- *Corresponding Author:
- Tao Yao
Department of Neurology, Renmin Hospital of Wuhan University, Wuchang, Wuhan, Hubei 430060, China
E-mail: yaotao@whu.edu.cn
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “133-143” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Circular RNA is crucial for the development of multiple malignant tumors including colorectal cancer. Circular RNA lamin B1 has recently been verified to accelerate colorectal cancer progression. However, it is not clear whether circular RNA lamin B1 can be regulated by the main methylase (methyltransferaselike 3) in colorectal cancer. This study was to explore the mechanism of methyltransferase-like 3-mediated epigenetic reactivation of circular RNA lamin B1 in colorectal cancer. We first analyzed the expression and correlation between methyltransferase-like 3 and circular RNA lamin B1 in colorectal cancer tissues, and then assessed the effects of methyltransferase-like 3 and circular RNA lamin B1 on the malignant properties of colorectal cancer cells by conducting loss-of-function and restoration studies. We also examined the growth and pathological structures in colorectal cancer xenograft tumors, as well as the levels of methyltransferase-like 3, circular RNA lamin B1 and caspase 3 expression in colorectal cancer xenograft tumors by reverse transcription-quantitative polymerase chain reaction, western blotting, hematoxylin and eosin staining and immunohistochemistry. Our results showed that the levels of circular RNA lamin B1 and methyltransferase-like 3 expressions were elevated and positively correlated in colorectal cancer tissues. Functionally, methyltransferase-like 3 silencing could prevent colorectal cancer cell proliferation, migration and invasion, and facilitate apoptosis in colorectal cancer cells by downregulating circular RNA lamin B1. Moreover, we found that knockdown of methyltransferase-like 3 could prevent tumor growth and disrupt the histological structure of colorectal cancer xenograft tumors by reducing circular RNA lamin B1 expression. Our data confirmed that methyltransferase-like 3 knockdown could attenuate the development of colorectal cancer cells by lowering circular RNA lamin B1 expression. This suggests circular RNA lamin B1 as a novel oncogene in colorectal cancer, which could be regulated by methyltransferaselike 3. Methyltransferase-like 3 silencing and circular RNA lamin B1 might be of great significance in colorectal cancer therapy.
Keywords
Colorectal cancer, circular RNA lamin B1, methyltransferase-like 3, apoptosis
Colorectal Cancer (CRC) is a common malignant tumor[1]. According to statistics, CRC causes nearly 700 000 deaths each year and the yearly number is increasing[2]. CRC is a modern disease with its highest incidence in developed countries. The current treatments for CRC still consist of surgery, supplemented by radiotherapy and chemotherapy[3]. Due to its insidious early symptoms, the vast majority of CRC patients are initially diagnosed at an advanced stage of their disease, where the surgery can longer provide a cure[4], the effects of chemotherapy is minimal and the overall prognosis is poor. Statistics show that the 5 y survival rate of patients with stage ? CRC is only 13.1 %[5]. CRC is caused by multiple factors, including intestinal inflammation, colorectal adenomas and gut microbiota[6]. The pathogenesis of CRC involves multiple mechanisms including excessive cell proliferation, increased angiogenesis, the acquisition of aggressive cell phenotypes, a decrease in cell apoptosis and the maintenance of CRC stem cells[7,8]. CRC progression typically involves the upregulation of oncogenes and downregulation of cancer suppressor genes, and similarly to other solid tumors, CRC is characterized by dysregulation in both oncogenes and tumor suppressor genes[9]. Therefore, it is imperative to search for novel molecular targets for treating CRC.
Circular Ribonucleic Acid (RNA) (circRNA) is a new type of RNA that is commonly generated from pre-messenger RNA (mRNA) molecules by variable shearing and has a closed circular structure[10]. CircRNA is highly conserved and is abundant and stable in mammals. As endogenous RNA molecules, circRNAs are mainly distributed in the cytoplasm[11]. Growing evidence indicates that circRNA molecules can sponge micro RNAs (miRNAs) to prevent the inhibitory effect of miRNA on mRNA[12,13]. Studies have shown that circRNA plays a key role in cellular functions, including protein synthesis, gene expression and post-transcriptional modification[14,15]. The study of circRNA has become a hot area of research in the field of oncology. CircRNAs contribute to tumorigenesis and the development of several cancers, including cervical cancer[16], lung cancer[17], breast cancer[18] and CRC[19], and could possibly serve as a latent prognostic markers and have significant clinical value in CRC. Several recent studies have examined the circRNA expression profiles in CRC tissues by the use of circRNA microarray methods[20,21]. Based on a literature review, we speculated that circular RNA Lamin B1 (circLMNB1) might be a latent biomarker for CRC[22]. Furthermore, research studies have confirmed that circLMNB1 contributes to CRC progression[23]. A review of a circRNA database revealed that has_circ_0127801 (circLMNB1) is located on chromosome 5 (chr5):126153227-126153886 with a 659 base pair (bp) spliced length. Therefore, we speculated that circLMNB1 might be a critical molecule involved in CRC progression. However, the mechanism of circLMNB1 in CRC has not been elucidated in previous studies.
To explore the function of circLMNB1 on malignant biological properties on CRCs and to confirm the regulatory effect of Methyltransferase-Like 3 (METTL3) on circLMNB1 expression, here in this study, we confirmed the changes in expression and the relevant functions of circLMNB1 in CRC cells and the xenograft tumors of CRC model mice. We also examined whether the influence of circLMNB1 on CRC progression could be regulated by N6-methyladenosine (m6A) modification, which can play an indispensable role in the function of circRNA[24,25].
Materials and Methods
Patients and samples:
Samples of CRC tissue were obtained from 20 CRC patients (age range, 42 to 78 y) who underwent surgery at the Renmin Hospital of Wuhan University from April 2019 to May 2020. Tissues at the tumor margins (3-5 cm) were also collected to serve as non-cancerous tissues. The patient inclusion criteria were as follows-A verified case of CRC; no other type of colorectal disease; no other tumors; no preoperative history of chemoradiotherapy. The exclusion criteria were pathological diagnosis excluded CRC; complication with other tumors; preoperative radiotherapy and chemotherapy. The study protocol was approved by the Ethics Committee of Renmin Hospital of Wuhan University and all patients provided their signed informed consent for study participation.
