- *Corresponding Author:
- G. Mustafa Bioactive Molecules Research Lab, Department of Biochemistry, University of Agriculture, Faisalabad, Punjab 38000, Pakistan E-mail: gmustafa_uaf@yahoo.com
| Date of Received | 08 October 2019 |
| Date of Revision | 01 January 2021 |
| Date of Acceptance | 21 August 2021 |
| Indian J Pharm Sci 2021;83(4):856-864 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Fibrinolytic therapy progressed by the evolution of different strategies that helped in enhancing its efficacy and specificity. The use of microbial fibrinolytic enzymes is leading to a promising direction for the treatment of cardiovascular diseases. Subtilisin, a member of subtilases is a fibrinolytic enzyme abundantly found in Bacillus species. The isolation of subtilisin gene (quaking homolog) from Bacillus subtilis was attempted in the present work. The genomic deoxyribonucleic acid extraction was done following Yamada protocol and used as a template for polymerase chain reaction amplification of the target gene using specifically designed primers. The polymerase chain reaction product was ligated into cloning vector pTZ57R/T followed by its transformation into Escherichia coli top 10 strain. A 1090 base pair, partial gene sequence was amplified coding for subtilisin protein of 363 amino acids with molecular weight of 37550.7 Daltons. The nucleotide sequence revealed significant evolutionary relationships with subtilases from other strains of Bacillus subtilis. Our study confirms the presence of subtilisin (quaking homolog) gene in local Bacillus species suggesting economical way to produce important thrombolytic agents of commercial and pharmaceutical worth.
Keywords
Fibrinolytic enzymes, subtilisin, Bacillus subtilis, vector pTZ57R/T, thrombolytic agentst
Cardiovascular diseases (CVDs) have become the leading cause of mortality throughout the world[1], including acute myocardial infarction, peripheral vascular disease, ischemic heart disease, high blood pressure, arrhythmias and stroke. Myocardial infarction and CVDs stem from the thrombosis in the coronary artery leading to fatal medical complications[2]. Various factors contribute to the thrombus formation in blood vessels however, fibrin is the main component of the blood clot which is produced after trauma or injury from fibrinogen by the action of thrombin[3].
Anticoagulants such as heparin and coumarin were used for the treatment of thrombosis in earlier days which have been substituted by the enzyme-based therapies involving in vivo lysis of fibrin[1]. The use of thrombolytic agents including streptokinase, urokinase and tissue-plasminogen activators for thrombosis confronted some limitations that confine their usage such as bleeding complications, resistance to reperfusion, incidence of acute coronary reocclusion and the high cost[2,4]. Research for improved thrombolytic agents with enhanced efficacy is being pursued involving the construction of genetically modified plasminogen activators and bacterial fibrinolytic enzymes.
In countries like China, Korea, India, Japan and West Africa, soybean-fermented foods had been used as a staple food for a long time. The significance of these foods lies in their potential of being thrombolytic that helps in preventing diseases like heart attacks, senility, strokes and osteoporosis[5]. These enzymes also have potential to be used in food fortification and nutraceutical applications as they can prevent CVDs[6].
Fibrinolytic enzymes have been identified from many organisms such as Aeromonas hydrophila, Serratia E15, Streptococcus pyogenes, Bacillus subtilis natto, Bacillus amyloliquefacens, Fusarium oxysporum, Actinomycetes and fungi[7].
Microorganisms are considered as important natural producers of different enzymes and other useful products due to their abundance in nature[8-10]. Microorganisms are also thought to be good source of thrombolytic agents due to the benefits like production in large quantity and oral administration. They had been used for curing the thrombotic disorders including acute myocardial and cerebral infarctions however, their use is limited due to the possible side effects[2,3]. On the basis of mechanism of action, the types of fibrinolytic enzymes include serine proteases, metallo proteases and mixture of both proteases. These enzymes dissolve fibrin clots through proteolytic activity and hydrolysis of the degradation products occurs by plasmin in the body[11]. One class of microbial fibrinolytic enzymes is subtilisin that is of substantial research and medical value because of its favorable medicinal role for thrombolytic therapy. Subtilisin belongs to the subtilases which is a group of serine proteases, with the serine residue in the active site for nucleophilic attack. Its confirmed efficacy, prolonged and preventive effects, stability in the gastrointestinal tract, low-cost and safety to be used as oral drug made subtilisin an eminent fibrinolytic enzyme[3]. Subtilisin is the major extracellular protease belonging to the serine proteases produced by Bacillus species.
Although various fibrinolytic enzymes have been broadly used for thrombolytic therapy but still scientistis are searching for some safer and cheaper sources of these enzymes. Considering this, the current study was designed for the isolation and cloning of a gene encoding subtilisin from an indigenous strain of Bacillus species for its gene expression studies. This study would be useful in future in pharmacological and clinical research involving in vivo fibrinolytic therapy studies, substrate specificity, protein engineering, stability, safety and many other characteristics to be evaluated.
