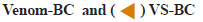- *Corresponding Author:
- P. Dongre
Department of Biophysics,
University of Mumbai,
Vidyanagari,
Santacruz,
Mumbai,
Maharashtra 400098,
India
E-mail: drpmdongre@yahoo.co.in
| Date of Received | 26 July 2021 |
| Date of Revision | 11 February 2022 |
| Date of Acceptance | 22 July 2022 |
| Indian J Pharm Sci 2022;84(4):938-949 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Russell’s viper being one of the most venomous snake species in Asia causes significant amount of snake bites and deaths due to neurotoxicity, coagulopathy, haemorrhage, cardiotoxicity, myotoxicity, hypotension, oedema, tissue damage and convulsion. Treatment with monoclonal antibodies, anti-snake venom is the only medically approved treatment available for snake envenomation and has many limitations and side effects such as anaphylactic shock, skin rashes, nausea to vomiting and fever, chills, hypotension, seizures, serum sickness, pruritus, urticaria, arthralgia, lymphadenopathy, encephalopathy etc. Recently, nanoscience and nanotechnology are playing an important role in medical field and has proven to be quite beneficial. In present study, chemically synthesized silver nanoparticles (40±3 nm) are used to inhibit Russell’s viper venom activity. The silver nanoparticles and albumin conjugated with curcumin was prepared and characterized using spectroscopic techniques. Venom inhibitory potency of the prepared conjugate was analysed using acidimetry, protease assay and whole blood clotting test. The ultravioletvisible spectra showed enhancement in absorbance of venom with significant hyperchromic shift and thus confirmed modification of venom by silver nanoparticles. This also led to the enhanced interaction with bovine serum albumin-curcumin conjugates. Fluorescence data also showed strong interaction between the conjugates and venom. Interestingly the activity of Russell’s viper venom was reduced to 75 %-96 %. The possible mechanism of action of bovine serum albumin-curcumin complex on venom treated with silver nanoparticles is explained at molecular level and biocompatibility of the same was also analysed. Blood agglutination by bovine serum albumin-curcumin complex showed its in vitro biocompatibility.
Keywords
Russell’s viper venom, silver nanoparticle, curcumin, fluorescence spectroscopy, blood clotting
Snake envenomation is common and unfortunate event in rural areas of tropical and subtropical countries of Asia and Africa[1]. Populations that suffer from this hazard mainly include farmers, plantation workers, forest guards, labourers etc. India accounts for one of the highest number of fatalities due to snake bites with 11 000 deaths per year[2]. This problem ensues due to the lack of knowledge of first aid measures, reliance on harmful tradition practices and myths, lack of medical emergency facilities, untimely administration of appropriate Anti-Snake Venom (ASV) dose and finally non-specific reactions of ASV.
Russell’s viper, one of the most venomous snake found in South-East Asia[3], accounts for 50 % of snake bites[4]. Russell’s Viper Venom (RVV) varies in composition depending on geographical variation. Despite this variation, basic composition remains the same. Analysis by tandem mass spectrometry has revealed the presence of approximately 63 different proteins belonging to 12 families. Amongst these, few have been proven to be responsible of venom intoxication. This includes acidic, basic and neutral Phospholipase A2 (basic PLA2 being the most toxic), serine proteases, metalloproteases, RVV factor X activator (RVV-X), RVV factor V activator (RVV-V), thrombin-like enzymes, cysteine-rich secretory proteins, L-Amino Acid Oxidases (LAAO), C-type lectin-like proteins, Kunitz-type serine protease inhibitor, disintegrin, nucleotidase, phosphodiesterase, vascular endothelial growth factor and vascular nerve growth factor families[5].
Irrespective of the presence of lot of many enzymes and components, PLA2 gains major attention owing to its ability to induce a wide range of pharmacological effects, which includes neurotoxicity, coagulopathy, haemorrhage, cardiotoxicity, myotoxicity, hypotension, oedema, tissue damage and convulsion[6]. However,metalloprotease, RVV-X and serine protease, RVV-V are the prime lead of coagulopathy induced by viper venom. They may act as pro-coagulant, anticoagulant, platelet aggregating and fibrinolytic proteinases[7]. LAAO is also one of the major components of viper venom. LAAO contains flavin, which contributes to the yellow color of venom, generates oxidative stress producing Hydrogen peroxide (H2O2). It induces apoptosis, cytotoxicity, interferes with platelet aggregation, haemorrhage, haemolysis, oedema etc.[8]. Nucleotidase acts by hydrolyzing Adenosine Triphosphate (ATP) to Adenosine Diphosphate (ADP) and ADP to Adenosine Monophosphate (AMP). This action of nucleotidases induces shock symptoms by depleting ATP abundance in the prey[9]. Vascular nerve growth factor has been suggested to induce hypotension through the liberation of nitric oxide and histamine[10]. It also enhances the expression of vascular endothelial growth factor which increases the permeability of blood vessels for venom[7,11].
ASV is medically approved treatment against snake envenomation. ASV is enzyme (pepsin) refined Fragment antigen-binding (Fab) fragments of Immunoglobulin G (IgG) antibodies isolated from serum of horse or sheep that has been immunized with venom of one or more species. ASV can be monovalent neutralizing venom of only one species or polyvalent neutralizing several species of snake venom[12]. The fundamental principle behind the interaction of venom with ASV is antigen- antibody reaction. Usually, antibodies form immune complexes with antigens and are rapidly phagocytized. Venom components in absence of ASV interact with surface antigen present on host cell forming agglutinins. However, antibodies present in ASV forms immune complexes with venom components, thereby inhibiting its contact with cell surface antigens. So formed immune complexes are either eliminated through renal route or phagocytised[13]. Unfortunately 20 % of ASV treated patients suffer early or late reactions of ASV[14]. Adverse reactions of ASV are usually seen in those who are not previously exposed to equine proteins[15]. These reactions of ASV include both acute and delayed responses. Acute adverse reactions are anaphylactic starting from rashes, nausea to vomiting and also pyrogenic i.e. fever, chills, hypotension, seizures etc. Delayed reaction includes serum sickness, pruritus, urticaria, arthralgia, lymphadenopathy, encephalopathy etc.[16].