Cell culture:
Human Colon Adenocarcinoma cell line, LoVo (CCL-229) and Human Colorectal Carcinoma cell line (HCT116 (CCL-247)) cells were obtained from the American Type Culture Collection (ATCC) (Manassas, Virginia, United States of America (USA)). The HCT116 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Gibco, Waltham, Massachusetts (MA), USA) and the LoVo cells were cultured in Ham’s F-12K (Kaighn's) medium (Invitrogen, Waltham, MA. USA). Both culture media were supplemented with 10 % Fetal Bovine Serum (FBS) (Sigma, St. Louis, Missouri (MO), USA) and 1 % Penicillin-Streptomycin (Solarbio, Beijing, China), and both cell lines were grown at 37° in a 5 % Carbon dioxide (CO2) atmosphere.
Cell transfection:
An Empty Vector (EV), circLMNB1 overexpression plasmid, METTL3 short hairpin RNA (shRNA) (sh METTL3) and Negative Control (NC) shRNA were obtained from HanBio Biotechnology (HanBio, Shanghai, China). METTL3 small interfering (siRNA) (si-METTL3) and NC siRNA were obtained from GenePharma (Shanghai, China). LoVo and HCT116 cells were transfected with the plasmids (EV or circLMNB1 overexpression plasmid) or oligonucleotides (NC siRNA or si-METTL3) by using LipofectamineTM 3000 (Invitrogen) for 48 h. METTL3-silencing or/and circLMNB1- overexpressing HCT116 cells were stably constructed by lentivirus infection.
Quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR):
TRIzol reagent (Invitrogen) was used to extract the total RNA from processed CRC cells or specimens of ground tumor tissue. Next, the total RNA was reverse transcribed into Complementary Deoxyribonucleic Acid (cDNA) using a PrimeScriptTM RT reagent kit (Takara, China). PCR amplification was performed using SYBR Green qPCR master mix (DBI Bioscience) and relative levels of gene expression were calculated using the 2-ΔΔCT method[23].
Western blotting:
Processed CRC cells or samples of ground tumor tissue were lysed in Radioimmunoprecipitation Assay (RIPA) buffer (Beyotime, China) containing protease inhibitors and the amount of total protein in each lysate was quantitated. The extracted proteins were then denatured at 100° and separated by Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE). Next, the separated protein bands were transferred onto the membranes that were subsequently blocked with 5 % skim milk for 2 h.
The membranes were then incubated with anti- METTL3 (Abcam, Cambridge, United Kingdom (UK)) or anti-Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) (Abcam) antibodies overnight at 4°, followed by incubation with a secondary antibody (Abcam) for 1 h. The immunostained protein bands were detected with an Enhanced Chemiluminescence (ECL) kit (Thermo Fisher Scientific, Waltham, MA, USA). GAPDH was set as the internal reference. Western blot assay method was performed as described by Liang et al.[26].
Transwell assay:
Cell migration and invasion assay was performed as described by Cui et al.[27]. Briefly, for migration experiments, processed CRC cells were adjusted to a density of 5×105/ml; after which, 200 μl of suspended cells were added to the upper compartment of a transwell chamber, while 600 μl of growth medium supplemented with 10 % FBS was added to the lower compartment. After 48 h of culture, the cells that were penetrated into the lower compartment were fixed with 4 % formaldehyde, stained with 0.1 % crystal violet (Sigma) and counted under a light microscope. For invasion experiments, Matrigel was diluted in the medium at a ratio of 1:4 in advance. Next, the Matrigel (40 μl) was added to the transwell chamber and let sit at 37° for 2 h. The other steps were the same as those used for the cell migration experiments.
Cell Counting Kit-8 (CCK-8) assay:
Treated CRC cells were harvested and inoculated into 96-well plates (100 μl/well) and the number of cells in each well was adjusted to 3000. Next, CCK- 8 solution (10 μl, Dojindo, Tokyo, Japan) was added to each well and incubated for 0, 24, 48 and 72 h, respectively, in a 37° incubator. After an additional 1 h of incubation, the absorption of each well at 450 nm was read with a microplate reader (Bio-Tek Epoch, Santa Clara, California, USA).
Flow cytometry:
An Annexin V-Fluorescein Isothiocyanate (FITC)/ Propidium Iodide (PI) apoptosis kit (Best Bio, Shanghai, China) was used to confirm changes in apoptosis, as described by Li et al.[28]. Processed cells were adjusted to a density of 1×106 cells/ml and then treated with Annexin V-FITC (10 μl) and PI (5 μl) for 15 min in the dark. The cell apoptosis rates were then confirmed by flow cytometry.
Enzyme-Linked Immunosorbent Assay (ELISA):
A caspase-3 ELISA kit (Abcam, ab285337) was used to test the caspase 3 activity according to instructions provided by the manufacturer.
Tumor xenograft model:
A tumor xenograft model was used to evaluate the effect of METTL3/circLMNB1 on tumor formation, as described by Liang et al.[29]. Male Bagg and Albino (BALB)/c nude mice (Specific Pathogen Free (SPF) grade; n=32; age=4 w, weight range=20±2 g) were purchased from the Shanghai Slack Laboratory and reared in separate cages in a barrier system (temperature of 22°-25°, humidity of 45 %-55 % and a 12 h light/dark cycle). The mice were fed SPF grade feed and autoclaved purified water on a daily basis. 1 w of adaptive feeding was performed before the mice were used in any experiments. The 1 ml suspension containing 1×106 HCT116 cells was subcutaneously inoculated into the right anterior axillary fold of each mouse. The length of each tumor was determined with Vernier calipers once every 7 d and a tumor growth curve were plotted. At the end of the study, the mice were sacrificed by cervical dislocation and the tumors were removed and fixed in 10 % formaldehyde for 24 h. After routine dehydration and paraffin embedding, the tumors were cut into 4 μm thick sections for subsequent staining and microscopic investigation.