Materials and Methods
Bacterial strain selection:
A strain of Bacillus species used in this study was isolated locally to isolate subtilisin gene. The strain was confirmed from Microbiology Department, University of Agriculture, Faisalabad.
Preparation of sporulation medium for bacterial culture:
Dubous salts medium[12] was employed to grow isolated Bacillus species. The composition of Dubous salts medium used was: Sodium nitrate (NaNO3) (2 %), dipotassium phosphate (K2HPO4) (1 %), magnesium sulfate (MgSO4) (1 %), potassium chloride (KCl) (1 %), yeast extract (0.2 %) as w/v and sterilized glucose. The pH of the medium was maintained 7.2 and the medium was autoclaved. The slants of sporulation medium and plates were prepared and bacterial culture was added aseptically. Incubation of 24 h was provided to the slants and plates for the sporulation at 40°.
Deoxyribonucleic acid (DNA) extraction:
Genomic DNA of the Bacillus species was extracted using the method described by Yamada[13] and 1 % agarose gel was employed to confirm DNA. The sample was stored at 20° till further procedure. A spectrophotometer was used to quantify isolated DNA along with purity determination through A260/280 ratio.
Polymerase chain reaction (PCR):
PCR (GeneAmp PCR System 2400, Perkin Elmer, USA) was employed for the amplification of partial gene of subtilisin quaking homolog (QK) from genomic DNA of Bacillus sp. using specifically designed primers having specific restriction sites. Primers were designed using PrimerQuest an online bioinformatics tool and are given below.
Forward primer (QKF): 5'-GTGAGAAGCAAAAAATTGTGGATCAGCTTG-3'
Reverse primer (QKR): 5'-CTTAAGGTGAGAAGCAAAAAATTGTGGATC-3'
The reaction mixture (50 µl) with optimized conditions was included 50 ng of genomic DNA, 2.5 mM deoxyribonucleotide triphosphates (dNTPs), 1.5 mM magnesium chloride (MgCl2), 10× PCR buffer (Fermentas), 0.1-1 µM primers (forward and reverse) and 1.25 U/50 μl of Taq DNA polymerase. PCR cycling conditions involved initial denaturation at 94° for 1 min followed by 35 cycles of repeated denaturation at 94° for 1 min, annealing at 56° for 1 min, polymerization at 72° for 2 min and extended polymerization of 10 min at 72°. The amplified products were checked on 1 % agarose gel. Gel extraction kit (Fermentas) was used to obtain the increased volume of the product from agarose gel required for cloning.
Cloning of subtilisin gene (QK):
The cloning vector pTZ57R/T was used to clone the amplified QK gene. Escherichia coli top 10 strain was used for the transformation and both Escherichia coli top 10 strain and Bacillus were propagated at 37° on Luria-Bertani (LB) ampicillin agar plates for 16 h. Clone analysis was done through PCR and restriction digestion by selecting the colonies with transformed cells. The insert released from the recombinant plasmid after digestion was visualized electrophoretically.
Bioinformatics analysis:
Sequence analysis and physicochemical properties: Physicochemical properties such as molecular weight (MW), theoretical isoelectric point (pI), amino acid composition, atomic composition, extinction coefficient, estimated half-life, instability index, aliphatic index and grand average of hydropathicity index (GRAVY) were calculated using ProtParam (http://web.expasy.org/protparam/). For further confirmation of theoretical pI and molecular weight calculations an online tool compute pI/MW was used.
Phylogenetic tree construction: Subtilisin protein sequence of B. subtilis was analyzed using position- specific iterative basic local alignment search tool (PSI-BLAST)[14]. Along with query sequence, forty distantly related sequences of subtilisin from different bacterial species were also retrieved from protein database provided by NCBI for phylogenetic analysis. Multiple sequence comparison by log-expectation (MUSCLE) was used to align all sequences and imported into the molecular evolutionary genetics analysis (MEGA) 7 program[15] for manual curation. Maximum-likelihood (ML) phylogenetic tree was reconstructed with 100 bootstrap replicates.
Homology modeling: To predict three-dimensional (3D) structure of subtilisin, homology or comparative modeling was employed which is the most appropriate method to build protein 3D models[16]. The best template for homology modeling was selected from cox proportional hazards (CPH) model server[17]. The alignment of template and query was performed using Modeller v9.11[18] using align2d command and output file in protein information resource (PIR) format was employed to build 5 models against our query. The 3D structure evaluation is important to reveal the accuracy levels and reliability of the predicted model[19]. The model evaluation and quality for all models was checked through ProSA-web Z-score[20] and a program to check the stereochemical quality of protein structures (PROCHECK) Ramachandran plot[21]. Root mean squared deviation (RMSD) for superimposition of subtilisin with its template was performed using University of California, San Francisco (UCSF) Chimera 1.10 workbench[22].