Many attempts have been taken to develop alternative therapy in order to antagonize snake venom. Folk medicines which are used in the preliminary treatments of snake bite in rural areas have been surveyed for its venom neutralizing potency. Herbs including Aristolochia bracteolata, Tylophora indica, Leucas aspera have been proven to be potent anti-venom against Russell’s viper and Cobra snake venom[17]. Postsynaptic neurotoxin of Thai Cobra venom was neutralized by rhizomes of Curcuma spp.[18]. Such herbal compounds have been hypothesized to act by following mechanisms viz. protein precipitation, enzyme inactivation hypothesis, chelation hypothesis, adjuvant action hypothesis, antioxidant action hypothesis, protein folding hypothesis and combination hypothesis, among which protein precipitation-inactivation hypothesis being most accepted one[19]. Nanotechnology has shown promising outcomes in the field of biomedicine. Nanoparticle C60 fullerene prolonged survival of Acheta domesticus (Cricket) envenomed with Crotalus venom[20]. Also green synthesized Silver Nanoparticles (SNPs) showed neutralizing efficiency against Cobra venom[21]. In recent study, SNPs have been shown to be potent against RVV[22]. Moreover, conjugation of nanoparticles to biomolecules outcomes the production of conjugates with altered functions (of both nanoparticles and biomolecules being conjugated to it). This was proved in a study where soy protein nanoparticles conjugated with ASV immunoglobulins Fab’2 fragment showed efficient neutralization of protease, phospholipase, hyaluronidase enzymes of Bungarus caeruleus than free ASV[23].
Present investigation explores the antisnake venom activity of Curcumin (CUR) isolated from Curcuma longa. This study aims to understand ASV activity of CUR and SNP-Bovine Serum Albumin (BSA) conjugates. Biophysical (Ultraviolet (UV)-visible spectroscopy, fluorescence spectroscopy) and biochemical (acidimetric assay, protease assay, whole blood clotting assay) aspects were used to investigate ASV activity of these conjugates.
Materials and Methods
Chemicals:
Lyophilized Doboia russelli (Russell’s viper) crude venom was purchased from Haffkine institute, Parel, Mumbai.
BSA (HiMedia laboratories), CUR (from Curcuma longa) (Sigma-Aldrich Company Ltd), silver nitrate (Qualigens Pharma Pvt. Ltd.), casein (SDFCL Sd Fine Chem Limited), tri-sodium citrate (SRL Chemicals), calcium chloride (SDFCL Sd Fine Chem Limited), sodium chloride (HiMedia laboratories), Sodium deoxycholate (HiMedia laboratories), Trichloroacetic Acid (TCA) (Loba Chemie Pvt. Ltd.), Sodium hydroxide (NaOH) (SDFCL Sd Fine Chem Limited), Sodium carbonate (Loba Chemie Pvt. Ltd.), copper sulphate (SRL Chemicals), sodium potassium tartarate (SDFCL Sd Fine Chem Limited), Folin-Ciocalteu reagent (SDFCL Sd Fine Chem Limited) were used. All chemicals used were of Analytical Reagent (AR) grade.
10 mg/ml of stock solution of RVV was prepared in double distilled water and 200 µg/ml working solution was used. CUR (1 mM) was prepared in ethanol. Phosphate buffer (pH 7.4) was used as a diluent for the experiments; however whole blood clotting test was performed using saline. All experiments were performed in triplicates.
Synthesis and characterization of SNPs:
Chemical reduction method was used for synthesis of SNPs[24]. 1 mM silver nitrate was heated to 80°, with continued stirring on hot plate magnetic stirrer. At 80°, 1 % tri-sodium citrate was added drop wise until the colorless solution tuned pale yellow. Synthesized SNPs were cooled and stored in dark. Characterization was carried out using UV-visible spectroscopy (NanoPhotometer®, Implen) and dynamic light scattering system (Zetasizer nano ZS90, Malvern).
UV-visible spectroscopy:
UV measurements were recorded for following interactions: CUR alone; venom-CUR; Venom-SNP (VS) and equimolar BSA-CUR (BC). Optimized VS conjugate was further studied for its interaction with varying CUR concentration and various equimolar BC conjugates [1.875 µM (BC1); 3.75 µM (BC2); 7.5 µM (BC3) and 15 µM (BC4)]. UV-visible measurements were recorded in the range of 200-600 nm.
Fluorescence spectroscopy:
The fluorescence spectroscopy (Varian, Cary Eclipse) was performed using 10 mm path length cuvette.
Fluorescence quenching of venom was studied for venom-CUR interaction and VS interaction. Following parameters were set to understand venom-CUR interaction and VS interaction. Excitation wavelength (of venom) was set at 280 nm. Emission spectra were recorded in the range of 310-500 nm. Excitation and emission slit width was set to 10 nm with Photomultiplier Tubes (PMT) voltage at 530 V.
Fluorescence enhancement of CUR was studied by exciting it at 425 nm. The emission spectra were recorded in the range of 450-700 nm. Excitation as well as emission slit width was set to 10 nm and PMT voltage at 570 V. VS conjugate was also studied for its interaction with varying CUR concentrations (0-5 µM); and BC conjugates (BC1-BC4). Venom was also studied for its interaction with BC conjugates.
Venom neutralizing assay:
Acidimetric assay: Acidimetric assay was performed using a method described by Tan et al.[25] with slight modification. PLA2 activity was studied using egg yolk which is a rich source of phospholipids. Egg yolk suspension was prepared by mixing equal volumes of egg yolk, calcium chloride (18 mM) and sodium deoxycholate (8.1 mM). The pH of this suspension was adjusted to 8.00 using 0.1 N NaOH.
15 ml of egg yolk suspension was added to 100 µl of reaction mixture containing venom in order to initiate hydrolysis and pH change was recorded as a function of time. Unit decrease in pH corresponds to 133 µM fatty acid release. Enzyme activity was calculated using equation 1.
Enzyme activity (µM/min=Fatty acid release/Time) (Eq. 1)
Appropriate control was used (100 µl saline) in place of reaction mixture. Inhibition studies were carried for all complexes containing venom.
Protease assay: Proteolysis of casein protein by venom proteases[26] was performed at pH 7.4. Casein (200 µg/ ml) and venom (200 µg/ml) were incubated in 2 ml phosphate buffer for an hour at 37°. The reaction was terminated by adding 100 µl TCA. The mixture was filtered and 1 ml of filtrate was used to determine the concentration of tyrosine released in the mixture. Tyrosine concentration was determined by Folin-Lowry method using L-tyrosine as a standard. Enzyme activity was expressed in unit/h where 1 unit corresponds to 0.02 µM tyrosine release. Assay was performed for all venom complexes to understand their inhibition profile.
Whole blood clotting test: Pro-coagulant activity of viper venom was studied using whole blood clotting test by determining recalcification time[27]. Blood was collected from a healthy volunteer in 3.8 % sodium citrate containing vacutainer tube. About 100 µl whole blood was incubated with 100 µl venom (200 µg/ml) (pH=7.4) and clotting time was recorded upon addition of 50 µl calcium chloride (0.25 M). Inhibition studies were performed for all venom complexes.
Results and Discussion
SNPs were synthesized using bottom-up approach of nanomaterials synthesis. The method involves reduction of silver ions by citrate ions forming particle seeds to which more and more citrate ion accumulates. This results in decrease in citrate ions availability, due to which larger seeds take the help of smaller one in order to grow via Ostwald ripening[28]. Here, tri-sodium citrate works as reducing agent as well as capping agent, preventing nanoparticles from aggregation. Change in colour of the solution indicates the formation of SNPs which were further characterized by UV-visible spectroscopy, showing an absorbance maximum at 420 nm. Hydrodynamic size of SNP was found to be 40±3 nm. Using equation 2, the concentration of SNP was found to be 0.50 nM.
