- *Corresponding Author:
- Diksha Sharma
Department of Medicinal Chemistry, Institute of Medical Sciences, Banaras Hindu University, Varanasi, Uttar Pradesh 221005, India
E-mail: diksha.mridu1991@gmail.com
| Date of Received | 19 March 2021 |
| Date of Revision | 26 September 2021 |
| Date of Acceptance | 04 August 2022 |
| Indian J Pharm Sci 2022;84(4):1026-1040 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Traditionally, Lentinula edodes are used in many Asian countries for the treatment of various chronic diseases. In the current study, ethyl acetate fraction of fruit body of Lentinula edodes was obtained by standard bioassay-guided fractionation procedure. This fractionation resulted in the isolation of three bioactive compounds (compound 1 already reported, compound 2 and 3 first time reported from this mushroom) and their structures were characterized using various spectroscopic techniques. Further, all the compounds were studied using molecular docking analysis. Compounds 1, 2, 3 and standard active ingredient used for the treatment of hyperpigmentation i.e. arbutin was prepared as program database files, was docked with the target receptor (tyrosinase, program database ID: 5M8L) which plays a vital role in melanogenesis pathway. The study mainly focuses on better results of compounds as potent tyrosinase inhibitors to down-regulate the melanogenesis pathway. The present study revealed that due to the presence of compound 1 and 3 in the fruit body of Lentinula edodes, it may be preferred as a cosmetic product in extract form to obtain effective skin lightening properties or treatment of melisma. Compounds 2 and 3 was first isolated from Lentinula edodes. The study further concludes that compound 3 could act as a potential lead molecule for the target gene tyrosinase with slight modification or optimization of chemical structure to obtain more effective, less toxic lead molecule and further exhibits remarkable skin lightening properties via inhibiting melanin production.
Keywords
Lentinula, tyrosinase inhibitors, pharmacokinetics, drug-likeness, PatchDock, melanogenesis
Lentinula edodes (L. edodes) (Berk.) Pegler belongs to the genus Lentinus of the family Marasmiceae, which generally grows on gregarious on fallen wood of a wide variety of deciduous trees particularly shii, oak, maple, beech, sweet gum, poplar, hornbeam, ironwood, mulberry and chinquapin in a warm and moist climate[1]. L. edodes which is also known as shiitake mushroom, after Agaricus bisporus, is the second most cultivated variety among edible and medicinal mushrooms cultivated throughout the world[2].
Mushrooms are considered to be a very useful natural source because of their edibility, their functional food properties and their medicinal value imparting various types of pharmacological activities. Medicinal mushrooms as part of drug therapy from ancient times and claimed to be a traditional folklore medicine[3]. Medicinal mushroom plays a vital role in the treatment of cancer, respiratory diseases and various cardiovascular disorders when used in the form of extract or specifically identifying the active ingredient, and further therapeutically acting for different disorders[4].
L. edodes can be used as a functional food, have high nutritive value and exhibits pharmacological activities. During the last few decades many secondary metabolites mainly polysaccharides, terpenes, glycosides, alkaloids, steroids, phenolic compounds and flavonoids have been isolated from L. edodes, reported to have therapeutic properties. Many new biologically active compounds have been isolated from L. edodes mainly, lentinan, eritadenine, lentinamycin and KS-2[5-7].
There are few active constituents called to be aroma components isolated from L. edodes namely, ketones, sulfides, alkanes and fatty acids. Among these major volatile flavor bearing compounds are matsutakeol (octen-1-ol-3) and ethyl-n-amyl ketone[8]. The characteristic aroma of L. edodes mushroom was named as 1,2,3,5,6-pentadiene. Generally, constituents bearing the delicious taste of shiitake are monosodium glutamate, free amino acids, organic acids, lower molecular weight peptides and sugars[9].
Several well-studied preparations, extracts and active compounds from L. edodes show remarkable medicinal properties including, anti-viral, anti-microbial, antibacterial, anti-fungal, anti-cancer, anti-oxidative, hepatoprotective, immunomodulatory, hypolipidemic and anti-neoplastic activities[10-13]. Polysaccharides and polysaccharide peptide exhibit no adverse effect, therefore they are considered as biological response modifiers for their anticancer and immunomodulation activity[14,15].
Melanogenesis is a multistage process of melanin production via cells called melanocytes and distribution in response to epidermal units present in the skin. Melanin which is known as skin pigment is helpful in the determination of skin, eye as well as hair color and protects the skin from absorption of Ultra-Violet Radiation (UVR)[16]. However, aberrant aggregation of melanin may give rise to various hyper-pigmentary disorders such as Addison’s disease, freckles, melisma, age spots, and hypo-pigmentary disorders including albinism, vitiligo, pityriasis alba which is mainly caused by various intrinsic and extrinsic factors such as hormonal changes, genetic disturbances, postinflammatory conditions, UV exposure and drugs[17]. There are primarily two types of melanin i.e. eumelanin (brownish-black synthesized from L-dopachrome) and pheomelanin (reddish-yellow whose synthesis depends upon the presence of sulfhydryl compounds); both of them are synthesized within melanosomes of melanocytes and thus catalyzed by specific melanogenic enzymes involved in series of reactions. In human skin pigmentation, eumelanin to pheomelanin ratio and overall melanin density contributes to the differences seen in the form of darker skin color[18].
There are mainly more than 125 distinct genes involved in the process of pigmentation regulation. Those genes further regulate the foremost activities that are crucial to melanoblasts (precursor cells of melanocytes, un pigmented cells that originate from embryonic neuronal crest cells) i.e. cell survival and differentiation, also plays important role in pathways involved in biogenesis and pigmentation of melanosomes[19]. Three signaling pathways are primarily involved in the regulation of melanogenesis such as cyclic Adenosine Monophosphate/Protein Kinase A (cAMP/PKA)- dependent signaling pathway is also known as alpha Melanocyte Stimulating Hormone-Melanocortin-1 Receptor (α-MSH-MC1R), Wingless-related integration site (Wnt)/Beta (β)-catenin signaling pathway and Mitogen-Activated Protein Kinases/Extracellular Signal- Regulated Kinase (MAPK/ERK) signaling pathway[20]. The gene expressions of these signaling pathways are controlled by specific melanocyte markers such as Tyrosinase-Related Protein (TRP) 1, 2 and Tyrosinase (TYR) and hence regulated by different transcription factors i.e. Receptor Tyrosine Kinase (cKIT), Stem Cell Factor (SCF, a ligand of tyrosine kinase) as well as Melanocyte Inducing Transcription Factor (MITF). MITF is the master regulator of melanin biosynthesis whose overexpression stimulates and under expression suppresses melanin biosynthesis[21].
PKA/cAMP-dependent signaling pathway regulates melanogenesis via a peptide derived from Proopiomelanocortin (POMC) namely α-MSH bound to its receptor MC1R. α-MSH-MC1R ligand-receptor binding results in activation of PKA by an increased level of intracellular cAMP via G-Protein Coupled Receptor (GPCR) type activation, further phosphorylation of cAMP Response Element-Binding Protein (CREB) activates the gene expressions of MITF and lastly, MITF stimulates melanogenesis by activating melanogenesisrelated enzymes[22]. MC1R/GPCR, is one of the major determinant of pigment phenotype of the skin that regulates the quality and quantity of melanins produced. An agonist stimulates the expression of melanogenic cascade by the activation of MC1R[18].
The second signaling pathway, the Wnt/β-catenin signaling pathway plays a vital role in melanocyte differentiation, pigmentation process and for melanocyte stem cells. Three types of proteins i.e. Wnt1, Wnt3a and β-catenin helps in the development process of neuronal cells into pigment cells, where Wnt1 signals melanoblasts to increase the number of melanocytes and Wnt3a, as well as β-catenin, are responsible for the promotion of differentiation of melanoblasts into melanocytes, thus maintaining MITF gene expression[23]. β-catenin is sequentially phosphorylated by Glycogen Synthase Kinase-3β (GSK-3β) in the absence of Wnt signals, thus this phosphorylated β-catenin was recognized by ubiquitin ligase complex which undergoes ubiquitindependent mechanism and resulted in degradation of β-catenin[24]. GSK-3β was negatively regulated after activation of the Wnt pathway which accumulates cytoplasmic β-catenin which translocates to nuclei by forming a complex with T-Cell Factor (TCF) and Lymphocyte Enhancer Factor-1 (LEF-1) and up regulates the MITF gene expression to stimulate melanogenesis[25].