Results and Discussion
Amplification of the subtilisin gene (QK) is carried out. The isolation of subtilisin gene was done by designing the primers using the already reported gene sequences from GenBank. Primer 3 online primer designing tool (http://primer3.ut.ee/) was employed for the confirmation of the designed pair of primers. Ko et al.[5] also amplified subtilisin encoding QK-2 and aprN gene from Bacillus subtilis through PCR using specific primers. They amplified 1473 base pairs (bp) aprN gene with an open reading frame of 1143 bp. The concentrations for PCR were optimized as 3.0 mM MgCl2, 20 picomoles primers and 50 ng DNA. The thermal cycling conditions have been mentioned in materials and methods. PCR results were confirmed through gel electrophoresis (1 % agarose gel). The amplified partial gene was ligated into pTZ57R/T cloning vector and transformed into freshly prepared competent cells of E. coli top 10 strain. The growth of recombinants was obtained at 37° on LB-Ampicillin agar plates. The isolated partial subtilisin gene was sequenced and submitted in GenBank under accession number JQ927217.
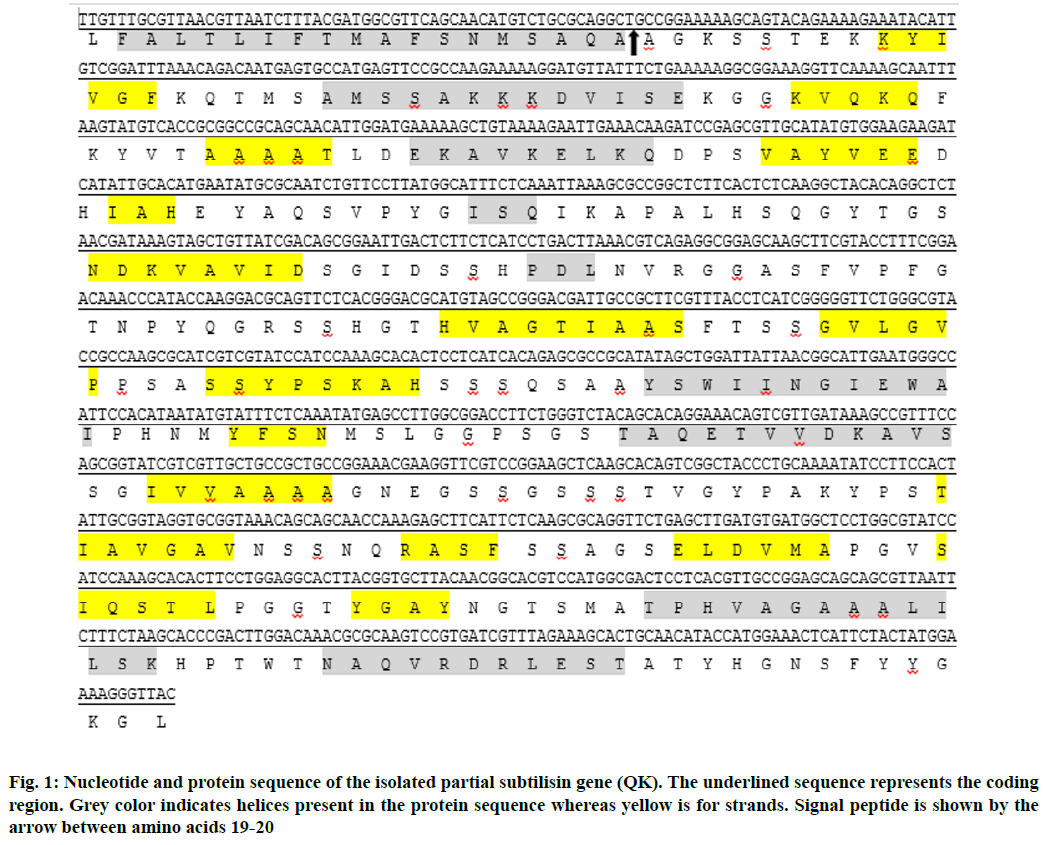
Analyses of translated protein sequence are done. ProtParam was employed to reveal physicochemical properties of deduced protein sequence. The subtilisin protein was found to have 363 amino acids, with molecular weight of 37550.7 Daltons (chemical formula: C1655H2567N451O532S8). The theoretical pI was 8.76 designating it as a positively charged protein. The instability index (II) was found to be 37.77 classifying the protein as stable. The estimated half-life was 5.5 h (mammalian reticulocytes, in vitro), 3 min (yeast, in vivo) and 2 min (Escherichia coli, in vivo). The N-terminal of the sequence considered is L (Leu). The value (-0.175) of negative GRAVY showed that the protein is hydrophilic and soluble in nature. The cellular functions are localized in definite compartments and finding of subcellular localization provides information regarding functions of novel proteins and also serves in the understanding of disease mechanisms and to develop new drugs. The prediction of subcellular localization by CELLO v.2.5 showed that query protein is extracellular. Other important properties of the protein are summarized in Table 1.