Concentration of SNP= NTotal/NVNA (Eq. 2)
Where NTotal is the total number of silver atoms added to the solution, N=Number of silver atoms per nanoparticle, V=Volume of the solution and NA=Avogadro’s number.
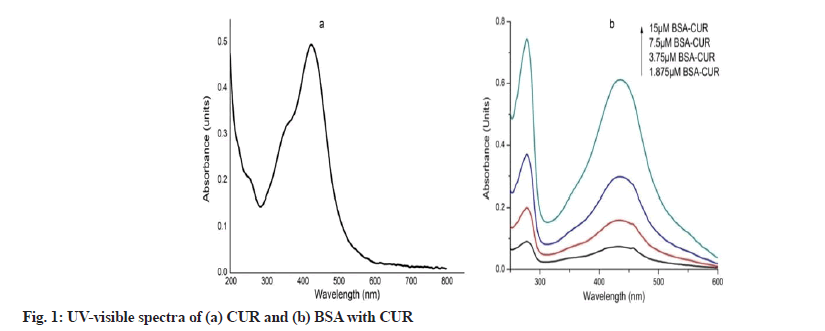
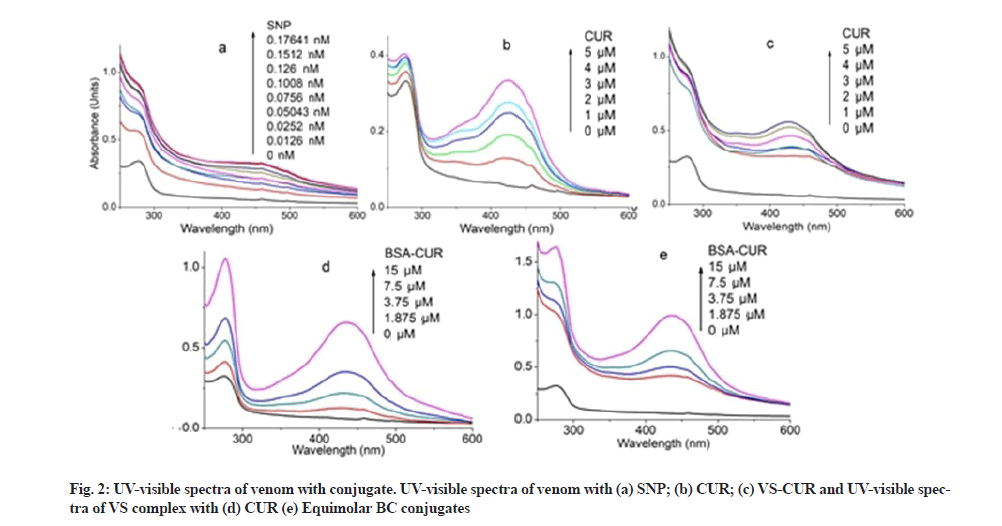
Spectral characteristics of equimolar BC conjugates are shown in fig. 1. CUR shows absorption maxima at 424 nm (fig. 1a). However in the presence of BSA, CUR becomes stabilize as evident by its peak shift to 434 nm (fig. 1b). Peak at 278 nm represents BSA peak. UV characteristic of venom complexes are shown in fig. 2. Venom shows a peak at 277 nm which upon SNP addition undergoes hyper chromic shift with prominent widening (fig. 2a). This hyper chromic shift reveals the formation of ground state complexes of venom components with SNP[29]. Fig. 2b represents the interaction between venom and CUR. An increase in the absorption of venom at 277 nm with a blue shift of 2 nm was observed with increasing CUR concentration, indicating hydrophobic interaction between CUR and venom components. Unlike BC interaction, venom-CUR interaction shows CUR peak at around 424 nm. This indicates that venom provide less stability to CUR as compared to BSA.
Absorption spectra of VS conjugate with increasing concentrations of CUR exhibited an increase in absorbance intensity of venom (fig. 2c). Absorbance intensity of CUR at 432 nm was obtained; however at high concentration the peak shifts towards blue region i.e. at 428 nm. This indicates the incapability of venom to provide stability to CUR.
Venom-BC conjugate interaction and VS-BC conjugate interaction is shown in fig. 2d and fig. 2e respectively. Unlike venom-CUR, CUR peak (430 nm) in both these absorption spectra confirms its stability in the hydrophobic core of the BSA. Stability was further improved as suggested by the red shift of 6 nm of CUR peak as its concentration was increased. Similar shifts were observed by Barik et al.[30] and Yang et al.[31]. Such a small spectral shifts owe majorly to the ionic interactions between protein and CUR along with some hydrophobic interactions[32]. Significant hyper chromic shift in the intensities of the protein and CUR peak was observed in VS-BC conjugate interaction when compared to corresponding venom-BC conjugate interaction. This could be attributed to the SNP modified venom that leads to an enhanced interaction between two conjugates.
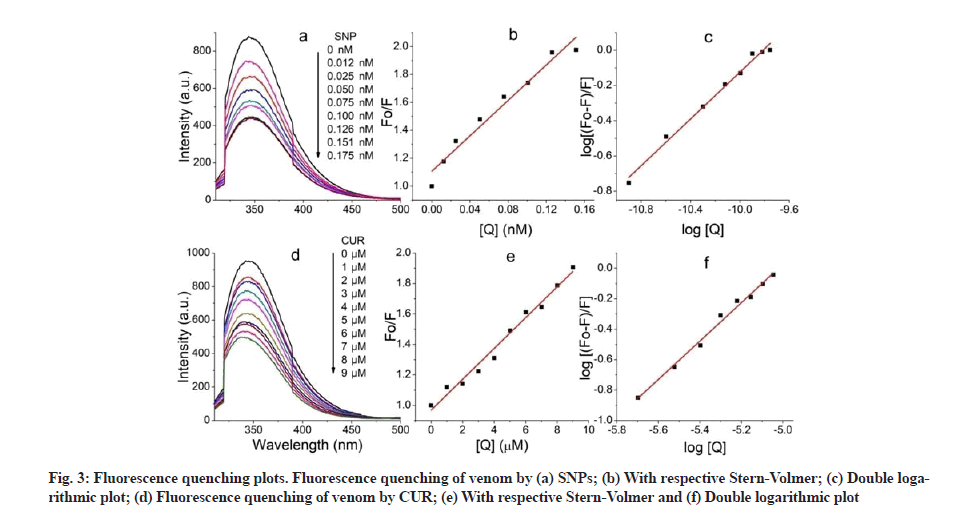
Fluorescence quenching plots were shown in fig. 3. Fluorescence characteristics of venom-CUR interaction (fig. 3a) and VS interaction (fig. 3d) were studied. Fluorescence emission intensity of venom at 341 nm was significantly quenched by SNP. Fluorescence spectral shift gives knowledge about the microenvironment of the fluorescence[33]. Here the red shift can be attributed to the exposure of fluorophore to the aqueous environment. Similar quenching was observed in venom-CUR interaction; however the blue shift indicates the hydrophobic interaction between venom and CUR.