ERK/MAPK signaling transduction pathway plays important role in the proliferation and differentiation of melanocytes, regulates melanogenesis via the degradation of the MITF protein. This pathway involves two types of kinases i.e. MEK and ERK which further activates melanocyte receptor via binding ligand to their extracellular domain (cKIT) resulting in up regulation of MITF gene expression by following complex mechanism namely Rat sarcoma virus/Rapidly accelerated fibrosarcoma-Mitogen-activated protein kinase/ERK kinase-ERK (Ras-Raf-MEK-ERK), leading to downstream signaling of ERK signaling pathway[26,27].
In the previous reports, various activators and inhibitors of melanogenesis signaling pathways have been studied to obtain a potential lead compound isolated from either natural sources or chemically synthesized which are available in the market as commercial skin-whitening agents such as arbutin, hydroquinone, kojic acid, liquorice extract, aloesin, azelaic acid, nicotamide and soyabean extract[28-30].
Adenosine is a naturally occurring purine nucleoside that shows anti-wrinkle properties and also used as functional ingredients in many cosmetic products. Previous study reported that adenosine inhibits melanogenesis pathway through negative regulation of TRY[31]. Adenosine is used intravenously for the treatment of certain form of supraventricular tachycardia (that do not show any improvement with vagal maneuvers), also regulates blood flow through vasodilation to various organs. adenosine is mainly considered as neuromodulator which leads to its active participation in suppression of arousal and promotion towards sleep[32].
From last decades, studies have been undertaken specifically on the extract of fruiting bodies or culture mycelium of mushroom to evaluate its therapeutic values, but still, lots of bioactive compounds are unexplored and the mechanism of already reported compounds is still unknown. Few mushroom preparations are available in the market in the form of combination drug treatment or as herbal preparation which gives synergistic effect while treatment of specific diseases. There’s a high demand for bioactive or lead compounds to be isolated from L. edodes and also to evaluate their medicinal properties.
The present study completely focuses on isolation and characterization of compounds isolated from ethyl acetate extract of fruit body of L. edodes and further in silico approach towards a ligand-receptor binding affinity inhibiting specific target (TYR) for down regulation of melanogenesis signaling pathway.
Materials and Methods
Collection of fruit body of L. edodes:
L. edodes mushroom fruit body was obtained from Indian Council of Agricultural Research (ICAR)-Directorate of Mushroom Research, National Research Centre for Mushroom located in District Solan, Himachal Pradesh. The fruit body of mushroom L. edodes (Strain No- DMRO 34) was authenticated by Dr. Sudheer Kumar Annepu, Scientist, Indian Council of Agricultural Research-Directorate of Mushroom Research (ICARDMR), Solan, Himachal Pradesh.
Extraction and isolation of compounds:
Hot continuous Soxhlet extraction: Fresh fruiting bodies of L. edodes were collected, sliced into the small portion, shade dried and grounded to obtain powdered material. Soxhlet apparatus was set up with a condenser and then a thimble (thick filter paper) was prepared and dried powder (2 kg) of fruit body of L. edodes was packed in a thimble. Then thimble was loaded in the inner tube of the Soxhlet and fitted with a round bottom flask containing solvent successively from n-hexane to methanol. 5 l of n-hexane was boiled gently at 40° for 72 h and afterward successively with chloroform, ethyl acetate, and methanol and was obtained to be 27 g, 150 g, 36 g and 650 g respectively.
Isolation of compounds using column chromatography: Ethyl acetate fraction of dried powdered fruit body of L. edodes was subjected to normal phase column chromatography over silica gel G (Thermo Fisher Scientific). All fractions were examined by Thin-Layer Chromatography (TLC) using aluminumcoated silica gel 60 F254 (Merck KGaA, Darmstadt, Germany). Aluminum coated TLC was used as solidphase and the mixture of solvents starting from non-polar to polar was followed to run the column i.e. hexane:ethyl acetate (v:v) as mobile phase. Further different fractions obtained from the column were collected; TLC was prepared for each and pooled according to the Retention factor (Rf) value calculated by TLC observation. The excessive solvent of various column fractions was kept for evaporation using a rotary vacuum evaporator under reduced pressure and further recrystallized using methanol.
Chemical analysis:
The structure of compounds isolated by column chromatography of the ethyl acetate fraction of L. edodes was identified by different spectroscopic techniques. Nuclear Magnetic Resonance (NMR) i.e. 1H NMR, 13C NMR and Distortionless Enhancement by Polarization Transfer (DEPT) 135 spectra were recorded on Bruker Avance II (400 MHz) NMR spectrometer with TMS as an internal solvent. Deuterated chloroform (CDCl3) and Deuterium oxide (D2O) were used as the solvent and chemical shift values, coupling constant values were given in (δ ppm) and (J Hz). Mass spectra were analyzed by Q-TOF micro mass (Electrospray Ionization-Mass Spectrometry (ESI-MS)).
In silico study:
Molecular docking analysis was done for the compounds isolated from shiitake mushroom considering arbutin as a standard active ingredient used for hyperpigmentation targeting a specific target gene.
Ligand preparation:
All the compounds Three Dimensional (3D) structure was obtained from PubChem https://pubchem.ncbi.nlm. nih.gov/ having specific Compound Identification (CID) number for each of them i.e. arbutin (CID No. 440936), ergosterol (CID No. 444679), cytosine(CID No. 597) and adenosine (CID No. 60961) and thus downloaded in Spatial Data File (SDF) file format. Afterward, all the SDF files were visualized using UCSF Chimera 1.11 version software and hence saved as Protein Data Bank (PDB) file format[33].
Target preparation:
The best target for arbutin which is used as a standard drug was obtained using Swiss Target Prediction software, Simplified Molecular-Input Line-Entry System (SMILES) of compound arbutin obtained from PubChem data, entered so as to run for target prediction and list of a potential target with their score will be displayed. For this study, ‘TYR’ (inhibiting enzyme) 3D crystal structure was obtained from Research Collaboratory for Structural Bioinformatics (RCSB) PDB https://www.rcsb.org/ structure/5m8l with ‘PDB ID: 5M8L’ (crystal structure of human TRY-related protein), hence downloaded and saved as a PDB file format. Further visualized using Chimera software, then crystal structure was prepared for docking by removing water molecules present of the surface of the protein and also various chains (B, C and D) was removed, as the crystal structure of TYR is of tetramer formation. Chain-A of TYR was considered as a receptor file in PDB format for further docking analysis.
Physicochemical parameters:
Various physicochemical parameters were evaluated using SwissADME software http://www.swissadme.ch where molecules to be estimated for Absorption, Distribution, Metabolism and Excretion (ADME), physicochemistry, drug-likeness, pharmacokinetics and medicinal chemistry friendliness properties can be input. Bioavailability radar is displayed for a rapid appraisal of drug-likeness[34]. Simple molecular and physicochemical descriptors like Molecular Weight (MW), Molecular Refractivity (MR), count of specific atom types and Polar Surface Area (PSA) are compiled in this section. The values are computed with OpenBabel9, version 2.3.0. The PSA is calculated using the fragmental technique called Topological PSA (TPSA), considering sulfur and phosphorus as polar atoms. Water solubility, lipophilicity, lead-likeness and synthetic accessibility were evaluated also using SwissADME[35].
Pharmacokinetics properties ADME and Toxicity (ADMET):
ADMET properties plays important role in drug discovery and development. Pharmacokinetics parameters were evaluated using the admetSAR webserver http://lmmd.ecust.edu.cn/admetsar2/ freely available online. Besides, this server database includes 22 qualitative classification and 5 quantitative regression models with high predictive accuracy, allowing estimating ecological/mammalian ADMET properties for novel chemicals[36].