Hematoxylin and Eosin (H&E) staining:
H&E staining was performed as previously described in the literature[30,31]. Slices of tissue were first baked for 2 h at 60° and then treated with hematoxylin for 5 min. Next, the slices were treated with hydrochloric acid-ethanol solution, stained with eosin for 3 min, dehydrated with gradient alcohol, made transparent with xylene and fixed with neutral glue. Pathological changes were confirmed by light microscopy.
Immunohistochemistry (IHC) assay:
The tissue slices in each group was first underwent routine dewaxing. After cleaning, the slices were repaired with sodium citrate (pH=6.0) at high temperature and high pressure for 8 min, allowed to cool and then immersed in 3 % Hydrogen peroxide (H2O2) for 30 min. The slices were then sealed with 10 % Bovine Serum Albumin (BSA) and incubated with anti-caspase 3 (Abcam) overnight at 4°. On the next day, the slices were further incubated with the primary antibody against caspase 3 for 1 h and subsequently incubated with a secondary antibody (Abcam) for an additional 1 h. After cleaning, the slices were subjected to multiple procedures, including 3, 3’-Diazminobenzidine (DAB) staining for 10 s, water flushing, hematoxylin staining for 30 s, routine dehydration, neutral gum sealing and natural drying. Finally, caspase 3 expression was detected using a light microscope and assessed by two pathologists.
Statistical analysis:
All data were analyzed using International Business Machines (IBM) Statistical Package for Social Sciences (SPSS) statistics for Windows, Version 21 software (IBM Corp., Armonk, New York, USA) and results are presented as a mean value±Standard Deviation (SD). GraphPad Prism 8.0 software was used for mapping. Statistical methods consisted of the paired Student’s t test and one-way Analysis of Variance (ANOVA). A p-value<0.05 was considered to be statistically significant.
Results and Discussion
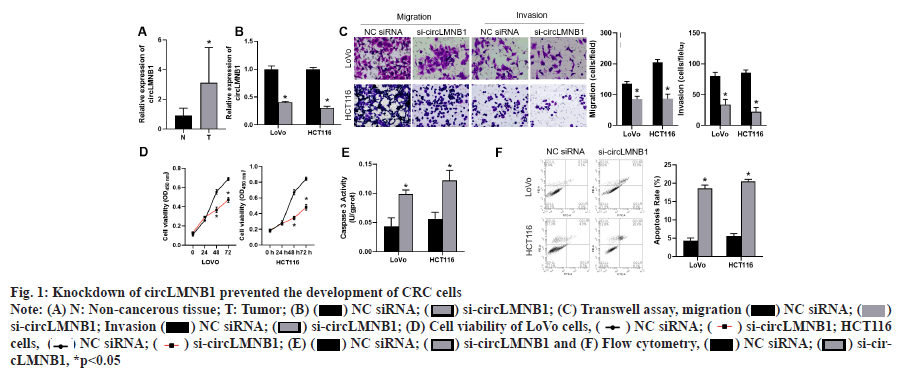
Knockdown of circLMNB1 prevented the proliferation, migration and invasion of CRC cells and accelerated apoptosis as shown in fig. 1. We first investigated the role of circLMNB1 in the pathogenesis of CRC. qRT-PCR was used to monitor circLMNB1 expression in CRC tissues and non-cancerous tissues (n=20). Results of qRTPCR studies showed that circLMNB1 expression was significantly increased in CRC tissues when compared to non-cancerous tissues (fig. 1A). A further analysis revealed that a high level of circLMNB1 expression was significantly correlated with a higher Tumor, Nodes and Metastases (TNM) stage (p=0.0011). However, there was no significant correlation between circLMNB1 expression and gender, age, tumor size, or distal metastasis (Table 1).
Fig. 1:Knockdown of circLMNB1 prevented the development of CRC cells
Note: (A) N: Non-cancerous tissue; T: Tumor; (B) ( ) NC siRNA; (
) NC siRNA; ( ) si-circLMNB1; (C) Transwell assay, migration (
) si-circLMNB1; (C) Transwell assay, migration ( ) NC siRNA; (
) NC siRNA; ( ) si-circLMNB1; Invasion (
) si-circLMNB1; Invasion ( ) NC siRNA; (
) NC siRNA; ( ) si-circLMNB1; (D) Cell viability of LoVo cells, (
) si-circLMNB1; (D) Cell viability of LoVo cells, ( ) NC siRNA; (
) NC siRNA; ( ) si-circLMNB1; HCT116 cells, (
) si-circLMNB1; HCT116 cells, ( ) NC siRNA; (
) NC siRNA; ( ) si-circLMNB1; (E) (
) si-circLMNB1; (E) ( ) NC siRNA; (
) NC siRNA; ( ) si-circLMNB1 and (F) Flow cytometry,
) si-circLMNB1 and (F) Flow cytometry,  ) NC siRNA; (
) NC siRNA; ( ) si-circLMNB1, *p<0.05
) si-circLMNB1, *p<0.05
| Features | Parameters | Number | Low | High | p-value |
|---|---|---|---|---|---|
| Gender | Female | 10 | 6 | 4 | NS |
| Male | 10 | 4 | 6 | ||
| Age | <60 | 15 | 9 | 6 | 0.3034 |
| =60 | 5 | 1 | 4 | ||
| Tumor size | <4.5 cm | 12 | 9 | 3 | 0.0988 |
| =4.5 cm | 8 | 1 | 7 | ||
| Distal metastasis | Absent | 9 | 7 | 2 | 0.0698 |
| Present | 11 | 3 | 8 | ||
| TNM stage | I+II | 10 | 9 | 1 | 0.0011 |
| III+IV | 10 | 1 | 9 |
Note: NS: Not Significant
Table 1: Correlation Between circLMNB1 Expression Levels and the Clinicopathological parameters of CRC
Next, circLMNB1 siRNA was used to knock down circLMNB1 expression in LoVo and HCT116 cells, and the efficiency of knockdown was verified by qRT-PCR. As shown in fig. 1B, the siRNA significantly suppressed circLMNB1 expression in both cell lines (fig. 1B). The migration and invasion capabilities of LoVo and HCT116 cells transfected with si-cirlLMNB1 were examined by transwell assays. Transwell assays showed that silencing of circLMNB1 could significantly reduce the migration and invasion capabilities of CRC cells (fig. 1C). CCK8 assays were performed to examine the viability of circLMNB1 siRNA-treated CRC cells. Subsequently, CCK-8 assay results showed that cell viability was also reduced in the circLMNB1 siRNA group when compared with the NC siRNA group (fig. 1D, while an ELISA assay for caspase 3 activity was performed where the caspase 3 activity was increased in the circLMNB1 siRNA group (fig. 1E). Cell apoptosis was measured by flow cytometry. Flow cytometry results indicated that circLMNB1 knockdown could markedly increase the apoptosis rate of CRC cells (fig. 1F). These findings revealed that circLMNB1 was upregulated in CRC tissues and circLMNB1 knockdown could prevent the malignant processes of CRC cells.