Quenching of fluorescence relates the decrease in fluorescence intensity of fluorophore due to number of reasons including ground state complex formation, excited state reactions, molecular rearrangements, energy transfer and collisional encounters etc. This quenching is mainly due the quenching of tryptophan fluorescence in proteins by polar or charged quencher. Since quencher molecules do not freely penetrate the hydrophobic core of proteins, thus only those tryptophan residues which are present on the surface of the protein are quenched. Quenching is of two types: Dynamic quenching, where the fluorescence is ceased upon the contact of fluorophore and quencher at excited state and static quenching, where non-fluorescent complex is formed between fluorophore and quencher. For both interactions to occur, it needs contact between fluorophore and quencher. The possible mechanism of quenching was studied using Stern- Volmer plot (fig. 2b and fig. 2e) and KSV was calculated with the help of following Stern-Volmer equation.
F0/F=1+kq τ0 [Q]=1+KSV [Q] (Eq. 3)
Where F0 and F are fluorescence intensities in absence and presence of quencher respectively, kq is the biomolecular quenching rate constant, τ0 is the life time of fluorophore, [Q] is concentration of the quencher and KSV is constant. Average fluorescence life time i.e. τ0 for biopolymers is 10-8 s. The Stern-Volmer quenching constant is given by KSV=kqτ0 thus biomolecular quenching rate constant can be calculated using formula, kq=KSV/τ0.
Generally a linear Stern-Volmer plot indicates the presence of only one class of fluorophore, all having equal access to the quencher. Deviation towards X-axis from linearity infers the presence of more than one class of fluorophore with unequal accessibility. In case of dynamic quenching the biomolecular quenching constant is near 1×1010 M-1s-1 and for static quenching this value becomes higher. Current investigation shows the quenching mechanism of venom components by both CUR and SNP is static. This was confirmed by the value of biomolecular quenching constant, which was found to be 1.01×1014 M-1s-1 and 6.35×1017 M-1s-1 for CUR and SNP respectively (Table 1).
| Samples | KSV | kq (M-1s-1) | Binding sites (n) | Binding constant |
|---|---|---|---|---|
| Venom-CUR VS |
(1.01±0.09)×105 (6.35±0.26)×109 |
(1.01±0.09)×1014 (6.35±0.26)×1017 |
1.261±0.12 0.664±0.04 |
(2.14±0.23)×106 (3.28±0.15)×106 |
Note: Stern-Volmer quenching constant, binding constant and number of binding sites of venom for SNP and CUR. The results are expressed as mean±Standard Deviation (SD)
Table 1: Binding Properties of Venom Conjugates
The number of binding sites (n) and binding constant (K) of venom for both CUR and SNP was calculated by plotting a graph of Log [(F0–F)/F] vs. log [Q]. This graph gives a straight line with an equation given below.
Log [(F0–F)/F]=logK+nlog [Q] (Eq. 4)
Where n is number of binding sites and K is binding constant.
The values of binding number and binding constants suggest that, though venom have less binding sites for SNP, their binding affinity is more.
Fluorescence data showed that the fluorescence of venom was greatly quenched by CUR at the concentration range of 1-9 μM. This result infers that the microenvironment of the fluorophore was affected by CUR. Also venom fluorescence was found to be completely quenched by SNP, showing a saturation point at 0.175 nM concentration of SNP. Thus 20 μM CUR and 0.175 nM SNP were used to study biochemical activities of venom.
Also, fluorescence enhancement of CUR was studied for following interaction: CUR; equimolar BC conjugates; venom-BC conjugates; VS-CUR conjugates and VS-BS conjugates.
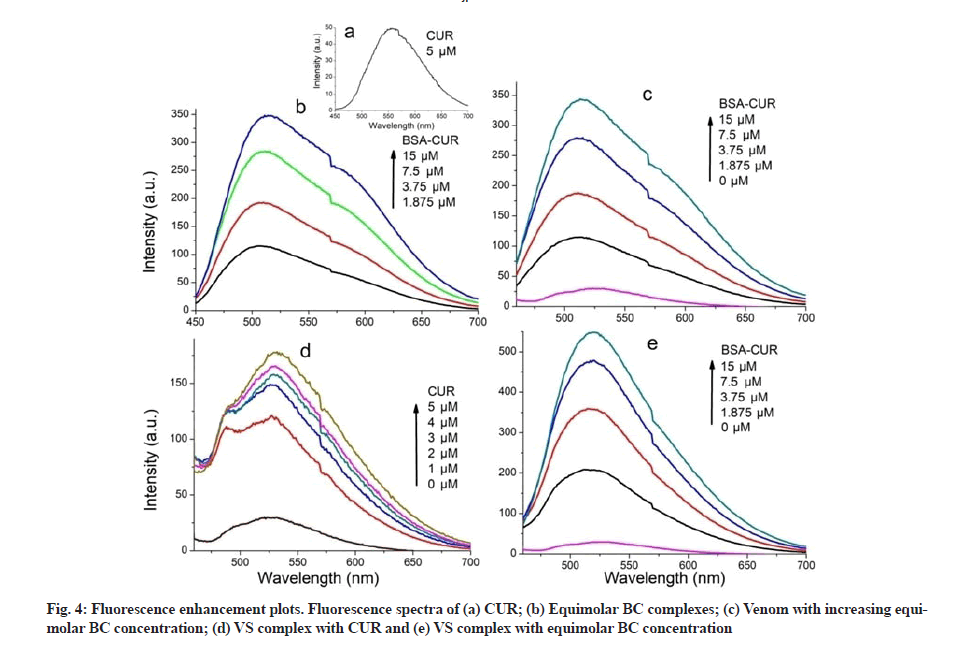
Fluorescence enhancement plots were shown in fig. 4. Fluorescence emission intensity of CUR was observed at 567 nm when excited at 425 nm (fig. 4a); it undergoes a blue shift upon BSA interaction (fig. 4b). This suggests that CUR is being localized in hydrophobic pockets of BSA, resulting in increased stability of CUR in aqueous environment. This localization can be attributed to the interaction of aryl group of CUR with hydrophobic spaces of BSA[31].
Fluorescence characteristics of VS complex with CUR (fig. 4d) was studied by exciting CUR at 425 nm. Enhancement in the fluorescence intensity was seen when CUR was added to the VS complex. Fluorescence intensity was further enhanced upon addition of more and more CUR. In comparison with the fluorescence spectra of CUR with venom, increased fluorescence enhancement was observed in the spectra of CUR with VS. This observation expects increased protection against venom in presence of SNP and CUR.
