cyclopropane from N. cassia (fig. 3d and fig. 4d) also showed good binding score but not interacted by hydrogen bonds with two target proteins (Table 1 and Table 2). It interacted through hydrophobic, electrostatic and Van der Waals interactions with both the target proteins.
Likewise, androstane from W. tinctoria (fig. 3e and fig. 4e) also is not interacted through hydrogen bond and electrostatic force; it is related to hydrophobic and Van der Waals interactions (Table 1 and Table 2). The lowermost value of free binding energy generates a useful binding molecule and then produces a potent biological activity. Free energy binding and surface interaction between ligands and target protein influence the inhibitory activity of selected bioactive compounds on α-amylase and DPP IV. The in silico analysis exposed the prominence and presence of oxygen, nitrogen and carbonyl groups in the compound structure was essential and amplified the tendency of hydrogen bond interactions [29]. Molecular docking studies are broadly exercising methods to analyze the promising drugs in the pharmaceutical industry. The binding pose of ligands or bioactive compounds to their target proteins divulges their possible affinity and activity of drugs.
The current study was aimed at finding novel drug like molecules as antidiabetic compounds using in silico approach. Intermolecular interactions between target proteins and different antidiabetic compounds were observed. The protein-ligand interaction shows that 6 urs-12-en-24-oic acid from P. zeylanica having the best interactions compared to the other identified compounds from different medicinal plants toward the enzymes human pancreatic α-Amylase and human DPP IV. From the above observations, we can assume that P. zeylanica can be a prominent resource for antidiabetic compounds.
Conflict of interests:
The authors declared no conflicts of interest.
References
- Kenchappa R, Bodke YD, Chandrashekar A, Sindhe MA, Peethambar SK. Synthesis of coumarin derivatives containing pyrazole and indenone rings as potent antioxidant and antihyperglycemic agents. Arab J Chem 2017;10:S3895-906.
- Kavitha S, Kannan K, Gnanavel S. Synthesis, characterization and biological evaluation of novel 2,5 substituted-1,3,4 oxadiazole derivatives. Saudi Pharm J 2017;25(3):337-45.
- Datar PA, Deokule TA. Design and synthesis of thiadiazole derivatives as antidiabetic agents. Med chem 2014;4(4):390-9.
- Taha M, Rahim F, Imran S, Ismail NH, Ullah H, Selvaraj M, et al. Synthesis, α-glucosidase inhibitory activity and in silico study of tris-indole hybrid scaffold with oxadiazole ring: As potential leads for the management of type-II diabetes mellitus. Bioorg Chem 2017;74:30-40.
- Nair SS, Kavrekar V, Mishra A. In vitro studies on alpha amylase and alpha glucosidase inhibitory activities of selected plant extracts. Eur J Exp Biol 2013;3(1):128-32.
- Balasubramaniam V, Mustar S, Khalid NM, Rashed AA, Noh MF, Wilcox MD, et al. Inhibitory activities of three Malaysian edible seaweeds on lipase and α-amylase. J Appl Phycol 2013;25(5):1405-12.
- Singh AK. Dipeptidyl peptidase-4 inhibitors: Novel mechanism of actions. Indian J Endocrinol Metab 2014;18(6):753.
- Ábel T. A new therapy of type 2 diabetes: DPP-4 inhibitors. Hypoglycemia. Everlon Rigobelo, INTECH 2011;1-4.
- Duez H, Smith AC, Xiao C, Giacca A, Szeto L, Drucker DJ, et al. Acute dipeptidyl peptidase-4 inhibition rapidly enhances insulin-mediated suppression of endogenous glucose production in mice. Endocrinology 2009;150(1):56-62.
- Sangeetha KN, Sujatha S, Muthusamy VS, Anand S, Shilpa K. Current trends in small molecule discovery targeting key cellular signaling events towards the combined management of diabetes and obesity. Bioinformation 2017;13(12):394.