| S. No. | Parameter | Minimum value | Maximum value |
|---|---|---|---|
| 1 | Bulkiness | 8.274 | 18.693 |
| 2 | Hydrophobicity (Kyte & Doolittle) | -2.178 | 2.567 |
| 3 | Recognition factors | 81.444 | 98.333 |
| 4 | Average flexibility | 0.373 | 0.513 |
| 5 | Percentage (%) buried residues | 3.333 | 11.422 |
| 6 | Refractivity | 5.769 | 21.613 |
| 7 | Relative mutability | 58.000 | 106.667 |
| 8 | Ratio hetero end/side | 0.082 | 1.203 |
| 9 | Polarity (Zimmerman) | 0.043 | 33.816 |
| 10 | Number of codon | 2.333 | 5.111 |
| 11 | Transmembrane tendency | -1.908 | 1.237 |
| 12 | Percent (%) accessible residues | 3.700 | 8.411 |
| 13 | Average area buried | 83.144 | 151.767 |
Table 1: Important Properties Calculated Using Protscale
PredictProtein was used to predict the secondary structure of subtilisin protein sequence and three structures (i.e. helices, strands and loops) were found in the sequence (Fig. 1). The predicted percentages of these structures include 20.66 % helices, 24.24 % strands and 55.10 % loops.
Subtilisin protein can play several roles in organisms producing it as predicted by statistical inference of function through evolutionary relationships (SIFTER) protein function prediction (sifter.berkeley.edu). The anticipated activities for subtilisin are serine-type endopeptidase activity, serine-type endopeptidase inhibitor activity, tripeptide aminopeptidase activity and tripeptidyl-peptidase activity. Along with these, subtilisin also has roles in peptide binding, nerve growth factor binding, heparin binding and protease binding.
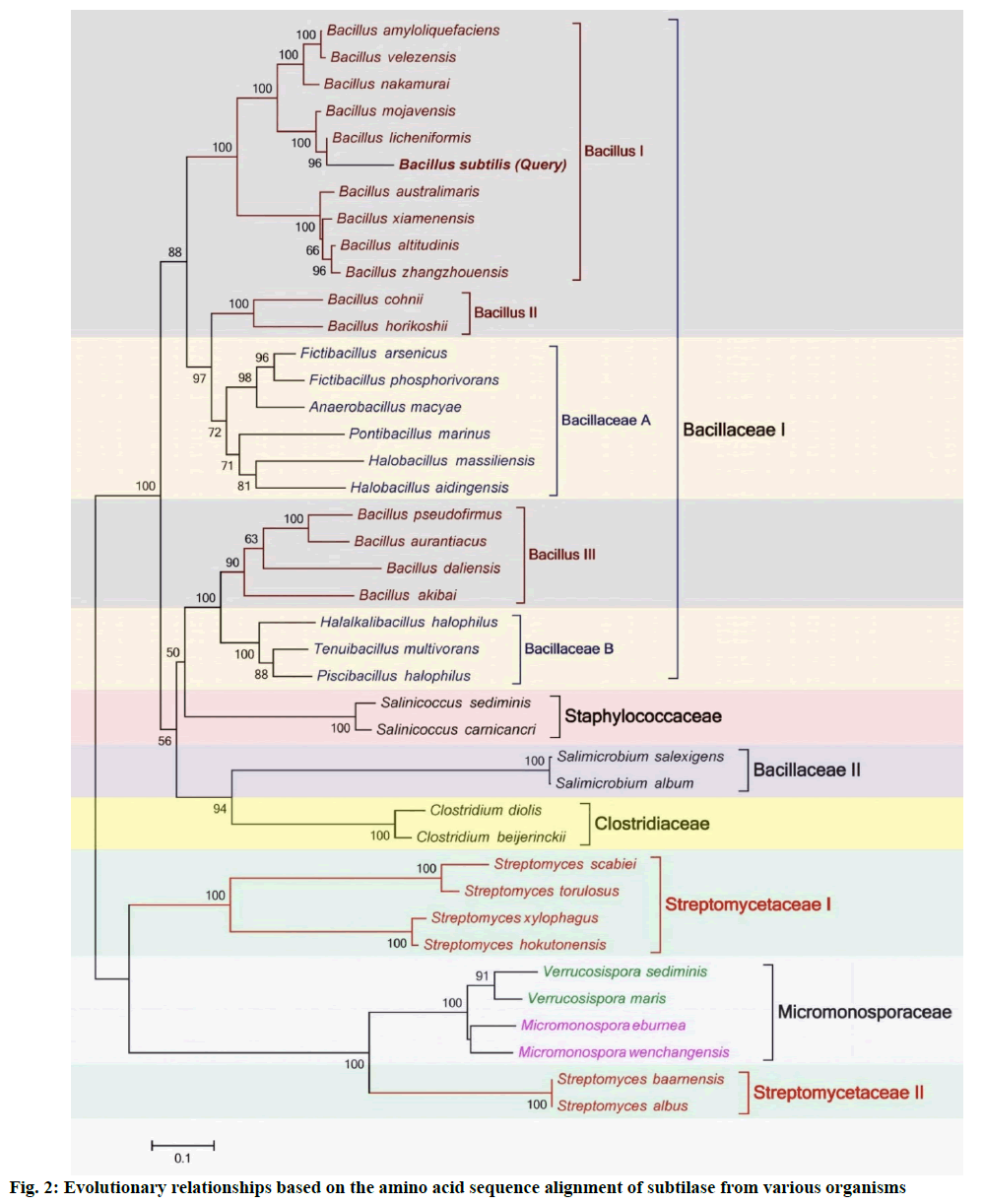
Phylogenetic relationship of Bacillus subtilis subtilase with other subtilases is described below. Molecular phylogenetic analyses are performed to show evolutionary relationships of different genes and/or proteins[23,24]. A phylogenetic tree was constructed using ML method in MEGA7 (fig. 2). Total 41 subtilisin producing Gram-positive bacterial