Drug-likeness and bioactivity score prediction:
Drug-likeness assesses qualitatively the chance for a molecule to become an oral drug concerning bioavailability. Drug likeness property was evaluated using SwissADME software. The Lipinski (Pfizer) filter is the pioneer rule-of-five implemented and afterward; the Ghose (Amgen), Veber (GSK), Egan (Pharmacia) and Muegge (Bayer) methods respectively were calculated[35].
Bioactivity prediction score was calculated using Molinspiration webserver http://www.molinspiration.com which is freely available in the public domain. This webserver helps the calculation of bioactivity score having various descriptors such as GPCR ligand, ion channel blockers, kinase inhibitors, enzyme inhibitors, protease inhibitors and nuclear receptor ligands[36].
Molecular docking analysis:
PatchDock server https://bioinfo3d.cs.tau.ac.il/ PatchDock/php.php was used for docking of ligand and receptor prepared using Chimera software. PatchDock server uses a molecular docking algorithm based on shape complementarity principles. PatchDock server resulted in the tabulated form output including solution number, dock score, transformations, Atomic Contact Energy (ACE), interface area of the complex and PDB file of the complex[37]. Further FireDock server http://bioinfo3d.cs.tau.ac.il/FireDock/php.php was used for fast interaction refinement of ligand-receptor interaction. FireDock results output in the form of a table including solution number, global binding energy (ΔG), attractive Van Der Waal (VDW) forces, repulsive VDW, ACE and zipped files containing the 3D structure of various 10 best solutions of a ligand-receptor binding complex[38].
After downloading the best structure complex file, it was visualized using UCSF Chimera 1.11 version software and hydrophobic surface interaction of ligand molecule with receptor was saved as an image to find out better fitting of a ligand in receptor binding pocket[39]. The Two- Dimensional (2D) structure of ligand and 3D structure of ligand interaction with amino acids with specific bond distance (Aº) was labeled and visualized using BIOVIA discovery studio visualizer 4.0 version[40].
Results and Discussion
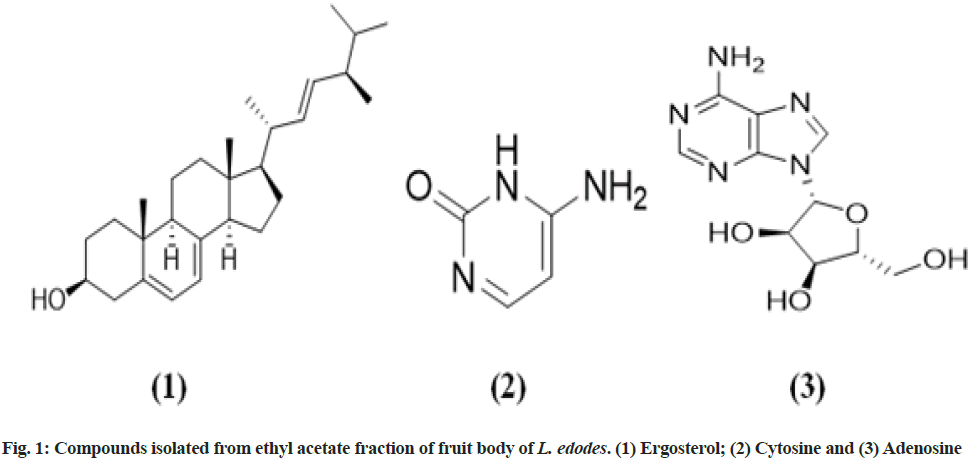
The fruit body of mushroom L. edodes was collected, dried and powdered, and further packed in a thimble for successive hot continuous soxhlet extraction. After soxhlet extraction of dried powder of fruit body of L. edodes, different fractions of the extract was obtained to be 27 g (Hexane fraction), 150 g (chloroform fraction), 36 g (ethyl acetate fraction) and 650 g (methanol fraction). Afterward, ethyl acetate fraction was chromatographed by column chromatography and thus obtaining three compounds designated as compounds 1, 2 and 3 (fig. 1). Compound 2 (cytosine) and compound 3 (adenosine) was first time isolated from L. edodes.
Column chromatography was performed by using different ratios of mobile phase starting from non-polar to polar solvent and silica gel G as a stationary phase. In the ratio 9.5:0.5 (hexane:ethyl acetate) the first compound was isolated after purifying it using methanol i.e. obtained to be a white-colored powder (20 mg). TLC was performed using Hexane:ethyl acetate (9:1) and found a single spot of Rf value 0.3 and it is UV visible. Liebermann reagent was sprayed onto TLC and the spot appeared to be light blue after 5 min of heating. Characterization and structure of compound 1 (C28H44O) was confirmed by using 1H and 13C NMR spectra at 400 MHz using CDCl3 as a solvent, where ‘J’ represents coupling constant.
1H NMR corroborated sterol Δ5,7 structure by signals δH5.58 dd (5.56, J=1.5, 1H) and 5.38 dd (5.46, J=1.75, 1H) diagnostic for olefin hydrogen’s H-6 and H-7, besides multiple in δH 3.49 (H-3) indicate the presence of hydrogen linked to carbolic carbon. Double bonds were observed at signal 5.20 (m) relative to H-22 and H-23. Still, signals at region δH 0.8 and 1.1 relative to methyl groups identified two singlet hydrogen in δH 0.94 (CH3-C-18) and 0.63 (CH3-C-19), and four duplets in δH 0.81 (CH3-27), 0.84 (CH3-26); 0.92 (CH3-28) and 1.04 (CH3-21) respectively.
13C NMR spectra reveals C28-sterol ergostane skeleton, including signals of six unsaturated carbons at δC 116.28-141.39 corresponding to C-5 (δC 139.79), C-6 (δC 119.59), C-7 (δC 116.28), C-8 (δC 141.39), C-22 (δC 135.57) and C-23 (δC 131.97). Methyl carbons were observed in C-18 (δC 12.06), C-19 (δC 16.29), C-21 (δC 21.11) C-26 (δC 19.97), C-27 (δC 19.65) and C-28 (δC 17.61), whereas hydroxyl group was observed in C-3 (δC 70.47).
Thus, based on above information obtained from different spectral data, compound 1 was elucidated as ergosterol, already reported from L. edodes fruit body and the structure was further confirmed by comparison of data with those reported in the literature.
Compound 2 was isolated after repeated column chromatography was done and observed in the ratio 7:3 (hexane:ethyl acetate), purified using methanol, obtained as a white crystalline powder (28 mg). The molecular formula of the compound was established as C4H5N3O based on the results of elemental analysis, UV (Water) λmax 259 nm and was further corroborated by its ESI-MS data which exhibited molecular ion peak at experimental calculated value i.e. m/z 112.95 [M-H]+.
The Fourier Transform Infrared (FT-IR) spectrum of compound 2 showed absorption bands νmax at 3104.82 cm-1 (symmetric N-H stretch), 2929.33 cm-1 (N-H stretching vibration), 1640.19 cm-1 (C=O stretching mode group), 1417.42 cm-1 and 1232.22cm-1 (ring modes appears), 765.60 cm-1 indicated the ring breathing mode in the molecule.
1H NMR spectrum showed δH 7.45 (J=7.60 Hz, d, 1H) and δH 5.72 (J=7.84 Hz, d, 1H) diagnostic for aromatic protons signals at H-10 and H-9 respectively. Singlets at regions δH 1.0-1.27 were identified as two singlet hydrogen i.e. δH 1.00 (C-H-C1) and δH 1.27 (C-H-C2) positions.
13C NMR spectra reveal the signals of four carbon atoms present in the ring moiety of a compound at δC ranging 101.01-173.63 corresponding to C-4 (δC 173.63), C-3 (δC 167.45), C-2 (δC 143.43) and C-1 (δC 101.01) respectively, whereas ketone (C=O) was observed at C-4 (δC 173.63). DEPT-135 NMR spectra confirm the degree of carbon molecule protonation implies the presence of a 2(C-H) signal in an aromatic ring structure of compound 2. Further, compound 2 was compared to earlier reported data and confirmed as cytosine.
Compound 3 was isolated immediately after fractions of compound 2 were collected from column chromatography in the solvent system of hexane:ethyl acetate (7:3), when purified using methanol and obtained as 32 mg white crystalline powder. The molecular formula of the compound was established as C10H13N5O4 based on the results of elemental analysis and was further corroborated by its ESI-MS data which exhibited a molecular ion peak at m/z 268.14 [M-H]+.