- Hamdan II, Afifi F, Taha MO. In vitro alpha amylase inhibitory effect of some clinically-used drugs. Int J Pharm Sci 2004;59(10):799-801.
- Lebovitz HE. α-Glucosidase inhibitors as agents in the treatment of diabetes. Diabetes Rev 1998;6:132-45.
- Farzaei F, Morovati MR, Farjadmand F, Farzaei MH. A mechanistic review on medicinal plants used for diabetes mellitus in traditional Persian medicine. J Evid Based Complement Alternat Med 2017;22(4):944-55.
- Sathasivampillai SV, Rajamanoharan PR, Heinrich M. Siddha Medicine in Eastern Sri Lanka today–Continuity and change in the treatment of diabetes. Front Pharmacol 2018;9:1022.
- Hamza N, Berke B, Umar A, Cheze C, Gin H, Moore N. A review of Algerian medicinal plants used in the treatment of diabetes. J Ethnopharmacol 2019;238:111841.
- Kalhotra P, Chittepu VC, Osorio-Revilla G, Gallardo-Velazquez T. Phytochemicals in garlic extract inhibit therapeutic enzyme DPP-4 and induce skeletal muscle cell proliferation: A possible mechanism of action to benefit the treatment of diabetes mellitus. Biomolecules 2020;10(2):305.
- Khadayat K, Marasini BP, Gautam H, Ghaju S, Parajuli N. Evaluation of the alpha-amylase inhibitory activity of Nepalese medicinal plants used in the treatment of diabetes mellitus. Clin Phytoscience 2020;6:1-8.
- Unuofin JO, Lebelo SL. Antioxidant effects and mechanisms of medicinal plants and their bioactive compounds for the prevention and treatment of type 2 diabetes: an updated review. Oxid Med Cell Longev 2020;2020.
- Faleyimu OI, Mohammed AA, Akinyemi O. Efficacy of forest plants in the treatment of diabetes in Birnin Gwari Local Government Area, Kaduna State, Nigeria. Int J Biomed Health Sci 2021;6(2).
- Choubey M, Pattanaik AK, Baliyan S, Dutta N, Jadhav SE, Sharma K. Dietary supplementation of a novel phytogenic feed additive: effects on nutrient metabolism, antioxidant status and immune response of goats. Anim Prod Sci 2016;56(10):1612-21.
- Mooers BH. Shortcuts for faster image creation in PyMOL. Protein Sci 2020;29(1):268-76.
- Paul A, Adnan M, Majumder M, Kar N, Meem M, Rahman MS, et al. Anthelmintic activity of Piper sylvaticum Roxb.(family: Piperaceae): In vitro and in silico studies. Clin Phytoscience 2018;4(1):17.
- Abdulfatai U, Uzairu A, Uba S. Quantitative structure-activity relationship and molecular docking studies of a series of quinazolinonyl analogues as inhibitors of gamma amino butyric acid aminotransferase. J Adv Res 2017;8(1):33-43.
- Boukarai Y, Khalil F, Bouachrine M. QSAR study of flavonoid derivatives as in vitro inhibitors agents of Aldose Reductase (ALR2) enzyme for diabetic complications. J Mater Environ Sci 2017;8(5):1532-45.
- Mills, N. ChemDraw Ultra 10.0 CambridgeSoft, 100 CambridgePark Drive, Cambridge, MA 02140; 2006.
- Utomo DH, Widodo N, Rifa'i M. Identifications small molecules inhibitor of p53-mortalin complex for cancer drug using virtual screening. Bioinformation 2012;8(9):426.
- Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminformatics 2009;1(1):1-6.
- Wade RC, Goodford PJ. The role of hydrogen-bonds in drug binding. Prog Clin Biol Res 1989;289:433-44.
- Jung SH, Lee JM, Lee HJ, Kim CY, Lee EH, Um BH. Aldose reductase and advanced glycation endproducts inhibitory effect of Phyllostachys nigra. Biol Pharm Bull 2007;30(8):1569-72.