species from five different families (i.e. Bacillaceae, Staphylococcaceae, Clostridiaceae, Micromonosporaceae and Streptomycetaceae) were added in the analysis to reveal their evolutionary relationships (labeled with square brackets). Bacillaceae family was divided into two clades (i.e. Bacillaceae I and II). Bacillaceae I was further divided into five sub-clades (Bacillus I, II and III and Bacillaceae A and B). Interestingly, all Bacillus species were not grouped into a single clade. Sub-clade Bacillus II has cladded with Bacillaceae A, showing stronger evolutionary relationships than Bacillus I and III. Similarly, Bacillus III has cladded with Bacillaceae B and showing strong evolutionary relationships. The phylogenetic analysis revealed that subtilase from Bacillus subtilis has high phylogenetic relationship with Bacillus species of Bacillus I and appeared with Bacillus licheniformis. Two bacterial species of Staphylococcaceae family have shown strong evolutionary relationships with Bacillaceae family as they have cladded with it. The bacterial species Salimicrobium salexigens and Salimicrobium album of Bacillaceae family have not cladded with other bacterial species of this family and grouped with Clostridiaceae family that is another interesting observation. The bacterial species of Streptomycetaceae family have also not grouped into a single clade and appeared under clades of Streptomycetaceae I and II. Surprisingly, Micromonosporaceae family has appeared in between both Streptomycetaceae clades showing strong evolutionary relationships with Streptomycetaceae II.
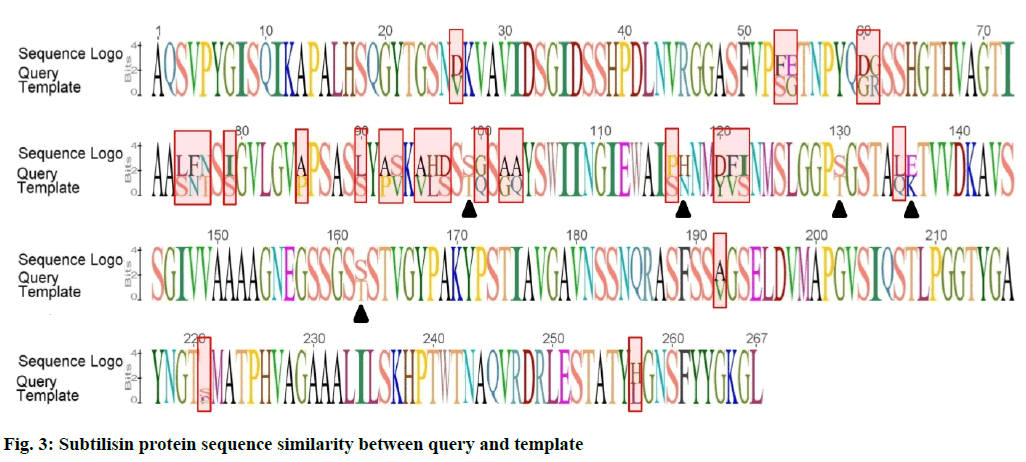
Modeller v9.11 was used for homology modeling and template was selected from CPH server. Nattokinase from Bacillus subtilis natto was taken as a template and its 3D structure was downloaded from protein databank (PDB). Fig. 3 is showing comparison of subtilisin protein sequences between query and template as sequence logo.