The FT-IR spectrum of compound 3 showed absorption bands νmax at 3340.10 cm-1 (OH Stretch in D-ribose moiety attached to the compound), 3174.32 cm-1 (NH2 stretch), 2935.12 cm-1 (C-H stretch), 2852.19 cm-1 (CH2 stretch of D-ribose ring), 1305 cm-1 (bending mode of CH2), 1051 cm-1 (bending mode of N-C-H) and absorbance 640.37 cm-1 observed at bending mode of N-C-C in the adenine ring structure attachment in the molecule.
1H NMR spectrum (D2O, 400 MHz), δH (ppm)) showed at 8.15 (1H, s, H-2), 8.24 (1H, s, H-8), 5.99 (1H, d, J=5.64 Hz, H1ʹ), 4.35 (1H, dd, J=3.32, 3.2 Hz, H-2ʹ), 4.22 (1H, dd, J=3.12, 3.16 Hz, H-3ʹ), 3.77 (1H, dd, J=3.6, 3.48 Hz, Ha-5ʹ) and 3.85 (1H, dd, J=2.76, 2.8 Hz, Hb-5ʹ).
13C NMR spectrum (D2O, 400 MHz, δC (ppm)) confirms the presence of signals of 10 carbon present in the molecule at 152.51 (C-2), 148.40 (C-4), 119.59 (C-5), 155.61 (C-6), 140.56 (C-8), 88.30 (C-1ʹ), 73.63 (C-2ʹ), 70.77 (C-3ʹ), 85.79 (C-4ʹ), 63.47 (C-5ʹ) respectively. The DEPT-135 spectrum (D2O, 400 MHz) confirms the degree of carbon molecule protonation. Compound 3 was confirmed to be ‘adenosine’ after comparison of NMR spectral analysis.
Ergosterol was also earlier reported from shiitake mushroom but after this study, it was confirmed cytosine and adenosine was the first time isolated from the ethyl acetate fraction of the fruit body of L. edodes.
Previous studies revealed that ergosterol-based compounds isolated from L. edodes extract show remarkable whitening effects when used in external cosmetic preparation as an active ingredient in a specific composition. The composition according to the previous invention resulted in whitening effects which are safe and remarkable to the skin by using 3β, 5 alpha (α)- dihydroxy-6β-methoxy-(22E, 24R)-ergosta-7,22-diene or 3β, 5α, 9α-trihydroxy-(22E, 24R)-ergosta-7,22- diene-6-one as an effective ingredient. L. edodes 70 % ethanolic extract with its isolated compounds was compared to positive control arbutin (commercially available active ingredient topically used in cosmetic preparations for hyperpigmentation or skin lightening properties) via 3-(4,5-Dimethylthiazol-2-yl)-2,5- Diphenyl-2H-Tetrazolium Bromide (MTT) assay to observe its cytotoxicity, thus extract did not affect the cell growth and the compound isolated exhibits the effect of inhibiting melanin production and identified as being concentration-dependent[41].
The present study was performed for the compounds 1, 2 and 3 isolated from ethyl acetate fraction of fruit body of L. edodes using in silico approach considering TYR target gene as a receptor to observe binding affinity and potency of the test ligands towards inhibition of melanin production. TYR enzyme plays a vital role in the generation of melanin (black pigment) contributing to the melanogenesis pathway via the TRY signaling inhibiting pathway. The present work focus on whether the compounds (1, 2, 3) possess better binding affinity as compare to arbutin to a specific target (TYR) and could act as a potential drug candidate for melanogenesis pathway modulation or down regulation.
The compounds isolated from ethyl acetate fraction of L. edodes i.e. ergosterol (1), cytosine (2) and adenosine (3) were further studied thoroughly against specific targets via computational tools. It was previously reported that due to the presence of bioactive compounds such as ergosterol and its derivatives in L. edodes extract, it shows potent skin lightening properties when compared to commercially approved drug i.e. arbutin (used in various cosmetic formulations like creams or gels for treatment of hyperpigmentation). Arbutin was analyzed against different target protein optimized using Swiss Target Prediction online software, which implies its better binding with the ‘TYR’ target gene. In a recent study, it was found that arbutin inhibits TYR via TRY inhibiting signaling pathway which further resulted in down regulation of melanogenesis.
The present study involves evaluation of various parameters such as physicochemical properties, lipophilicity, water-solubility, pharmacokinetics (ADMET), drug-likeness and bioactivity score prediction of the above-mentioned compounds isolated from L. edodes along with arbutin. The molecular docking was performed for the above-mentioned compounds against the ‘TYR’ (PDB ID: 5M8L) target gene to observe binding affinity and their signaling pathway modulation involved in melanin production.
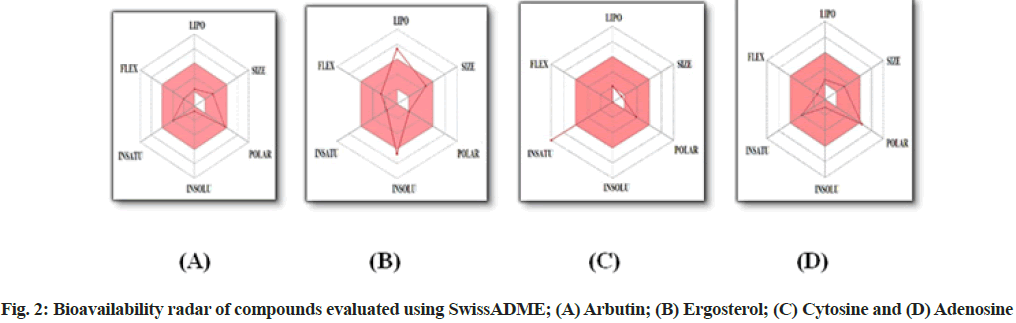
Physicochemical properties of arbutin, compounds 1, 2 and 3 were tabulated using SwissADME online software (Table 1). The bioavailability radar that provides a graphical snapshot of the drug-likeness parameters of an orally available bioactive drug was obtained by SwissADME. The drug-likeness graph is presented as a hexagon (fig. 2) with each of the vertices representing a parameter that define a bioavailable drug. The pink area within the hexagon represents the optimal range for each property (lipophilicity: XLOGP3 between -0.7 and +5.0, size: MW between 150 and 500 g/mol, polarity: TPSA between 20 and 130 Å2, solubility: log S not higher than 6, saturation: the fraction of carbons in the sp3 hybridization not less than 0.25 and flexibility: no more than 9 rotatable bonds).
| Physiochemical parameters | Name of compounds | |||
|---|---|---|---|---|
| Arbutin | 1 (Ergosterol) | 2 (Cytosine) | 3 (Adenosine) | |
| Molecular formula | C12H16O7 | C28H44O | C4H5N3O | C10H13N5O4 |
| Molecular weight | 272.25 g/mol | 396.65 g/mol | 111.10 g/mol | 267.24 g/mol |
| Number of heavy atoms | 19 | 29 | 8 | 19 |
| Number of aromatic heavy atoms | 6 | 0 | 6 | 9 |
| Fraction Csp3 | 0.5 | 0.79 | 0 | 0.5 |
| Number of rotatable bonds | 3 | 4 | 0 | 2 |
| Number of H-bond acceptor | 7 | 1 | 2 | 7 |
| Number of H-bond donor | 5 | 1 | 2 | 4 |
| Molar refractivity | 62.61 | 127.47 | 29.26 | 62.67 |
| TPSA | 119.61 Å2 | 20.23 Å2 | 71.77 Å2 | 139.54 Å2 |
Table 1: Physiochemical Properties of Arbutin, Compound 1, 2 And 3 Using SwissADME
The physicochemical parameters results have shown (Table 1) that arbutin (272.25 g/mol), compound 1 (396.65 g/mol) and 3 (267.24 g/mol) all fall in the range of molecular weight further fulfill the Lipinski requirement for drug develop-ability, whereas compound 2 possess MW lower than 150 g/mol which describes the compound as a poor drug candidate. Further, all the compounds have fraction carbon-sp3 hybridization of more than 0.25 except compound 2 (0) resulting in unsaturation in the bioavailability radar. The number of rotatable bonds in arbutin, compound 1 and compound 3 was 3, 4 and 2 respectively which shows these compounds have good molecular flexibility, an exception is only compound 2 which are having zero rotatable bonds.