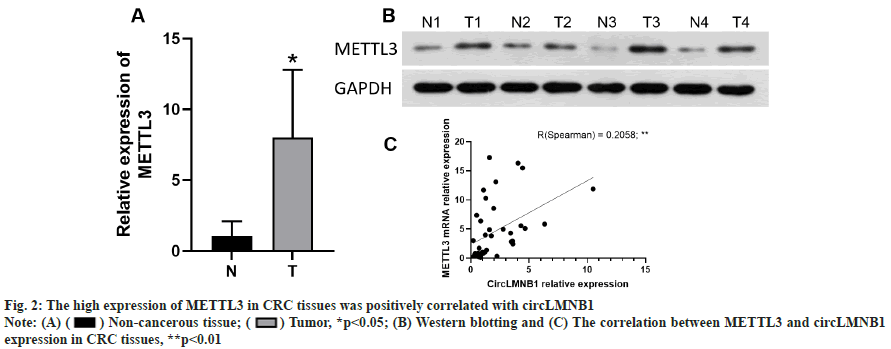
The high expression of METTL3 in CRC tissues was positively correlated with circLMNB1. METTL3 expression in clinical tissues was measured by qRTPCR. We found that METTL3 mRNA expression was notably upregulated in the CRC tissues when compared to the non-cancerous tissues (fig. 2A). Furthermore, Western blotting was performed to detect METTL3 expression in CRC tissues. Western blot assays verified that the METTL3 protein was more highly expressed in the CRC tissues than in the non-cancerous tissues (fig. 2B). A further analysis revealed that circLMNB1 expression was positively correlated with METTL3 expression in CRC tissues (fig. 2C, R2=0.2058, p<0.01). In summary, our data showed that METTL3 expression was related to circLMNB1 in CRC cells and METTL3 might participate in the CRC development process by regulating circLMNB1.
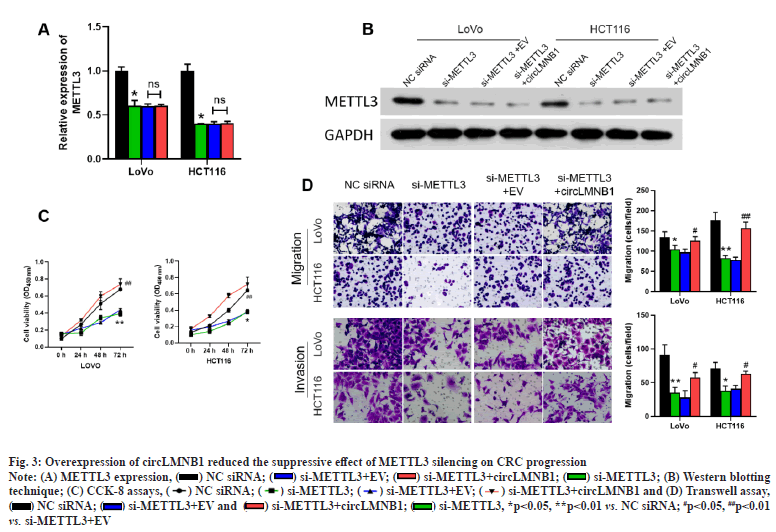
Overexpression of circLMNB1 attenuated the suppressive effect of METTL3 silencing on CRC progression. Based on the positive correlation between METTL3 and circLMNB1, we decided to further verify the effects of METTL3 and circLMNB1 on the viability and metastasis of CRC cells by conducting rescue experiments with METTL3 siRNA and the circLMNB1 overexpression plasmid. METTL3 siRNA or/and the circLMNB1 overexpression plasmid was transfected into CRC cells. qRT-PCR results showed that METTL3 siRNA suppressed METTL3 expression, but had no effect on circLMNB1 expression (fig. 3A). METTL3 expression was measured by Western blotting. The trend in METTL3 expression shown in Western blot studies was basically consistent with the trend shown by qRT-PCR results (fig. 3B). CCK-8 assays were performed to assess the viability of CRC cells. Functionally, CCK-8 assays showed that CRC cell viability could be significantly reduced by METTL3 knockdown and the decline in cell viability could be reversed by circLMNB1 overexpression (fig. 3C). Meanwhile, transwell data revealed that METTL3 knockdown contributed to significant reductions in the migration and invasion capabilities of CRC cells, and those reductions caused by METTL3 siRNA could also be weakened by circLMNB1 overexpression (fig. 3D). Overall, these findings showed that METTL3 knockdown could reduce the viability, migration and invasion of CRC cells by downregulating circLMNB1.