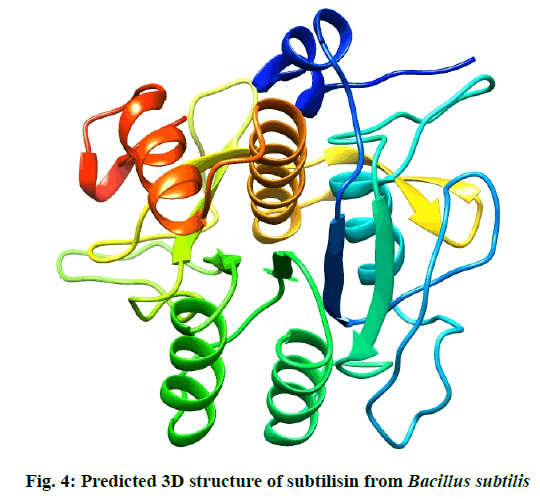
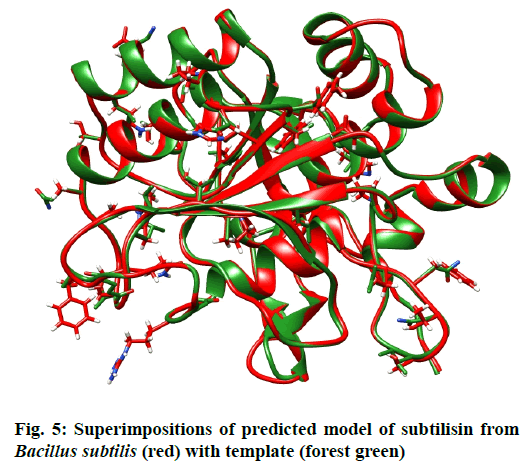
The residues in red rectangles have same physicochemical properties while black triangles are showing residues with different physicochemical properties. Total 5 models for query sequence were made by Modeller v9.11 and the best model was chosen on the basis of its structural evaluations by ProSA-web Z-scores and PROCHECK Ramachandran plots. Values of RMSD and percent identity were determined from superimpositions of query model with template (reference structure) using UCSF Chimera v1.10 program. It was revealed that there is >88 % identity between query and template. The 3D predicted structure of subtilisin from B. subtilis is shown in fig. 4.
When the 3D structure of query was superimposed with its template, an overall RMSD of 0.208 Å and Q-score 0.966 were found. Although both structures adopt similar backbone conformations but there were so many residue differences observed in both structures. fig 5. is showing superimposition of query with template. Atoms have been shown as sticks of residues differ in both structures at same positions. The complete information regarding differences in amino acids at same positions in both 3D structures is given in Table 2.
| Sr. No. | Position | Query | Template |
|---|---|---|---|
| 1 | 26 | Asp | Val |
| 2 | 53 | Phe | Ser |
| 3 | 54 | Gly | Glu |
| 4 | 60 | Gly | Asp |
| 5 | 61 | Arg | Gly |
| 6 | 75 | Ser | Leu |
| 7 | 76 | Phe | Asn |
| 8 | 77 | Thr | Asn |
| 9 | 79 | Ser | Ile |
| 10 | 85 | Pro | Ala |
| 11 | 90 | Ser | Leu |
| 12 | 92 | Pro | Ala |
| 13 | 93 | Ser | Val |
| 14 | 95 | Ala | Val |
| 15 | 96 | His | Leu |
| 16 | 97 | Ser | Asp |
| 17 | 99 | Ser | Thr |
| 18 | 100 | Gln | Gly |
| 19 | 102 | Ala | Gly |
| 20 | 103 | Ala | Gln |
| 21 | 116 | Pro | Ser |
| 22 | 117 | His | Asn |
| 23 | 120 | Tyr | Asp |
| 24 | 121 | Phe | Val |
| 25 | 122 | Ser | Ile |
| 26 | 130 | Ser | Thr |
| 27 | 135 | Gln | Leu |
| 28 | 136 | Glu | Lys |
| 29 | 162 | Ser | Thr |
| 30 | 192 | Ala | Val |
| 31 | 221 | Ser | Seb |
| 32 | 257 | His | Leu |
Table 2: Differences of Residues at Same Positions between Query and Template
Natural products obtained from different organisms are a very useful resource for improving health care issues and treating different ailments[25-28]. Fibrinolytic enzymes have been used in clinical treatments including prevention and cure of thrombotic diseases, regulation of cellular fibrinolysis where they act as antimicrobial agents[29]. Fibrinolytic enzymes are promising therapeutic agents for treating CVDs that are categorized into two groups i.e. plasminogen activators and plasmin like protein type fibrinolytic enzyme. Commercially available plasminogen activators such as tissue plasminogen activator (t-PA), urokinase and reteplase have drawbacks of reocclusion, short plasma half-life, antigenic reactions and bleeding complications[30]. However, plasmin like protein type fibrinolytic enzymes (nattokinase, lumbrokinase) are capable of directly degrading fibrin and rapidly dissolving blood clot and hence thought to be a superior therapy for treating CVDs[31].