The polarity of compounds was inferred by several hydrogen bond acceptors, the number of hydrogen bond donors and TPSA. The TPSA mean values were obtained to be 119.61 Å2 of arbutin, 20.23 Å2 of compound 1 (which represents the compound is having very low polarity), 71.77 Å2 of compound 2 and 139.54 Å2 of (compound 3 which represents it as higher polarity as compare to polarity range and bit out from bioavailability radar).
All these physicochemical parameters reveal that arbutin and compound 3 almost follow all the properties mentioned as per bioavailability radar prepared by SwissADME, but only one exception is there i.e. compound 3 is slightly more polar as compare to descriptor values.
Lipophilicity was assessed using different methods obtained as log P value (Table 2), which could vary from method to method as all the methods were predicted using different formulas and comparative values. All the compounds were having a very low lipophilic value of XLOGP3 except compound 1 which is having high lipophilicity which represents that arbutin, compounds 2 and 3 were insoluble in oil or fats and compound 1 is very soluble in oil or fats.
| Lipophilicity | List of compounds | |||
|---|---|---|---|---|
| Arbutin | 1 (Ergosterol) | 2 (Cytosine) | 3 (Adenosine) | |
| Log Po/w (iLOGP) | 1.64 | 4.81 | 0.22 | 0.61 |
| Log Po/w (XLOGP3) | -1.35 | 7.43 | -1.73 | -1.05 |
| Log Po/w (WLOGP) | -1.43 | 7.33 | -0.64 | -2.3 |
| Log Po/w (MLOGP) | -1.49 | 6.33 | -0.85 | -2.32 |
| Log Po/w (SILICOS-IT) | -1.22 | 6.44 | 0.49 | -2.37 |
| Consensus Log Po/w | -0.077 | 6.47 | -0.5 | -1.49 |
| Water solubility | ||||
| Log S (ESOL) solubility | Very soluble | Poorly soluble | Highly soluble | Very soluble |
| Log S (Ali) solubility | Very soluble | Poorly soluble | Highly soluble | Very soluble |
| Log S (SILICOS-IT) solubility | Soluble | Moderately soluble | Soluble | Soluble |
Table 2: Lipophilicity and Water Solubility of Arbutin, Compound 1, 2 and 3 Using SwissADME
Water solubility (log S) is considered to be the most important parameter for drug discovery and development. It is said to be that lower water solubility can lead to poor absorption and oral bioavailability which could confer additional challenges in later development stages. The water solubility evaluation parameters using different assessment methods to obtain log S value resulted in accounts to log S Estimated Solubility (ESOL) value all the compounds such as arbutin, compound 2 and 3 were having high solubility in water, except compound 1 proven to be a poor water-soluble agent. All other descriptors of water solubility resulted in the same manner and were enlisted in Table 2.
Pharmacokinetics parameters (Table 3) were studied using admetSAR software including evaluation of intestinal absorption, oral bioavailability, Caco-2 permeability, plasma protein binding, blood-brain barrier permeation, Cytochrome P450 3A4 (CYP3A4) inhibitory effect, skin permeation, excretion via Uridine 5'-Diphospho-Glucoronosyltransferases (UGT) catalyzing property and toxicity study. The ADMET study reveals that except for arbutin all the other compounds i.e. 1, 2 and 3 had better human intestinal absorption which leads to better absorption of a compound through intestinal tract when administered orally. The subcellular localization of arbutin, as well as compound 1, is on mitochondria and of compounds 2 and 3 in the nucleus of a cell. All the compounds considered for the ADMET study were not Blood Brain Barrier (BBB) permeant and were concluded to be non-substrate of P-gp. In terms of metabolism, it was observed that compounds discussed above were non-inhibitor of CYP450 except compound 1 which is predicted as an inhibitor of CYP2C9 and CYP3A4. A non-inhibitor of CYP450 means that the molecule will not hamper the biotransformation of drugs metabolized by the CYP450 enzyme.
| Pharmacokinetics parameters | Name of compounds | |||
|---|---|---|---|---|
| Arbutin | 1 (Ergosterol) | 2 (Cytosine) | 3 (Adenosine) | |
| Absorption | ||||
| Human intestinal absorption | No | Yes | Yes | Yes |
| Human oral bioavailability | No | No | Yes | No |
| Caco-2 permeability | No | Yes | No | No |
| Distribution | ||||
| Subcellular localization | Mitochondria | Mitochondria | Nucleus | Nucleus |
| BBB permeant | No | No | No | No |
| Metabolism | ||||
| CYP2C9 inhibitor | No | Yes | No | No |
| CYP3A4, CYP2D6, CYP2C19 and CYP1A2 inhibitor | No | No | No | No |
| Log Kp (skin permeation) | -8.92 cm/s | -3.44 cm/s | -8.21 cm/s | -8.68 cm/s |
| Excretion | ||||
| UGT catalyzed | No | Yes | No | Yes |
| Toxicity | ||||
| Acute oral toxicity | III | I | III | III |
| Hepatotoxicity | No | No | Yes | Yes |
| Carcinogenicity (binary) | No | No | No | No |
| Carcinogenicity (ternary) | Not required | Not required | Not required | Not required |
Table 3: Pharmacokinetics Properties (ADMET) of Compounds using admetSAR
In case of excretion or elimination of drug candidate, UGT catalyzed reaction plays a vital role and in this study, we observed that the compound 2 and arbutin both are predicted as non-UGT catalyzed, whereas compound 1 and 3 were UGT-catalyzed products which make then easily excrete out from the body. After evaluation of various toxicity parameters, it was obtained that acute oral toxicity of all the compounds falls under category III except compound 1 it also possesses the highest oral toxicity, comes under category I as compared to other test ligands, in contrast, compound 3 exhibits the lowest oral toxicity. The carcinogenic profile revealed that all the test ligands were non-carcinogenic.
Drug-likeness is a key criterion in screening drug candidates at the earlier phase of drug discovery and development. Drug-likeness evaluation helps in a correlation of physicochemical aspects of a compound with its biopharmaceutical aspects in the human body, especially adhering to its bioavailability perioral route. Drug-likeness properties (Table 4) were predicted using SwissADME software for the specified test ligands i.e. compounds 1, 2, 3 and arbutin. Drug-likeness parameters include various algorithms which were used for the evaluation i.e. Lipinski rule, Ghosh, Veber, Egan and Muegge rule mainly considered for drug ability of potential lead molecule. It was revealed that compounds 2, 3 and arbutin follow Lipinski’s rule of five having zero violation, whereas compound 1 does follow the rule but does have one violation (Moriguchi Octanol-Water Partition Coefficient (MLOGP) >4.15). According to Ghosh’s rule of drug-likeness, none of the test ligands pass the criteria, but all of them pass the rule of Veber having zero violation.
| Drug-likeness Parameters | Name of Compounds | |||
|---|---|---|---|---|
| Arbutin | 1 (Ergosterol) | 2 (Cytosine) | 3 (Adenosine) | |
| Lipinski | Yes, 0 violation | Yes, 1 violation and MLOGP >4.15 | Yes, 0 violation | Yes, 0 violation |
| Ghose | No, 1 violation and WLOGP <-0.4 | No, 2 violations; WLOGP >5.6 and #atoms >70 | No, 4 violations; MW <160; WLOGP <-0.4; MR <40 and #atoms <20 | No, 1 violation and WLOGP <-0.4 |
| Veber | Yes | Yes | Yes | Yes |
| Egan | Yes | No, 1 violation and WLOGP >5.88 | Yes | No, 1 violation and WLOGP <-0.4 |
| Muegge | Yes | No, 2 violations; XLOGP3 >5 and Heteroatoms <2 | No, 2 violations; MW <200 and #C <5 | Yes |
| Bioavailability score | 0.55 | 0.55 | 0.55 | 0.55 |
| Lead likeness | Yes | No, 2 violations; MW >350 and XLOGP3 >3.5 | No, 1 violation and MW <250 | Yes |
| Synthetic accessibility | 4.18 | 6.58 | 1.47 | 3.86 |
Table 4: Drug-Likeness Properties of Compounds 1, 2, 3 and Arbutin using SwissADME
Besides, the evaluation was also carried out using A Bioavailability Score (ABS) criteria, where all the above-mentioned compounds obtained the value of 0.55. This criterion is based on the probability value of a compound to possess an optimum profile of bioavailability and permeability, where a value of 0.55 implies the obedience of Lipinski’s role of five and 55 % probability of rat bioavailability value higher than 10 %. The synthetic accessibility of all the three compounds has a better score to obtain the molecule synthetically but an exception is only compound 2 (1.47) which is having a very low synthetic accessibility score. Lead-likeness properties were positively achieved by arbutin and compound 3, whereas compound 1 and 2 exhibits two violations (MW>350, XLOGP3>3.5) and one violation (MW<250) respectively.