Fig. 3:Overexpression of circLMNB1 reduced the suppressive effect of METTL3 silencing on CRC progression
Note: (A) METTL3 expression, (![]() ) NC siRNA; (
) NC siRNA; ( ) si-METTL3+EV; (
) si-METTL3+EV; ( ) si-METTL3+circLMNB1; (
) si-METTL3+circLMNB1; ( ) si-METTL3; (B) Western blotting technique; (C) CCK-8 assays, (
) si-METTL3; (B) Western blotting technique; (C) CCK-8 assays, ( ) NC siRNA; (
) NC siRNA; ( ) si-METTL3; (
) si-METTL3; ( ) si-METTL3+EV; (
) si-METTL3+EV; ( ) si-METTL3+circLMNB1 and (D) Transwell assay, (
) si-METTL3+circLMNB1 and (D) Transwell assay, ( ) NC siRNA; (
) NC siRNA; ( ) si-METTL3+EV and (
) si-METTL3+EV and ( ) si-METTL3+circLMNB1; (
) si-METTL3+circLMNB1; ( ) si-METTL3, *p<0.05, **p<0.01 vs. NC siRNA; #p<0.05, ##p<0.01 vs. si-METTL3+EV
) si-METTL3, *p<0.05, **p<0.01 vs. NC siRNA; #p<0.05, ##p<0.01 vs. si-METTL3+EV
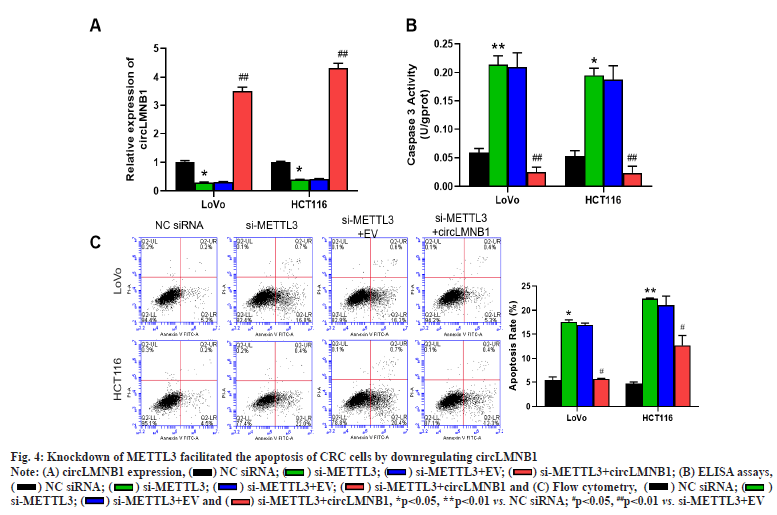
Knockdown of METTL3 facilitated the apoptosis of CRC cells by downregulating circLMNB1. Changes in circLMNB1 expression in CRC cells transfected with si-METTL3 or/and the circLMNB1 overexpression plasmid were detected by RT-qPCR. qRT-PCR results showed that knockdown of METTL3 dramatically downregulated circLMNB1 expression, which also could be attenuated by circLMNB1 overexpression in CRC cells (fig. 4A). ELISA assays were performed to confirm the change in caspase 3 activity in METTL3-silencing or/and circLMNB1- overexpressing CRC cells. Furthermore, METTL3 silencing markedly increased caspase 3 activities in CRC cells and those increases in caspase 3 activity could be reversed by circLMNB1 overexpression (fig. 4B). Flow cytometry was used to monitor cell apoptosis in the transfected CRC cells. Additionally, we found that knockdown of METTL3 significantly increased the apoptosis rate of CRC cells, while circLMNB1 overexpression could attenuate the increase in cell apoptosis mediated by METTL3 knockdown (fig. 4C). Generally, we found that METTL3 knockdown could increase the apoptosis of CRC cells through its effect on circLMNB1.
Fig. 4:Knockdown of METTL3 facilitated the apoptosis of CRC cells by downregulating circLMNB1
Note: (A) circLMNB1 expression, ( ) NC siRNA; (
) NC siRNA; ( ) si-METTL3; (
) si-METTL3; ( ) si-METTL3+EV; (
) si-METTL3+EV; ( ) si-METTL3+circLMNB1; (B) ELISA assays, (
) si-METTL3+circLMNB1; (B) ELISA assays, ( ) NC siRNA; (
) NC siRNA; ( ) si-METTL3; (
) si-METTL3; ( ) si-METTL3+EV; (
) si-METTL3+EV; ( ) si-METTL3+circLMNB1 and (C) Flow cytometry, (
) si-METTL3+circLMNB1 and (C) Flow cytometry, ( ) NC siRNA; (
) NC siRNA; ( ) si-METTL3; (
) si-METTL3; ( ) si-METTL3+EV and (
) si-METTL3+EV and ( ) si-METTL3+circLMNB1, *p<0.05, **p<0.01 vs. NC siRNA; #p<0.05, ##p<0.01 vs. si-METTL3+EV
) si-METTL3+circLMNB1, *p<0.05, **p<0.01 vs. NC siRNA; #p<0.05, ##p<0.01 vs. si-METTL3+EV
CircLMNB1 overexpression enhanced the growth and improved the histopathological structure of METTL3-silenced CRC xenograft tumors. Next, we further examined whether METTL3 siRNA could suppress the growth of the CRC xenograft tumors and improve their pathological structure by effecting circLMNB1. LoVo cells were transfected with METTL3 shRNA or/and circLMNB1 overexpression plasmids and then injected into nude mice. As shown in fig. 5A, the sizes and volumes of the tumors were significantly reduced in the sh-METTL3 group relative to those in the NC shRNA group and those reductions in tumor size and volume could be reversed by circLMNB1 overexpression (fig. 5A and fig. 5B). Additionally, METTL3 knockdown can also notably downregulate METTL3 expression in the CRC xenograft tumors. The nude mice were sacrificed, their bodies were examined, the tumors were removed and tumor volume growth curves were plotted. However, overexpression of circLMNB1 did not recover the loss of METTL3 expression (fig. 5B and fig. 5C). METTL3 and circLMNB1 expression was detected by qRT-PCR and Western blotting. METTL3 knockdown significantly downregulated circLMNB1 expression in the CRC xenograft tumors and the downregulation mediated by METTL3 silencing could be reversed by circLMNB1 overexpression (fig. 5D and fig. 5E). H&E staining was used to analyze the pathological structure of tumors and caspase 3 expression was detected by IHC. When examined by H&E staining, tissues in the sh-METTL3 group displayed signs of cell necrosis and nucleus fixation, while tissue necrosis in the sh-METTL3 group was somewhat ameliorated when circLMNB1 was overexpressed (fig. 5F, upper line). IHC results revealed that METTL3 knockdown significantly increased caspase 3 expressions in the CRC xenograft tumors and those increases could be markedly attenuated by circLMNB1 overexpression (fig. 5F, lower line). These findings verified that METTL3 knockdown could significantly suppress tumor growth and destroy the pathological structure of CRC xenograft tumors by effecting circLMNB1.