Fibrinolytic enzymes have been derived from various sources including animals, plants and microbes but amongst all, microbial sources have attracted the attention of scientists due to the ever increasing advancement in the fields of molecular biology and fermentation technology. Notably genus Bacillus is well regarded and most well studied species to produce fibrinolytic enzymes and various fibrinolytic enzymes have been isolated from it including nattokinase from Bacillus natto, subtilisin douchi fibrinolytic enzyme (DFE) from Bacillus amyloliqueficiens[32], Bacillokinase II (BKII) from Bacillus subtilis strain A1[2], fibrinolytic enzymes (QK-1 and QK-2) from Bacillus subtilis QK02, Brevithrombolase from Brevibacillus brevis[33], fibrinolytic enzyme from Paenibacillus sp. IND8[1].
Subtilisin is a fibrinolytic enzyme belonging to subtilase and is the second largest serine protease family reported so far[34]. It is an alkaline protease with narrow functional diversity as its presence has been reported only in Gram-positive Bacillus species so far. Regardless of the evolutionary and structural difference, subtilisin shares the common mechanism with trypsin- like family enzymes. There is considerable variation in Bacillus subtilisins but containing quite similar 3D fold and the stability of subtilisins is dependent on the metal ions present in their structure[35]. All subtilisins are well known for their conserved sequences around the three amino acids i.e. serine 221, histidine 64 and aspartic acid 32 and therefore, same mechanism of catalysis exists in all subtilisins[3]. The research for highly potent fibrinolytic agent with less probability of hemorrhage occurrence and more specificity is still going on. The need of active fibrinolytic enzymes with high efficacy laid the foundation of our research work. The main objective of our work was the isolation of subtilisin gene from locally isolated Bacillus species and the bioinformatics analysis for the detailed structural, functional and evolutionary studies of the gene.
Fibrinolytic enzymes such as streptokinase, nattokinase encountered the problems of being cost effective, susceptible to reocclusion, short plasma half-life and bleeding complications. To add to the ongoing search for potential fibrinolytic enzymes, the isolation of subtilisin gene (QK), a fibrinolytic enzyme gene from bacterial source was practiced and its extensive bioinformatics study was done. This study contributes to an enhanced knowledge of the structure and function of the subtilisin gene. Further work in future will help in elucidating the therapeutic benefits and to get access to the highly purified fibrinolytic enzymes from cheaper and safer sources like bacterial species.
Conflicts of interest:
The authors declare that they have no conflict of interest.
References
- Vijayaraghavan P, Vincent SG, Arasu MV. Purification, characterization of a novel fibrinolytic enzyme from Paenibacillus sp. IND8 and its in vitro thrombolytic activity. South Indian J Biol Sci 2016;2(4):434-44.
- Jeong YK, Kim JH, Gal SW, Kim JE, Park SS, Chung KT, et al. Molecular cloning and characterization of the gene encoding a fibrinolytic enzyme from Bacillus subtilis strain A1. World J Microbiol Biotechnol 2004;20(7):711-7.
- Ghasemi Y, Dabbagh F, Ghasemian A. Cloning of a fibrinolytic enzyme (subtilisin) gene from Bacillus subtilis in Escherichia coli. Mol Biotechnol 2012;52(1):1-7.
- uz Zaman MA, Al Mamun MA, Khan SN, Hoq MM, Mazid MA. Partial purification of alkaline protease as thrombolytic agent from mutant strain Bacillus licheniformis EMS250-O-1. Dhaka Univ J Pharm Sci 2016;15(2):135-41.
- Ko JH, Yan JP, Zhu L, Qi YP. Identification of two novel fibrinolytic enzymes from Bacillus subtilis QK02. Comp Biochem Physiol Part C: Toxicol Pharmacol 2004;137(1):65-74.
- Obeid AE, Alawad AM, Ibrahim HM. Isolation and characterization of Bacillus subtillus with potential production of nattokinase. Int J Adv Res 2015;3(3):94-101.
- Dubey R, Kumar J, Agrawala D, Char T, Pusp P. Isolation, production, purification, assay and characterization of fibrinolytic enzymes (Nattokinase, Streptokinase and Urokinase) from bacterial sources. Afr J Biotechnol 2011;10(8):1408-20.
- Mustafa G, Jamil A. Comparative analyses of gene clusters and Ks-alpha genes involved in the biosynthesis of chromomycin A3 and mithramycin. Indian J Pharm Sci 2017;79(5):707-14.
- Mushtaq A, Ansari TM, Mustafa G, Shad MA, Cruz-Reyes J, Jamil A. Isolation and characterization of nprB, a novel protease from Streptomyces thermovulgaris. Pak J Pharm Sci 2020;33:2361-9.