Moreover, interaction of venom and BC complexes was investigated. Fluorescence spectra were recorded by exciting CUR (fig. 4c). Emission intensity at 520 nm was not changed upon addition of increasing concentration of BC complexes to venom. Finally VS complex incubated with BC complex and fluorescence spectra were recorded by exciting CUR (fig. 4e). A great increase in emission intensity was observed at 520 nm with a blue shift of about 1 nm. Such a huge increase entails an effective interaction predicting higher protection against venom toxicity.
Venom neutralizing potential of CUR complexes is shown here. For all biochemical assays saturating concentration of CUR (20 μM), SNP (0.18 nM) and equimolar BC (15 μM) were used.
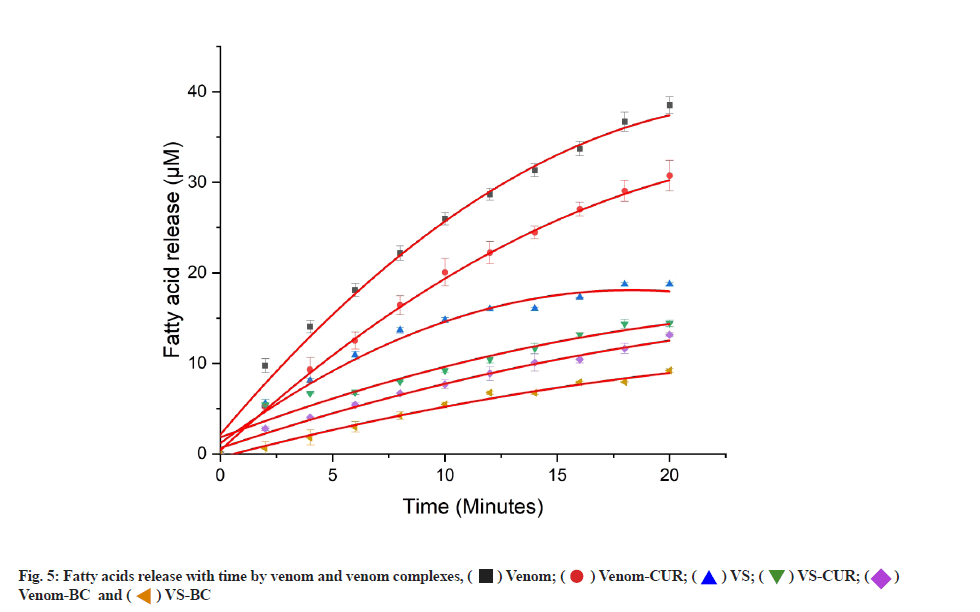
Acidimetry was performed to measure the activity of PLA2 of RVV. PLA2 with Asp49 at its active site hydrolyses ester bond at Nucleophilic Substitution bimolecular (SN2) position of phospholipids[34] present in the membrane of various cells. Upon the action of PLA2, lysophospholipids and fatty acids, mostly arachidonic acids[35] are released. Released fatty acids were measured acidimetrically on pH meter as a drop in pH of the egg yolk suspension (i.e. one unit change in pH corresponds to 133 μM fatty acids released). Egg yolk which is a rich source of lecithin wherein phospholipids are present in great amount[36] was used as a substrate.
Activity of viper venom and viper venom incubated with different test complexes was studied using this method (fig. 5).
Venom showed increased fatty acids released with time, whereas its activity with various complexes was found to be serially reduced in following order, VS-BC>Venom- BC>VS-CUR>VS>Venom-CUR. Activity of venom in presence of different inhibitory complexes is shown in Table 2.
| Samples | % Reduction |
|---|---|
| Venom-CUR | 21.37±1.59 |
| VS | 51.72±0.56 |
| VS-CUR | 62.07±0.93 |
| Venom-BC | 65.51±0.59 |
| VS-BC | 75.86±0.46 |
Note: The table shows percentage reduction in the PLA2 activity of various venom complexes. Venom treated with SNP and BC complex showed highest reduction in venom phospholipase activity. The results are expressed as mean±SD
Table 2: Percentage Reduction in the PLA2 Activity of Venom Complexes
In a report by Kadali et al.[23], soy protein nanoparticles were conjugated with anti-venom and neutralization potential of this formulation was studied. Complete neutralization of PLA2 activity of Bungarus caeruleus venom was achieved by this formulation at ratio 1:400 (venom conjugated soy nanoparticles). Also activity of Doboia russelli venom was found to be antagonized up to 40 folds by methanolic extract of Curcuma aromatica[37]. Polyphenols may inhibit snake venom either by precipitating venom proteins or by chelating cofactor required for these enzymes to work, however CUR interacts with active site of PLA2 [38]. Molecular docking experiment on PLA2 of RVV and compounds such as CUR suggests their interaction with amino acids and ions present in active site of PLA2. These inhibitory components have been shown to interact with Asp49, Gly30 substrate binding[39]. CUR exhibit strong antioxidant activity which can help to prevent oxidative damage caused by PLA2 by specifically interacting with conserved residues of PLA2 that is critical for its catalysis[40].
Protease activity was assessed using the method given by Greenberg with slight modification. Protease activity by this method involved incubation of protease with substrate, resulting in release of tyrosine residues which can be quantified spectrometrically using Folin- Lowry method. Viper venom contains many serine and metalloproteinases[41]. Venom metalloproteases are known to be major lead in the pathogenesis of venominduced local tissue damage[42]. These proteases cleave proteins releasing amino acids and several peptides. Zinc dependent metalloproteinases from viper venom are major cause of hemorrhagic damage[43], disrupting microvessels that further causes uncontrolled bleeding[44]. In current investigation, milk casein was used as substrate on which venom proteases acts, releasing tyrosine residues along with fragmented peptides in the system. Reaction terminator TCA stops the reaction and it also forms complexes with undigested casein which can be separated from free tyrosine by filtration. The filtrate containing released tyrosine residues were quantified using Folin’s method. Phenolic group of tyrosine residues complexes with copper increasing its reactivity with phosphomolybdate present in Folin’s reagent. Copper complexed phenol group reduces phosphomolybdate giving rise to a blue colored compound which absorbs maximum at 660 nm. This color intensity is directly proportional to the concentration of tyrosine residues. Enzyme activity of viper venom in presence of different inhibitory complexes was studied (fig. 6).
Protease activity of venom in presence of various CUR and SNP complexes was found to be reduced. Percentage reduction of venom activity by CUR conjugates are shown in Table 3.
| Samples | % Reduction |
|---|---|
| Venom-CUR | 10.99±2.35 |
| VS | 37.41±3.9 |
| VS-CUR | 40.20±4.5 |
| Venom-BC | 83.79±3.28 |
| VS-BC | 96.94±2.44 |
Note: The table shows percentage reduction in the protease activity of various venom complexes. Venom treated with SNP and BC complex showed highest reduction in venom protease activity. The results are expressed as mean±SD
Table 3: Percentage Reduction in the Protease Activity of Venom Complexes
CUR is a well-known protease inhibitor having ability to inhibit chymotrypsin-like activity of purified rabbit 20S proteasome and cellular 26S proteasome[45]. It is also known to inhibit matrix metalloproteinase activity[46]. It has a profound metal chelating ability[47] that can make it a good metalloprotease inhibitor. RVV proteases thus could be inhibited by chelation of metal ions that are required for enzyme catalysis.