The bioactivity score of test ligands; arbutin, compounds 1, 2 and 3 were calculated for different parameters such as binding to GPCR ligand, nuclear receptor ligand, ion channel modulation, kinase inhibition, protease inhibition and enzyme activity inhibition. In the present study the bioactivity score prediction was calculated using Molinspiration software (Table 5), which indicates that for organic molecules the probability is if the bioactivity score is (>0), then it is active; if (-5.0 to 0.0) then moderately active and if (<-5.0) then the compound said to be inactive. The bioactivity score prediction of the above mentioned compound revealed that compound 3 possesses a better score for each parameter of bioactivity evaluation, which further results in it as a potential drug with some chemical structure modifications.
| Bioactivity scoring parameters | Name of compounds | |||
|---|---|---|---|---|
| Arbutin | 1 (Ergosterol) | 2 (Cytosine) | 3 (Adenosine) | |
| GPCR ligand | 0.05 | 0.14 | -3.06 | 1.1 |
| Ion exchange modulator | 0.12 | -0.13 | -3.22 | 0.54 |
| Kinase inhibitor | -0.13 | -0.34 | -2.64 | 0.87 |
| Nuclear receptor ligand | 0.04 | 0.74 | -3.76 | -1.74 |
| Protease inhibitor | -0.09 | -0.08 | -3.31 | -0.01 |
| Enzyme inhibitor | 0.46 | 0.53 | -1.94 | 1.28 |
Table 5: Bioactivity Score Prediction using Molinspiration Software
Molecular docking analysis was performed using PatchDock software, which is generally a geometrybased molecular docking algorithm. PatchDock mainly aims at finding docking transformations that yield good molecular shape complementarity. PatchDock algorithm divides the Connolly dot surface representation of the molecule into concave, convex and flat patches. Then, these patches were matched to generate candidate transformations. Each candidate transformation was further evaluated by a scoring function that considers both geometric fit and atomic desolation energy. Finally, root mean square deviation 4Aº (Root-Mean-Square Deviation (RMSD)) clustering is applied to the candidate solutions to discard redundant solutions[37].
The present docking study was evaluated using PatchDock software (Table 6), in which PDB-files of ligands i.e. arbutin, compounds 1, 2 and 3 was prepared and uploaded one by one with the receptor TYR (PDB ID: 5M8L) file and results were sent via email as specified by the user. The results were in the form of top 20 solutions which could be visualized as well as downloaded as a compressed file. These results indicate that arbutin; compounds 1 and 3 possess a good docking score i.e. 3620, 5868, 3496 respectively after binding to the receptor TYR. The ACE of the best solution of each compound reveals that compound 1 has higher ACE and the interface area of the complex (ligand-receptor binding complex) was maximum covered by compound 1 (692.50) followed by compound 3 (434.00) then arbutin (429.70) and lowest was covered by compound 2 (227.90). Further, the automatic redirection of the PatchDock candidate solution to the FireDock server was performed to obtain the top 10 best-refined data with ΔG.
| Docking results | ||||||
|---|---|---|---|---|---|---|
| Compounds | Score | Area | ACE | ΔG (kcal/mol) | Attractive VDW | Repulsive VDW |
| Arbutin | 3620 | 429.7 | -215.53 | -35.97 | -13.14 | 5.05 |
| Ergosterol (1) | 5868 | 692.5 | -394.5 | -52.37 | -15.68 | 1.98 |
| Cytosine (2) | 2032 | 227.9 | -142.38 | -22.06 | -7.76 | 1.02 |
| Adenosine (3) | 3496 | 434 | -306.97 | -38.91 | -11.87 | 1.39 |
Table 6: Docking Result of Receptor TYR (PDB ID: 5M8l) With Compound 1, 2, 3 and Arbutin Using Patchdock and Firedock Software
FireDock server is a web-server for fast interaction refinement in molecular docking, which further provides flexible refinement, scoring of protein-protein and protein-ligand docking solutions. This method of docking refines each candidate and ranks all the candidates according to their binding energy. The FireDock method involves three main stages i.e. side-chain optimization, rigid body minimization and scoring as well as ranking according to their global binding energy[38].
In this study, the FireDock results revealed that compound 1 exhibits the highest global binding energy (-52.37 kcal/mol) followed by compound 3 (-38.91 kcal/mol) then arbutin (-35.97 kcal/mol) and lowest by compound 2 (-22.06 kcal/mol) was obtained to be lowest when compared to another test ligand. The global binding energy results of arbutin with its receptor TYR were slightly lower than that of compound 3 with TYR, which could further assume that compound 3 could work as a potential drug candidate via targeting the TYR target gene which plays a vital role in the melanogenesis pathway. It could be concluded from the docking analysis that compound 3 may exhibit skin lightening properties likewise arbutin via inhibiting TRY signaling pathway and down regulating the melanin production.
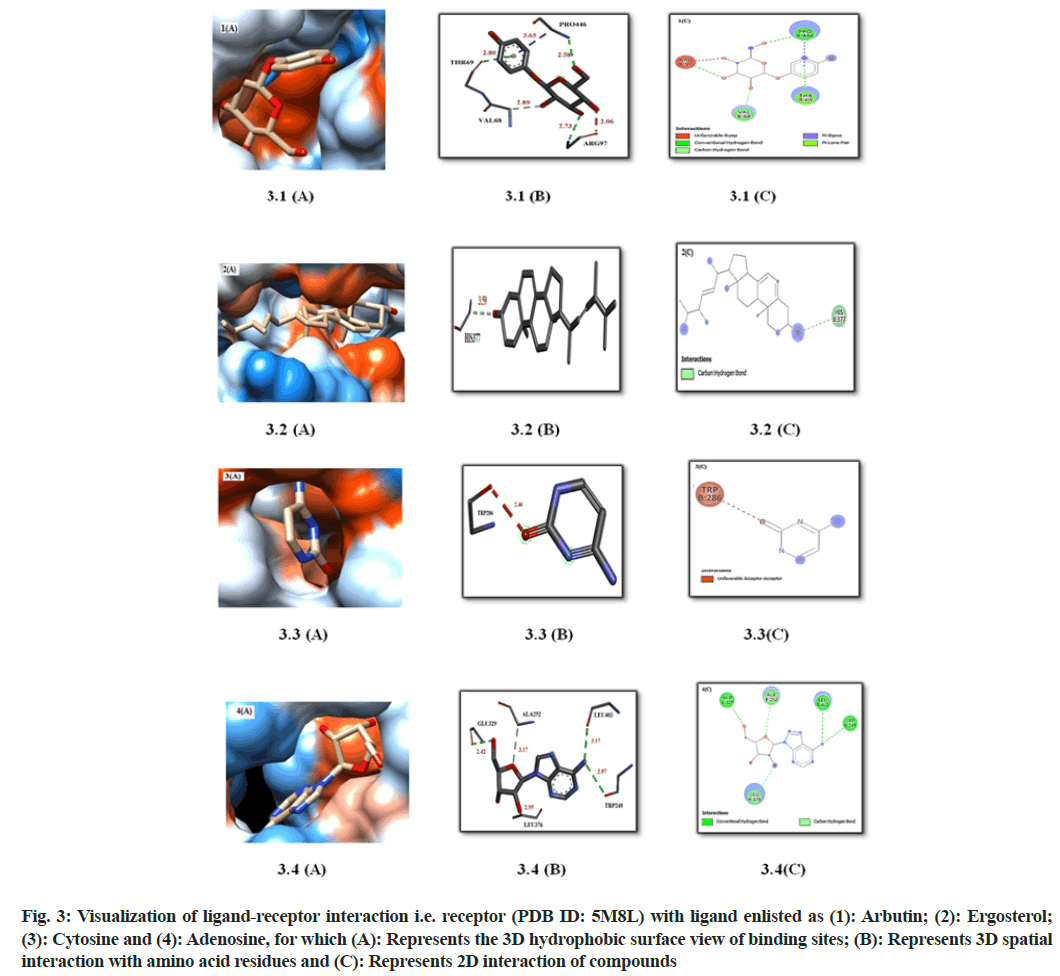
Visualization of a best-docked complex of receptor TYR and various test ligand were done using UCSF Chimera software and BIOVIA discovery studio visualizer to obtain interactions of ligand docked in the binding pocket of the hydrophobic surface of receptor; the 3D structure of ligand interacting with amino acids having a different type of bonding such as hydrogen bonding, hydrophobic bonding, pi-lone pair, pi-sigma bonding, with their bond distance (Aº) and 2D structure of ligand explaining the interaction of amino acids with different atoms of test ligand having a specific type of molecular bonding.