- Mustafa G, Arshad M, Bano I, Abbas M. Biotechnological applications of sugarcane bagasse and sugar beet molasses. Biomass Convers Biorefin 2020:1-3.
- Rajendran SP, Liji L, Jayachandran K, Anie Y. Bacteria from South Indian fermented foods and food-waste dump sites as sources of fibrinolytic enzymes. Imperial J Interdisciplin Res 2016;2(7).
- Rajoka MI, Bashir A. Cloning of multiple EgI genes conferring production of endoglucanase on Escherichia coli. Pak J Biol Sci 1998;1:179-82.
- Yamada Y, Makimura K, Merhendi H, Ueda K, Nishiyama Y, Yamaguchi H, et al. Comparison of different methods for extraction of mitochondrial DNA from human pathogenic yeasts. Jpn J Infect Dis 2002;55(4):122-5.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990;215(3):403-10.
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 2016;33(7):1870-4.
- Mustafa G, Arif R, Bukhari SA, Ali M, Sharif S, Atta A. Structural and functional annotation of citrate synthase from Aspergillus niger ANJ-120. Pak J Pharm Sci 2018;31: 709-17.
- Nielsen M, Lundegaard C, Lund O, Petersen TN. CPH models-3.0-remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res 2010;38(2):576-81.
- Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol 2003;374:461-91.
- Bukhari SA, Tahir MF, Akhter N, Anjum F, Anwar H, Mustafa G. Phylogeny and comparative modeling of phytochelatin synthase from Chlorella sp. as an efficient bioagent for detoxification of heavy metals. J Biol Regul Homeost Agents 2018;32(5):1191-7.
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007;35(2):407-10.
- Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 1996;8(4):477-86.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 2004;25(13):1605-12.
- Mustafa G, Tahir A, Asgher M, Rahman MU, Jamil A. Comparative sequence analysis of citrate synthase and 18S ribosomal DNA from a wild and mutant strains of Aspergillus niger with various fungi. Bioinformation 2014;10(1):1-7.
- Jabbir F, Irfan M, Mustafa G, Ahmad HI. Bioinformatics approaches to explore the phylogeny and role of BRCA1 in breast cancer. Crit Rev Eukaryot Gene Expr 2019;29(6):551-64.
- Mustafa GH, Ahmed S, Ahmed NI, Jamil AM. Phytochemical and antibacterial activity of some unexplored medicinal plants of Cholistan desert. Pak J Bot 2016;48(5):2057-62.
- Arif RA, Ahmed S, Mustafa G. In silico study to reveal annotation and significant interactions of human defensin with its isoforms and their phylogeny. Indian J Pharm Sci 2020;82(3):465-71.
- Bukhari SA, Salman M, Numan M, Javed MR, Zubair M, Mustafa G. Characterization of antifungal metabolites produced by Lactobacillus plantarum and Lactobacillus coryniformis isolated from rice rinsed water. Mol Biol Rep 2020;47(3):1871-81.
- Munir N, Mahmood Z, Yameen M, Mustafa G. Therapeutic response of Epimedium gandiflorum’s different doses to restore the antioxidant potential and reproductive hormones in male albino rats. Dose Response 2020;18(3):1-13.
- Saxena R, Singh R. MALDI-TOF MS and CD spectral analysis for identification and structure prediction of a purified, novel, organic solvent stable, fibrinolytic metalloprotease from Bacillus cereus B80. Biomed Res Int 2015.
- Balaraman K, Prabakaran G. Production & purification of a fibrinolytic enzyme (thrombinase) from Bacillus sphaericus. Indian J Med Res 2007;126(5):459.
- Avhad DN, Vanjari SS. A novel fibrinolytic enzyme from Bacillus sphaericus MTCC 3672: optimization and purification studies. Am J Curr Microbiol 2013;1(1):1-3.
- Zhang RH, Xiao L, Peng Y, Wang HY, Bai F, Zhang YZ. Gene expression and characteristics of a novel fibrinolytic enzyme (subtilisin DFE) in Escherichia coli. Lett Appl Microbiol 2005;41(2):190-5.
- Majumdar S, Sarmah B, Gogoi D, Banerjee S, Ghosh SS, Banerjee S, et al. Characterization, mechanism of anticoagulant action, and assessment of therapeutic potential of a fibrinolytic serine protease (Brevithrombolase) purified from Brevibacillus brevis strain FF02B. Biochimie 2014;103:50-60.
- Siezen RJ, Leunissen JA. Subtilases: the superfamily of subtilisin‐like serine proteases. Protein Sci 1997;6(3):501-23.
- Dohnalek J, McAuley KE, Brzozowski AM, Ostergaard PR, Svendsen A, Wilsona KS. Stabilization of enzymes by metal binding: structures of two alkalophilic Bacillus subtilases and analysis of the second metal-binding site of the subtilase family. In understanding enzymes. Jenny Stanford Publishing; 2016. p. 227-90.