Fig. 5:CircLMNB1 overexpression enhanced the growth and improved the histopathological structure of METTL3-silenced CRC xenograft tumors
Note: (A-C) ( ) NC shRNA; (
) NC shRNA; ( ) sh-METTL3; (
) sh-METTL3; ( ) sh-METTL3+EV; (
) sh-METTL3+EV; ( ) sh-METTL3+circLMNB1; (D) (
) sh-METTL3+circLMNB1; (D) ( ) NC shRNA; (
) NC shRNA; ( ) sh-METTL3; (
) sh-METTL3; ( ) sh-METTL3+EV; (
) sh-METTL3+EV; ( ) sh-METTL3+circLMNB1; (E) Western blotting and (F) H&E staining and IHC, *p<0.05, **p<0.01 vs. NC shRNA; ##p<0.01 vs. sh METTL3+EV; NS: Not Significant
) sh-METTL3+circLMNB1; (E) Western blotting and (F) H&E staining and IHC, *p<0.05, **p<0.01 vs. NC shRNA; ##p<0.01 vs. sh METTL3+EV; NS: Not Significant
In 2016, the American Cancer Society reported an increase in the number of new cases and deaths resulting from CRC[32]. The clinical manifestations of CRC are shaped by interactions at multiple levels[33]. However, its specific pathogenesis is not fully understood. CircRNA has gradually become a key RNA studied in malignant cancers due to its closed loop structure, high stability, strong specificity and ease of detection[34]. Increasing evidence suggests that circRNA has the characteristics of tissue-specific expression and plays a regulatory role in malignant tumors[35-37]. Therefore, circRNA is expected to be the most promising molecular marker for use in tumor diagnosis, assessment and prognosis[38]. Similar to other malignant tumors, CRC development requires a battery of complex biological processes that involve both coding and non-coding genes. In recent years, numerous circRNAs such as circular RNA Homeodomain-Interacting Protein Kinase 3 (circHIPK3)[39], circular RNA Erbb2 Interacting Protein (circERBIN)[40], circRNA_0084927[41], circSMARCA5[42] and circRNA_0000392[43]. have been found to play vital roles in CRC progression. The above research suggests that specific circRNAs are essential for the malignant progression of CRC. Our research revealed that circLMNB1 was notably downregulated in CRC tissues and knockdown of circLMNB1 could block the progression of CRC, including CRC cell proliferation, migration, invasion and apoptosis.
The interaction between circRNA and miRNA through sponge adsorption is the main direction of current related research. For example, in lung cancer, circ_101237 interacts with miR-490-3p to affect the expression of Mitogen-Activated Protein Kinase 1 (MAPK1), thereby promoting the malignant biological behavior of lung cancer cells and the clinical progression of lung cancer[44]. Similarly, Xu et al. found that circ_0000392 interacts with miR-193a-5p to promote the Phosphoinositide-3- Kinase (PI3K)/Protein Kinase B (AKT) pathway and ultimately promote tumor development in CRC[43]. In addition, circRNA can also be encapsulated in exosomes to achieve intercellular communication. Shang et al. found that exosomal circPACRGL has the ability to interact with both miR-142-3p and miR- 506-3p in CRC, promoting Transforming Growth Factor-beta (TGF-β1) expression, and promoting tumor metastasis ultimately[45]. In our previous study, we found that circLMNB1 can directly interact with miR-143 and participate in the regulation of malignant behavior of CRC cells[46]. However, the mechanism of circLMNB1 in depth remains to be further explored.
The field of genetics-epigenetics is developing very rapidly. Epigenetics mainly refers to changes in gene expression that result from gene modification rather than changes in the nucleotide sequence of genes[47,48]. The RNA modification can extensively alter the structure, function and stability of an RNA molecule[49]. The m6A modification mainly results from the action of a methyltransferase, which is removed by demethylase and is recognized by a binding protein[50]. The m6A modification mainly affects immune tolerance, mRNA stability, RNA export and mRNA splicing[51,52]. The m6A modification has been reported to play a significant role in multiple cancers, such as bladder cancer[53], prostate cancer[54], gastric cancer[55], CRC[56] and even hematological malignancies[57]. Studies have also proved that METTL3, as the most significant methylase, can malfunction and be carcinogenic in numerous malignant tumors. For instance, METTL3 was shown to accelerate the proliferation of bladder cancer[53], be conducive to the metastasis of gastric cancer[58] and enhance the tumorigenesis of cervical cancer[59]. Overall, METTL3 plays a critical role in cancer progression.
Prior to 2017, studies on m6A modification were limited to the mRNA level. It was not until March 2017 that the first circRNA m6A modification was published, thus filling in the previous information gap[25]. Subsequent studies proved that m6A modification can affect the nuclear localization of circRNA and that circRNA can also bind to the corresponding regulatory proteins of m6A to affect its stability[60]. The two can regulate and permeate each other[24]. Recent studies have shown that m6Amodified circRNA-Sorafenib (circ-SORE) can maintain the sorafenib resistance of hepatocellular carcinoma cells[61] and that m6A modification of circNSUN2 can induce the liver metastasis of CRC[62]. In our study, we found that METTL3 was highly expressed in CRC and positively correlated with circLMNB1. In addition, we also found that METTL3 silencing could prevent the proliferation and metastasis, and enhance the apoptosis of CRC cells, by downregulating circLMNB1. Our data also showed that METTL3 silencing could suppress the growth and destroy the pathological structure of CRC xenograft tumors by decreasing circLMNB1 expression.
Our current study demonstrated that circLMNB1, as an oncogene, might be involved in CRC progression. Additionally, we verified that circLMNB1 could be distinctly upregulated by METTL3 and METTL3 silencing could inhibit the malignant activities of CRC. These findings suggest METTL3-mediated circLMNB1 as a possible target for treating CRC.
Author’s contributions:
Chun-Ping He and Yuan-Jie Yu contributed equally to this work.
Funding:
This work was financially supported by the Hubei Province Key Laboratory Open Project [Grant number 2121KFY010].
Conflict of interests:
The authors declared no conflict of interest.
References
- Dekker E, Tanis PJ, Vleugels JL, Kasi PM, Wallace M. Colorectal cancer. Lancet 2019;394(10207):1467-80.