The present docking results were visualized (fig. 3 and Table 7), and resulted in good interaction with the receptor’s amino acid specifically that of compound 3 and arbutin. It was observed in fig. 3.1 (A) that arbutin have perfectly fitted in the pocket of receptor in the hydrophobic surface and thus interacting with various amino acid (3D structure and 2D structure with amino acid (fig. 3.1 (B) and fig. 3.1 (C)) describing conventional hydrogen bond with Pro446 at 2.58 Aº and Arg97 at 2.73 Aº, pi-sigma bond at a distance of 3.65 Aº of Pro446, pi-lone pair with Thr69 at 2.80 Aº, C-H bond with Val68 at 2.89 Aº and an unfavorable bump at a distance of 2.09 Aº with Arg97. The hydrophobic surface interaction of compound 3 with the receptor observed to be best fitted to the pocket of the receptor (fig. 3.4 (A)) which was better represented in the form of 3D-structure fig. 3.4 (B) as well as 2D-structure fig. 3.4 (C) describing ligand interaction with amino acids i.e. conventional hydrogen bond with Glu329 at 2.42Aº, Leu403 at 3.17 Aº, Trp249 at 2.97 Aº and C-H bond with Leu376 at 2.95 Aº and Ala252 at a distance of 3.17 Aº. All these ligand-receptor interactions concluded that compound 3 could act as the best ligand for targeting the specific receptor TYR to down regulate the melanogenesis pathway.
| Compounds | ΔG | Residues direct interaction with ligand through hydrogen bonds and their bond distance (Aº) | Residues interaction with ligand through other interactions and bond distance |
|---|---|---|---|
| Arbutin | -35.97 | Pro446 (1H at 2.58 Aº), Val68 (1H at 2.89 Aº), Arg97 (1H at 2.09 Aº and 2.73 Aº) |
Conventional hydrogen bond (Pro446, Arg97), pi-sigma bond (Pro446 at 3.65Aº), pi-lone pair (Thr69 at 2.80Aº), carbon hydrogen bond (Val68) and unfavorable bump at distance 2.09 Aº (Arg97) |
| Ergosterol (1) | -52.37 | His377 (1H at 3.59Aº) | Carbon hydrogen bond (His377) |
| Cytosine (2) | -22.06 | ----- | Unfavorable acceptor-acceptor interaction (Trp286) |
| Adenosine (3) | -38.91 | Ala252 (1H at 3.17Aº), Glu329 (1H at 2.42Aº) Leu403 (1H at 3.17Aº) Trp249 (1H at 2.97Aº) Leu376 (1H at 2.95Aº) |
Conventional hydrogen bond (Glu329, Leu403, Trp249) and carbon hydrogen bond (Leu376, Ala252) |
Table 7: Ligand Interaction Analysis with Different Amino Acid Residues with Their Binding Global Energy
Compounds 1 and 2 were observed to be weakly interacting with the amino acid present on the surface of receptor TYR as enlisted in (Table 7 and fig. 3).
Fig. 3: Visualization of ligand-receptor interaction i.e. receptor (PDB ID: 5M8L) with ligand enlisted as (1): Arbutin; (2): Ergosterol; (3): Cytosine and (4): Adenosine, for which (A): Represents the 3D hydrophobic surface view of binding sites; (B): Represents 3D spatial interaction with amino acid residues and (C): Represents 2D interaction of compounds
Docking analysis results revealed that compound 3 (adenosine) may act as a potential lead molecule as it possesses higher binding energy as compare to arbutin when completed with target gene TYR, for the treatment of hyperpigmentation or better skin lightening effects.
L. edodes was used as a traditional medicine in the form of functional foods, dietary supplements or naturally/ semi-synthetically isolated compounds from their fruit body extract imparting various types of pharmacological activities. Shiitake mushroom was commonly known to exhibit effectiveness regarding an action promoting blood circulation, anti-cancer effect, and anti-oxidant effect, further prevention of cholesterol, anemia, osteoporosis and hypertension[42]. This study was done to obtain bioactive compounds which could further be used for the treatment of the various type of disease. Adenosine and cytosine was first time isolated from ethyl acetate extract of L. edodes and thus evaluated via targeting TRY enzyme which plays important role in melanin production through the in silico approach. Adenosine was compared to arbutin and resulted in better global binding energy which could further conclude that adenosine can be used as skin lightening active ingredient which works as a potential inhibitor of the melanogenesis signaling pathway. Also, the presence of ergosterol, adenosine and cytosine in the fruit body of L. edodes which have shown good docking score and better binding affinity towards inhibition of melanin production, may conclude that Shiitake mushroom may also be used in extract form for topical preparations as effective skin lightening product. Further, there is a need for chemical structure modification and optimization of adenosine which could enhance its absorption, blood-brain barrier permeation, skin permeation, oral bioavailability and its drug-likeness properties.
Acknowledgements:
Diksha Sharma wishes to express gratitude to ICARDMR, Solan, Himachal Pradesh, India for providing the fruit body of L. edodes. Diksha Sharma would like to thank Avtar Singh, Manish Kumar and Anoop Patyal, Sophisticated Analytical Instrumentation Facility (SAIF), Punjab University, Chandigarh, India for providing NMR and ESI-MS spectral data.DS is grateful to Dr. Shreyans Kumar Jain, Assistant Professor, IIT, BHU, Varanasi, India for providing valuable help in the interpretation of Spectral data mainly 1HNMR, 13C NMR and ESI-MS. Diksha Sharma thanks the software vendors for their continuing support for academic research efforts, in particular, BIOVIA discovery studio visualizer, UCSF Chimera and various freely available web servers mainly SwissADME, ADMETSAR, Molinspiration, PatchDock and FireDock server. Diksha Sharma is thankful to Dr. Rajesh Kumar Singh, Banaras Hindu University, Varanasi, India for guidance in molecular docking. Diksha Sharma would like to thank the Indian Council of Medical Research (ICMR), New Delhi, India for providing financial assistance through ICMR-SRF (ICMR-Senior Research Fellowship).
Conflict of interests:
The authors declared no conflict of interest.
References
- Finimundy TC, Dillon AJ, Henriques JA, Ely MR. A review on general nutritional compounds and pharmacological properties of the Lentinula edodes mushroom. Food Nutr Sci 2014;5(12):1095-105.
- Roupas P, Keogh J, Noakes M, Margetts C, Taylor P. The role of edible mushrooms in health: Evaluation of the evidence. J Function Foods 2012;4(4):687-709.
- Blagodatski A, Yatsunskaya M, Mikhailova V, Tiasto V, Kagansky A, Katanaev VL. Medicinal mushrooms as an attractive new source of natural compounds for future cancer therapy. Oncotarget 2018;9(49):29259.