Viperidae family is very well known for its procoagulant and anticoagulant activities. Among these species Doboia russelli is a strong procoagulant owing to the presence of RVV-X[48] and RVV-V[49]. RVV-X, a calcium dependent metalloproteinase, digests heavy chain of factor-X of blood coagulation cascade and cleaves the bond between Arg52 and Ile53. However, serine protease RVV-V acts on factor-V which is a component of blood coagulation cascade. It is acted upon by RVV-V and gets cleaved at two positions i.e. at Arg1545 and Arg1018[50]. Normal blood coagulation cascade is classified in two pathways, intrinsic and extrinsic, both converging at factor-X activation. Extrinsic pathway starts with the exposure of Tissue Factor (TF) to coagulation cascade of plasma due to any injury to vascular system. TF interacts with factor-VIIa and calcium converting factor-X to Xa. Factor-X has a cofactor i.e. factor-V. Factor-Xa, factor-V,tissue phospholipids, platelet phospholipids and calcium form a catalytic complex called prothrombinase which convert prothrombin to thrombin. Thrombin intern converts fribrinogen to insoluble fibrins which are cross linked by activated factor-XIII forming a plug at of intertwined fibrin polymers. On the other hand intrinsic pathway starts with factor-XII which activates factor- XI. Factor-XI further stimulate factor-IX to activate its cofactor i.e. factor VIII. This activated cofactor acts on factor-X and the cascade is continued in similar way to that of extrinsic pathway[51]. Since RVV-X and RVV-V from viper venom directly activates factor-X and factor-V, lengthy extrinsic and intrinsic routes are skipped. This results in procoagulation of blood. Clotting time of venom with several inhibitory complexes was found to be prolonged (Table 4).
| Samples | Clotting time (min) |
|---|---|
| Venom | 0.47±0.035 |
| Venom-CUR | 1.08±0.02 |
| VS | 1.14±0.02 |
| VS-CUR | 1.20±0.09 |
| Venom-BC | 1.36±0.06 |
| VS-BC Saline control |
1.40±0.021 2.54±0.155 |
Note: The table shows reduction in the procoagulant activity of various venom complexes. Procoagulant activity of venom was reduced greatly when treated with SNP and BC complex. The results are expressed as mean±SD
Table 4: Procoagulant Activity of Venom and Venom Complexes
Stability of RVV-X is due to disulphide linkages between one heavy and two light chains. The proposed mechanism of interaction states that light chain recognizes Gamma (γ)-Carboxyglutamic acid (Gla) domain of factor-X and heavy chain cleaves it, leading to the activation of factor-X[52]. CUR on the other hand is supportive for hemostasis, anticoagulation and fibrinolysis[53]. It has been observed that CUR inhibits both intrinsic and extrinsic pathways of coagulation by inhibiting factor-Xa and thrombin generation[54]. Hence in current investigation inhibition of procoagulant activity may be due the disruption of this stable conformation of RVV-X by CUR and SNP complexes.
Venom PLA2 acts on membrane phospholipids by binding to its substrate through hydrophobic channels, which suggests that the activity of venom can be inhibited by blocking this hydrophobic channel[55]. Structural and molecular chemistry of CUR gives it an advantage to proteins with high affinity. Phenyl rings of CUR can make pi-pi (?-?) interaction with aromatic amino acids. Phenolic and carbonyl functional groups can further share hydrogen bond with proteins[56]. Simulation studies have also shown how CUR adopts different conformation by making hydrophobic contacts with protein. In docked complex of PLA2 and CUR it was shown that one half portion of the CUR was bound to the active site of the enzyme and the other half was protruding outside. This indicates that CUR might be blocking the substrate from entering the active site of PLA2 [57].
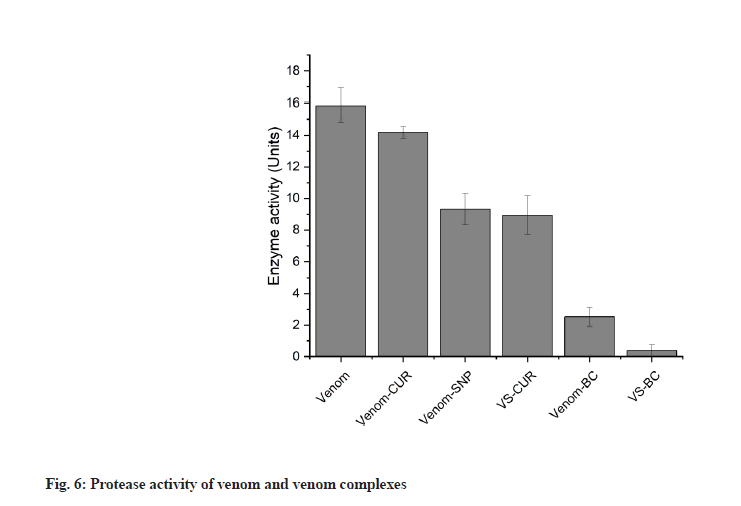
Direct agglutination test is one of the preliminary means to identify biocompatibility of any component to be used in medicine. It is a simple test that involves mixing of test compound with a drop of blood on a cavity slide and the results could be observed in few seconds. SNP+BC complex was analysed for its biocompatibility of all four blood group. Appropriate positive and negative controls were used. The complex showed no agglutination for any of the blood group (fig. 7) suggesting it’s in vitro biocompatibility.
In the present study, SNPs were synthesized and characterized using UV-visible and dynamic light scattering. RVV-SNP and BC conjugates were prepared and analysed for anti-venom activity. UV-visible and fluorescence spectra of BC conjugate showed hydrophobic and hydrophilic interaction between CUR and BSA, hence making CUR stable in aqueous environment. Fluorescence intensity of RVV was found to be significantly quenched by SNP and by CUR; however RVV was found to have more binding site for CUR. Enhancement of fluorescence intensity in VS-BC complex was significantly higher than other complexes. Inhibitory effect of these complexes was observed using biochemical assays. VS-BC complex showed major inhibition. PLA2 activity and protease activity of venom was reduced by 75 % and 96 % respectively. Procoagulant activity of venom was also inhibited and clotting time of blood was increased to normal. This complex was also confirmed to be biocompatible in vitro, which was studied by simple agglutination test.
The inhibitory effects of this complex can be attributed to the interaction of active sites of venom enzymes with SNP and CUR as well. Moreover BSA involved in BC conjugates improves solubility, stability of CUR and protect it from photodamage. CUR has greatest therapeutic properties and the current report suggests its anti-venom activity against RVV, This complex needs to be further analysed for the same in vivo.