[Crossref] [Google scholar] [PubMed]
- Hadjipetrou A, Anyfantakis D, Galanakis CG, Kastanakis M, Kastanakis S. Colorectal cancer, screening and primary care: A mini literature review. World J Gastroenterol 2017;23(33):6049-58.
[Crossref] [Google scholar] [PubMed]
- Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA 2021;325(7):669-85.
[Crossref] [Google scholar] [PubMed]
- Jin M, Frankel WL. Lymph node metastasis in colorectal cancer. Surg Oncol Clin N Am 2018;27(2):401-12.
[Crossref] [Google scholar] [PubMed]
- Phipps AI, Limburg PJ, Baron JA, Burnett-Hartman AN, Weisenberger DJ, Laird PW, et al. Association between molecular subtypes of colorectal cancer and patient survival. Gastroenterology 2015;148(1):77-87.
[Crossref] [Google scholar] [PubMed]
- Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci 2017;18(1):197.
[Crossref] [Google scholar] [PubMed]
- Gao R, Gao Z, Huang L, Qin H. Gut microbiota and colorectal cancer. Eur J Clin Microbiol Infect Dis 2017;36(5):757-69.
[Crossref] [Google scholar] [PubMed]
- La Vecchia S, Sebastián C. Metabolic pathways regulating colorectal cancer initiation and progression. Semin Cell Dev Biol 2020;98:63-70.
[Crossref] [Google scholar] [PubMed]
- Balasubramaniam SD, Balakrishnan V, Oon CE, Kaur G. Key molecular events in cervical cancer development. Medicina 2019;55(7):384.
[Crossref] [Google scholar] [PubMed]
- Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: Metabolism, functions and interactions with proteins. Mol Cancer 2020;19(1):1-9.
[Crossref] [Google scholar] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LV, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20(11):675-91.
[Crossref] [Google scholar] [PubMed]
- Panni S, Lovering RC, Porras P, Orchard S. Non-coding RNA regulatory networks. Biochim Biophys Acta Gene Regul Mech 2020;1863(6):194417.
[Crossref] [Google scholar] [PubMed]
- Su Q, Lv X. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis. Genomics 2020;112(2):1680-5.
[Crossref] [Google scholar] [PubMed]
- Huang A, Zheng H, Wu Z, Chen M, Huang Y. Circular RNA-protein interactions: Functions, mechanisms and identification. Theranostics 2020;10(8):3503-17.
[Crossref] [Google scholar] [PubMed]
- Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: Biogenesis, function and clinical significance. Trends Cancer 2020;6(4):319-36.
[Crossref] [Google scholar] [PubMed]
- Chen RX, Liu HL, Yang LL, Kang FH, Xin LP, Huang LR, et al. Circular RNA circRNA_0000285 promotes cervical cancer development by regulating FUS. Eur Rev Med Pharmacol Sci 2019;23(20):8771-8.
[Crossref] [Google scholar] [PubMed]
- Wang C, Tan S, Li J, Liu WR, Peng Y, Li W. CircRNAs in lung cancer-Biogenesis, function and clinical implication. Cancer Lett 2020;492:106-15.
[Crossref] [Google scholar] [PubMed]
- Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis 2019;10(2):1-4.
[Crossref] [Google scholar] [PubMed]
- Zeng K, Wang S. Circular RNAs: The crucial regulatory molecules in colorectal cancer. Pathol Res Pract 2020;216(4):152861.
[Crossref] [Google scholar] [PubMed]
- Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL, Yang Y. RNA sequencing reveals the expression profiles of circRNA and indicates that circDDX17 acts as a tumor suppressor in colorectal cancer. J Exp Clin Cancer Res 2018;37(1):1-4.
[Crossref] [Google scholar] [PubMed]
- Xu H, Wang C, Song H, Xu Y, Ji G. RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer 2019;18(1):1-6.
[Crossref] [Google scholar] [PubMed]
- Zhang J, Cai A, Zhao Y. Three CircRNAs function as potential biomarkers for colorectal cancer. Clin Lab 2020;66(12).
[Crossref] [Google scholar] [PubMed]
- He C, Huang C, Zhou R, Yu H. CircLMNB1 promotes colorectal cancer by regulating cell proliferation, apoptosis and epithelial-mesenchymal transition. Onco Targets Ther 2019;12:6349-59.
[Crossref] [Google scholar] [PubMed]
- Chen YG, Chen R, Ahmad S, Verma R, Kasturi SP, Amaya L, et al. N6-methyladenosine modification controls circular RNA immunity. Mol Cell 2019;76(1):96-109.
[Crossref] [Google scholar] [PubMed]
- Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res 2017;27(5):626-41.
[Crossref] [Google scholar] [PubMed]
- Liang M, Huang G, Liu Z, Wang Q, Yu Z, Liu Z, et al. Elevated levels of hsa_circ_006100 in gastric cancer promote cell growth and metastasis viamiR?195/GPRC5A signalling. Cell Prolif 2019;52(5):e12661.
[Crossref] [Google scholar] [PubMed]
- Cui H, Wang Q, Lei Z, Feng M, Zhao Z, Wang Y, et al. DTL promotes cancer progression by PDCD4 ubiquitin-dependent degradation. J Exp Clin Cancer Res 2019;38(1):1-3.
[Crossref] [Google scholar] [PubMed]
- Li H, Yan W, Suo X, Peng H, Yang X, Li Z, et al. Nucleus-targeted nano delivery system eradicates cancer stem cells by combined thermotherapy and hypoxia-activated chemotherapy. Biomaterials 2019;200:1-4.
[Crossref] [Google scholar] [PubMed]
- Liang Y, Song X, Li Y, Chen B, Zhao W, Wang L, et al. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol Cancer 2020;19(1):1-20.
[Crossref] [Google scholar] [PubMed]
- Yoon SJ, Park J, Shin Y, Choi Y, Park SW, Kang SG, Son HY, Huh YM. Deconvolution of diffuse gastric cancer and the suppression of CD34 on the BALB/c nude mice model. BMC cancer 2020;20(1):1-10.
- Chaowawanit W, Campbell V, Wilson E, Chetty N, Perrin L, Jagasia N, et al. Retrospective review of sentinel lymph node mapping in endometrial cancer using indocyanine green and near infra-red fluorescence imaging during minimally invasive surgery. J Obstet Gynaecol 2021;42(4):694-702.