[Crossref] [Google Scholar] [PubMed]
- Ganeshpurkar A, Rai G, Jain AP. Medicinal mushrooms: Towards a new horizon. Pharmacogn Rev 2010;4(8):127-35.
[Crossref] [Google Scholar] [PubMed]
- Sharma D, Singh VP, Singh NK. A review on phytochemistry and pharmacology of medicinal as well as poisonous mushrooms. Mini Rev Med Chem 2018;18(13):1095-109.
[Crossref] [Google Scholar] [PubMed]
- Yamaguchi Y, Miyahara E, Hihara J. Efficacy and safety of orally administered Lentinulaedodes mycelia extract for patients undergoing cancer chemotherapy: A pilot study. Am J Chin Med 2011;39(3):451-9.
[Crossref] [Google Scholar] [PubMed]
- Ina K, Kataoka T, Ando T. The use of lentinan for treating gastric cancer. Anticancer Agents Med Chem 2013;13(5):681-8.
[Crossref] [Google Scholar] [PubMed]
- Bisen PS, Baghel RK, Sanodiya BS, Thakur GS, Prasad GB. Lentinus edodes: A macrofungus with pharmacological activities. Curr Med Chem 2010;17(22):2419-30.
[Crossref] [Google Scholar] [PubMed]
- Yang X, Zhang Y, Kong Y, Zhao J, Sun Y, Huang M. Comparative analysis of taste compounds in shiitake mushrooms processed by hot-air drying and freeze drying. Int J Food Properties 2019;22(1):1100-11.
- Da Silva AC, Jorge N. Antioxidant properties of Lentinus edodes and Agaricus blazei extracts. J Food Quality 2011;34(6):386-94.
- Rincão VP, Yamamoto KA, Silva Ricardo NM, Soares SA, Paccola Meirelles LD, Nozawa C, et al. Polysaccharide and extracts from Lentinula edodes: Structural features and antiviral activity. Virol J 2012;9(1):37.
[Crossref] [Google Scholar] [PubMed]
- Hearst R, Nelson D, McCollum G, Millar BC, Maeda Y, Goldsmith CE, et al. An examination of antibacterial and antifungal properties of constituents of shiitake (Lentinula edodes) and oyster (Pleurotus ostreatus) mushrooms. Complement Ther Clin Pract 2009;15(1):5-7.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Zhang L. Physicochemical properties and antitumor activities for sulfated derivatives of lentinan. Carbohydr Res 2009;344(16):2209-16.
[Crossref] [Google Scholar] [PubMed]
- Israilides C, Kletsas D, Arapoglou D, Philippoussis A, Pratsinis H, Ebringerová A, et al. In vitro cytostatic and immunomodulatory properties of the medicinal mushroom Lentinula edodes. Phytomedicine 2008;15(6-7):512-9.
[Crossref] [Google Scholar] [PubMed]
- Lee HH, Lee JS, Cho JY, Kim YE, Hong EK. Study on immunostimulating activity of macrophage treated with purified polysaccharides from liquid culture and fruiting body of Lentinus edodes. J Microbiol Biotechnol 2009;19(6):566-72.
[Crossref] [Google Scholar] [PubMed]
- Videira IF, Moura DF, Magina S. Mechanisms regulating melanogenesis. An Bras Dermatol 2013;88(1):76-83.
[Crossref] [Google Scholar] [PubMed]
- Nicolaidou E, Katsambas AD. Pigmentation disorders: Hyperpigmentation and hypopigmentation. Clin Dermatol 2014;32(1):66-72.
[Crossref] [Google Scholar] [PubMed]
- Lin JY, Fisher DE. Melanocyte biology and skin pigmentation. Nature 2007;445(7130):843-50.
[Crossref] [Google Scholar] [PubMed]
- Yamaguchi Y, Brenner M, Hearing VJ. The regulation of skin pigmentation. J Biol Chem 2007;282(38):27557-61.
[Crossref] [Google Scholar] [PubMed]
- Park HY, Kosmadaki M, Yaar M, Gilchrest BA. Cellular mechanisms regulating human melanogenesis. Cell Mol Life Sci 2009;66(9):1493-506.
[Crossref] [Google Scholar] [PubMed]
- Vance KW, Goding CR. The transcription network regulating melanocyte development and melanoma. Pigment Cell Res 2004;17(4):318-25.
[Crossref] [Google Scholar] [PubMed]
- Wu D, Pan W. GSK3: A multifaceted kinase in Wnt signaling. Trends Biochem Sci 2010;35(3):161-8.
[Crossref] [Google Scholar] [PubMed]
- Dunn KJ, Brady M, Ochsenbauer-Jambor C, Snyder S, Incao A, Pavan WJ. WNT1 and WNT3a promote expansion of melanocytes through distinct modes of action. Pigment Cell Res 2005;18(3):167-80.
[Crossref] [Google Scholar] [PubMed]
- Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): Regulation, actions and diseases. Pharmacol Ther 2015;148:114-31.
[Crossref] [Google Scholar] [PubMed]
- Bellei B, Pitisci A, Izzo E, Picardo M. Inhibition of melanogenesis by the pyridinyl imidazole class of compounds: Possible involvement of the Wnt/ß-catenin signaling pathway. PLoS One 2012;7(3):e33021.
[Crossref] [Google Scholar] [PubMed]
- Halaban R. The regulation of normal melanocyte proliferation. Pigment Cell Res 2000;13(1):4-14.
[Crossref] [Google Scholar] [PubMed]
- D’Mello SA, Finlay GJ, Baguley BC, Askarian-Amiri ME. Signaling pathways in melanogenesis. Int J Mol Sci 2016;17(7):1144.
[Crossref] [Google Scholar] [PubMed]
- Chang TS. Natural melanogenesis inhibitors acting through the down-regulation of tyrosinase activity. Materials 2012;5(9):1661-85.
- Rendon MI, Gaviria JI. Review of skin-lightening agents. Dermatol Surg 2005;31(7):886-90.
[Crossref] [Google Scholar] [PubMed]
- Draelos ZD. Skin lightening preparations and the hydroquinone controversy. Dermatol Ther 2007;20(5):308-13.
[Crossref] [Google Scholar] [PubMed]
- Abella ML. Evaluation of anti-wrinkle efficacy of adenosine-containing products using the FOITS technique. Int J Cosmetic Sci 2006;28(6):447-51.
[Crossref] [Google Scholar] [PubMed]
- Mitchell J, Lazarenko G. Wide QRS complex tachycardia. CJEM 2008;10(6):572-8.
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7(1):42717.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. iLOGP: A simple, robust and efficient description of n-octanol/water partition coefficient for drug design using the GB/SA approach. J Chem Inf Model 2014;54(12):3284-301.
[Crossref] [Google Scholar] [PubMed]
- Yang H, Lou C, Sun L, Li J, Cai Y, Wang Z, et al. admetSAR 2.0: Web-service for prediction and optimization of chemical ADMET properties. Bioinformatics 2019;35(6):1067-9.
[Crossref] [Google Scholar] [PubMed]
- Molinspiration Cheminformatics. Nova Ulica, SK-900 26 Slovensky Grob, Slovak Republic.
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: Servers for rigid and symmetric docking. Nucleic Acids Res 2005;33(2):W363-7.
[Crossref] [Google Scholar] [PubMed]
- Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: A web server for fast interaction refinement in molecular docking. Nucleic Acids Res 2008;36(2):W229-32.
[Crossref] [Google Scholar] [PubMed]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera-A visualization system for exploratory research and analysis. J Comput Chem 2004;25(13):1605-12.
[Crossref] [Google Scholar] [PubMed]
- Singh RK, Ranjan A, Tripathi R, Verma SS, Sharma V, Singh M, et al. Semecarpus anacardium Linn. leaf extract exhibits activities against breast cancer and prolongs the survival of tumor bearing mice. BioRxiv 2020.
- Sejin YO, Park N, Baek S, Shin S, Lee J. External use skin composition, containing Lentinula edodes-derived ergosterol. United States patent, US 9901525: Amorepacific Corporation; 2018.
- Valverde ME, Hernández-Pérez T, Paredes-López O. Edible mushrooms: Improving human health and promoting quality life. Int J Microbiol 2015;2015.
[Crossref] [Google Scholar] [PubMed]