Acknowledgements:
The financial assistance of Chhatrapati Shahu Maharaj Research, Training and Human Development Institute (SARTHI), Pune is greatly acknowledged.
Conflict of interests:
The authors declared no conflict of interest.
References
- Gupta YK, Peshin SS. Snake bite in India: Current scenario of an old problem. J Clin Toxicol 2014;4(1):1-9.
- Kasturiratne A, Wickremasinghe AR, de Silva N, Gunawardena NK, Pathmeswaran A, Premaratna R, et al. The global burden of snakebite: A literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med 2008;5(11):1591-604.
[Crossref] [Google scholar] [PubMed]
- Jayanthi GP, Gowda TV. Geographical variation in India in the composition and lethal potency of Russell's viper (Vipera russelli) venom. Toxicon 1988;26(3):257-64.
[Crossref] [Google scholar] [PubMed]
- Pe T, Aye B, Myint AA, Swe TN, Warrell DA. Bites by Russell’s viper (Daboia russellisiamensis) in Myanmar: Effect of the snake’s length and recent feeding on venom antigenaemia and severity of envenoming. Trans R Soc Trop Med Hyg 1991;5(6):804-8.
[Crossref] [Google scholar] [PubMed]
- Sharma M, Das D, Iyer JK, Kini RM, Doley R. Unveiling the complexities of Daboiarusselii venom, a medically important snake of India, by tandem mass spectrometry. Toxicon 2015;107:266-81.
[Crossref] [Google scholar] [PubMed]
- Kini RM. Excitement ahead: Structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 2003;42(8):827-40.
[Crossref] [Google scholar] [PubMed]
- Tan NH, Fung SY, Tan KY, Yap MK, Gnanathasan CA, Tan CH. Functional venomics of the Sri Lankan Russell's viper (Daboia russelii) and its toxinological correlations. J Proteomics 2015;128:403-23.
[Crossref] [Google scholar] [PubMed]
- Costa TR, Burin SM, Menaldo DL, de Castro FA, Sampaio SV. Snake venom L-amino acid oxidases: An overview on their antitumor effects. J Venom Anim Toxins Incl Trop Dis 2014;20(23):1-7.
[Crossref] [Google scholar] [PubMed]
- Kalita B, Patra A, Jahan S, Mukherjee AK. First report of the characterization of a snake venom apyrase (Ruviapyrase) from Indian Russell's viper (Daboia russelii) venom. Int J Biol Macromol 2018;111:639-48.
[Crossref] [Google scholar] [PubMed]
- Aird SD, Watanabe Y, Villar-Briones A, Roy MC, Terada K, Mikheyev AS. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophisokinavensis and Protobothrops flavoviridis). BMC Genomics 2013;14(1):1-27.
- Julio-Pieper M, Lozada P, Tapia V, Vega M, Miranda C, Vantman D, et al. Nerve growth factor induces vascular endothelial growth factor expression in granulosa cells via a trkA receptor/mitogen-activated protein kinase-extracellularly regulated kinase 2-dependent pathway. J Clin Endocrinol Metab 2009;94(8):3065-71.
[Crossref] [Google scholar] [PubMed]
- Sani I, Umar RA, Hassan SW, Faruq UZ. Antisnake venoms and their mechanisms of action: A review. Saudi J Med Pharm Sci 2018;4(5):512-20.
- León G, Stiles B, Alape A, Rojas G, Gutiérrez JM. Comparative study on the ability of IgG and F (ab') 2 antivenoms to neutralize lethal and myotoxic effects induced by Micrurus nigrocinctus (coral snake) venom. Am J Trop Med Hyg 1999;61(2):266-71.
[Crossref] [Google scholar] [PubMed]
- Warrell DA. WHO/SEARO Guidelines for the clinical management of snakebite in the Southeast Asia region. SE Asian J Trop Med Public Health 1999;30(1):1-85.
[Google scholar] [PubMed]
- de Silva HA, Ryan NM, de Silva HJ. Adverse reactions to snake anti-venom and their prevention and treatment. Br J Clin Pharmacol 2016;81(3):446-52.
[Crossref] [Google scholar] [PubMed]
- Poovazhagi V, Ravikumar SA, Jagadeeshwari A, Arulganesh P, Prabhu RS, Anupama S. Anti-snake venom induces reactions among children with snake envenomation. Int J Contemp Pediatr 2017;4(2):629-34.
- Sakthivel G, Dey A, Nongalleima K, Chavali M, Rimal Isaac RS, Singh NS, et al. In vitro and in vivo evaluation of polyherbal formulation against Russell’s viper and cobra venom and screening of bioactive components by docking studies. Evid Based Complement Alternat Med 2013;2013:1-13.
- Ratanabanangkoon K, Cherdchu C, Chudapongse P. Studies on the cobra neurotoxin inhibiting activity in an extract of Curcuma sp. (Zingiberaceae) rhizome. Southeast Asian J Trop Med Public Health 1993;24(1):178-85.
[Google scholar] [PubMed]
- Gomes A, Das R, Sarkhel S, Mishra R, Mukherjee S, Bhattacharya S, et al. Herbs and herbal constituents active against snake bite. Indian J Exp Biol 2010;48:865-78.
[Google scholar] [PubMed]
- Karain BD, Lee MK, Hayes WK. C60 Fullerenes as a novel treatment for poisoning and envenomation: A proof-of-concept study for snakebite. J Nanosci Nanotechnol 2016;16(7):7764-71.
- Suman B, Eswari B, Divya BJ, Pallavi C, Venkataswamy M, Kemparaj K, et al. Effect of silver nano particles synthesized of Trichodesma indicum against Naja Naja (cobra) venom. Int J Pharm Sci Res 2018;9(8):3291-6.
- Hingane VC, Pangam D, Dongre PM. Inhibition of crude viper venom action by silver nanoparticles: A biophysical and biochemical study. Biophys Physicobiol 2018;15:204-13.
[Crossref] [Google scholar] [PubMed]
- Renu K, Gopi K, Jayaraman G. Formulation and characterisation of antibody-conjugated soy protein nanoparticles-Implications for neutralisation of snake venom with improved efficiency. Appl Biochem Biotechnol 2014;174(7):2557-70.
[Crossref] [Google scholar] [PubMed]
- Lee PC, Meisel D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J Phys Chem 1982;86(17):3391-5.
- Tan NH, Tan CS. Acidimetric assay for phospholipase A using egg yolk suspension as substrate. Anal Biochem 1988;170(2):282-8.
[Crossref] [Google scholar] [PubMed]
- Greenberg DM. Plant proteolytic enzymes. In: Colowick SP and Kalpan SP, editors. Methods in Enzymology. 2nd ed. USA: Academic press Inc; 1955.
- Vineetha MS, Janardhan B, More SS. Biochemical and pharmacological neutralization of Indian saw-scaled viper snake venom by Canthium parviflorum extracts. Indian J Biochem Biophys 2017;54:173-85.