[Crossref] [Google scholar] [PubMed]
- Navarro M, Nicolas A, Ferrandez A, Lanas A. Colorectal cancer population screening programs worldwide in 2016: An update. World J Gastroenterol 2017;23(20):3632-42.
[Crossref] [Google scholar] [PubMed]
- Simon K. Colorectal cancer development and advances in screening. Clin Interv Aging 2016;11:967-76.
[Crossref] [Google scholar] [PubMed]
- Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: A novel type of biomarker for cancer. Breast Cancer 2018;25(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Sarraf JS, Puty TC, da Silva EM, Allen TS, Sarraf YS, de Carvalho LE, et al. Noncoding RNAs and colorectal cancer: A general overview. Microrna 2020;9(5):336-45.
[Crossref] [Google scholar] [PubMed]
- Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev 2018;48:121-7.
[Crossref] [Google scholar] [PubMed]
- Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, et al. CircRNAs in cancer metabolism: A review. J Hematol Oncol 2019;12(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer 2021;21(1):22-36.
[Crossref] [Google scholar] [PubMed]
- Zeng K, Chen X, Xu MU, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis 2018;9(4):1-5.
[Crossref] [Google scholar] [PubMed]
- Chen LY, Wang L, Ren YX, Pang Z, Liu Y, Sun XD, et al. The circular RNA circ-ERBIN promotes growth and metastasis of colorectal cancer by miR-125a-5p and miR-138-5p/4EBP-1 mediated cap-independent HIF-1α translation. Mol Cancer 2020;19(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Liu F, Xiao XL, Liu YJ, Xu RH, Zhou WJ, Xu HC, et al. CircRNA_0084927 promotes colorectal cancer progression by regulating miRNA-20b-3p/glutathione S-transferase mu 5 axis. World J Gastroenterol 2021;27(36):6064-78.
[Crossref] [Google scholar] [PubMed]
- Miao X, Xi Z, Zhang Y, Li Z, Huang L, Xin T, et al. Circ-SMARCA5 suppresses colorectal cancer progression viadownregulating miR-39-3p and upregulating ARID4B. Dig Liver Dis 2020;52(12):1494-502.
[Crossref] [Google scholar] [PubMed]
- Xu H, Liu Y, Cheng P, Wang C, Liu Y, Zhou W, et al. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res 2020;39(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Zhang ZY, Gao XH, Ma MY, Zhao CL, Zhang YL, Guo SS. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci Rep 2020;10(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer 2020;19(1):1-5.
[Crossref] [Google scholar] [PubMed]
- He CP, Huang C, Zhou R, Yu HG, Wu PB. Effects of circLMNB1 on proliferation, apoptosis and cell cycle of colorectal cancer cells. Shandong Med J 2021;61(20):45-8.
- Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol 2020;1253:3-55.
[Crossref] [Google scholar] [PubMed]
- Rotondo JC, Mazziotta C, Lanzillotti C, Tognon M, Martini F. Epigenetic dysregulations in merkel cell polyomavirus-driven merkel cell carcinoma. Int J Mol Sci 2021;22(21):11464.
[Crossref] [Google scholar] [PubMed]
- Jonkhout N, Tran J, Smith MA, Schonrock N, Mattick JS, Novoa EM. The RNA modification landscape in human disease. RNA 2017;23(12):1754-69.
[Crossref] [Google scholar] [PubMed]
- Liu ZX, Li LM, Sun HL, Liu SM. Link between m6A modification and cancers. Front Bioeng Biotechnol 2018;6:89.
[Crossref] [Google scholar] [PubMed]
- He L, Li H, Wu A, Peng Y, Shu G, Yin G. Functions of N6-methyladenosine and its role in cancer. Mol Cancer 2019;18(1):1-5.
[Crossref] [Google scholar] [PubMed]
- Sun T, Wu R, Ming L. The role of m6A RNA methylation in cancer. Biomed Pharmacother 2019;112:108613.
[Crossref] [Google scholar] [PubMed]
- Han J, Wang JZ, Yang X, Yu H, Zhou R, Lu HC, et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol Cancer 2019;18(1):1-5.
[Crossref] [Google scholar] [PubMed]
- Ji G, Huang C, He S, Gong Y, Song G, Li X, et al. Comprehensive analysis of m6A regulators prognostic value in prostate cancer. Aging 2020;12(14):14863-84.
[Crossref] [Google scholar] [PubMed]
- Zhang C, Zhang M, Ge S, Huang W, Lin X, Gao J, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K?AKT signaling in gastric cancer. Cancer Med 2019;8(10):4766-81.
[Crossref] [Google scholar] [PubMed]
- Meng Y, Li S, Gu D, Xu K, Du M, Zhu L, et al. Genetic variants in m6A modification genes are associated with colorectal cancer risk. Carcinogenesis 2020;41(1):8-17.
[Crossref] [Google scholar] [PubMed]
- Zhao Y, Peng H. The role of N6-methyladenosine (m6A) methylation modifications in hematological malignancies. Cancers 2022;14(2):332.
[Crossref] [Google scholar] [PubMed]
- Yue B, Song C, Yang L, Cui R, Cheng X, Zhang Z, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer 2019;18(1):1-5.
[Crossref] [Google scholar] [PubMed]
- Wang Q, Guo X, Li L, Gao Z, Su X, Ji M, et al. N6-methyladenosine METTL3 promotes cervical cancer tumorigenesis and Warburg effect through YTHDF1/HK2 modification. Cell Death Dis 2020;11(10):1-10.
[Crossref] [Google scholar] [PubMed]
- Dai F, Wu Y, Lu Y, An C, Zheng X, Dai L, et al. Crosstalk between RNA m6A modification and non-coding RNA contributes to cancer growth and progression. Mol Ther Nucleic Acids 2020;22:62-71.
[Crossref] [Google scholar] [PubMed]
- Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N6-methyladenosine-modified circRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer 2020;19(1):1-6.
[Crossref] [Google scholar] [PubMed]
- Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun 2019;10(1):1-5.
[Crossref] [Google scholar] [PubMed]