- Pacioni NL, Borsarelli CD, Rey V, Veglia AV. Synthetic routes for the preparation of silver nanoparticles: A mechanistic perspective. In: Alarcon EI, Griffith M, Udekwwu KI, editors. Silver nanoparticle applications: In the fabrication and design of medical and biosensing devices; Cham: Springer 2015. p. 13-46.
- Kathiravan A, Renganathan R, Anandan S. Interaction of colloidal AgTiO2 nanoparticles with bovine serum albumin. Polyhedron 2009;28(1):157-61.
- Barik A, Priyadarsini KI, Mohan H. Photophysical studies on binding of curcumin to bovine serum albumin. Photochem Photobiol 2003;77(6):597-603.
[Crossref] [Google scholar] [PubMed]
- Yang M, Wu Y, Li J, Zhou H, Wang X. Binding of curcumin with bovine serum albumin in the presence of i-carrageenan and implications on the stability and antioxidant activity of curcumin. J Agric Food Chem 2013;61(29):7150-5.
[Crossref] [Google scholar] [PubMed]
- Mitra SP. Binding and stability of curcumin in presence of bovine serum albumin. J Surf Sci Technol 2007;23(3):91.
- Kar T, Basak P, Sen S, Ghosh RK, Bhattacharyya M. Analysis of curcumin interaction with human serum albumin using spectroscopic studies with molecular simulation. Front Biol 2017;12(3):199-209.
- Stafford RE, Dennis EA. Lysophospholipids as biosurfactants. Colloids Surf 1987;30(1):47-64.
- Urs NA, Yariswamy M, Joshi V, Nataraju A, Gowda TV, Vishwanath BS. Implications of phytochemicals in snakebite management: Present status and future prospective. Toxin Rev 2014;33(3):60-83.
- Blesso CN. Egg phospholipids and cardiovascular health. Nutrients 2015;7(4):2731-47.
[Crossref] [Google scholar] [PubMed]
- Alam MI. Inhibition of toxic effects of viper and cobra venom by Indian medicinal plants. Pharmacol Pharm 2014;5(8):828-37.
- Pereanez JA, Nunez V, Patino AC, Londono M, Quintana JC. Inhibitory effects of plant phenolic compounds on enzymatic and cytotoxic activities induced by a snake venom phospholipase A2. Vitae 2011;18(3):295-304.
- Nirmal N, Praba GO, Velmurugan D. Modeling studies on phospholipase A2-inhibitor complexes. Indian J Biochem Biophys 2008;45:256-62.
- Gómez-Betancur I, Gogineni V, Salazar-Ospina A, León F. Perspective on the therapeutics of anti-snake venom. Molecules 2019;24(18):3276.
[Crossref] [Google scholar] [PubMed]
- Matsui T, Fujimura Y, Titani K. Snake venom proteases affecting hemostasis and thrombosis. Biochim Biophys Acta 2000;1477(2):146-56.
[Crossref] [Google scholar] [PubMed]
- Aung HT, Nikai T, Komori Y, Nonogaki T, Niwa M, Takaya Y. Biological and pathological studies of rosmarinic acid as an inhibitor of hemorrhagic Trimeresurusflavoviridis (habu) venom. Toxins 2010;2(10):2478-89.
[Crossref] [Google scholar] [PubMed]
- Gutiérrez JM, Rucavado A, Escalante T, Díaz C. Hemorrhage induced by snake venom metalloproteinases: Biochemical and biophysical mechanisms involved in microvessel damage. Toxicon 2005;45(8):997-1011.
[Crossref] [Google scholar] [PubMed]
- Bjarnason JB, Fox JW. Hemorrhagic metalloproteinases from snake venoms. Pharmacol Ther 1994;62(3):325-72.
[Crossref] [Google scholar] [PubMed]
- Milacic V, Banerjee S, Landis-Piwowar KR, Sarkar FH, Majumdar AP, Dou QP. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and invivo. Cancer Res 2008;68(18):7283-92.
[Crossref] [Google scholar] [PubMed]
- Cheng TS, Chen WC, Lin YY, Tsai CH, Liao CI, Shyu HY, et al. Curcumin-targeting pericellular serine protease matriptase role in suppression of prostate cancer cell invasion, tumor growth and metastasis curcumin-suppressing matriptase, prostate cancer cell invasion, and metastasis. Cancer Prev Res 2013;6(5):495-505.
[Crossref] [Google scholar] [PubMed]
- Ferrari E, Benassi R, Sacchi S, Pignedoli F, Asti M, Saladini M. Curcumin derivatives as metal-chelating agents with potential multifunctional activity for pharmaceutical applications. J Inorg Biochem 2014;139:38-48.
[Crossref] [Google scholar] [PubMed]
- Suntravat M, Nuchprayoon I, Pérez JC. Comparative study of anticoagulant and procoagulant properties of 28 snake venoms from families Elapidae, Viperidae and purified Russell’s viper venom-factor X activator (RVV-X). Toxicon 2010;56(4):544-53.
[Crossref] [Google scholar] [PubMed]
- Kini RM, Rao VS, Joseph JS. Procoagulant proteins form snake venoms. Hemostasis 2001;31(3):218-24.
[Crossref] [Google scholar] [PubMed]
- Kalafatis M, Beck DO, Mann KG. Structural requirements for expression of factor Va activity. J Biol Chem 2003;278(35):33550-61.
[Crossref] [Google scholar] [PubMed]
- Palta S, Saroa R, Palta A. Overview of the coagulation system. Indian J Anaesth 2014;58(5):515-23.
[Crossref] [Google scholar] [PubMed]
- Liu X, Atwater M, Wang J, Huo Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids Surf B Biointerfaces 2007;58(1):3-7.
[Crossref] [Google scholar] [PubMed]
- Keihanian F, Saeidinia A, Bagheri RK, Johnston TP, Sahebkar A. Curcumin, hemostasis, thrombosis, and coagulation. J Cell Physiol 2018;233(6):4497-511.
[Crossref] [Google scholar] [PubMed]
- Kim DC, Ku SK, Bae JS. Anticoagulant activities of curcumin and its derivative. BMB Rep 2012;45(4):221-6.
[Crossref] [Google scholar] [PubMed]
- Dileep KV, Tintu I, Mandal PK, Karthe P, Haridas M, Sadasivan C. Crystal structure of porcine pancreatic phospholipase A2 in complex with 2-methoxycyclohexa-2-5-diene-1, 4-dione. Front Life Sci 2011;5(3-4):135-9.
- Gupta SC, Prasad S, Kim JH, Patchva S, Webb LJ, Priyadarsini IK, et al. Multitargeting by curcumin as revealed by molecular interaction studies. Nat Prod Rep 2011;28(12):1937-55.
[Crossref] [Google scholar] [PubMed]
- Dileep KV, Tintu I, Sadasivan C. Molecular docking studies of curcumin analogs with phospholipase A2. Interdiscip Sci 2011;3(3):189-97.
[Crossref] [Google scholar] [PubMed]