- *Corresponding Author:
- Ke Mei
Department of Radiology, Dahua Hospital, Xuhui District, Shanghai 200237, China
E-mail: lunasanger@126.com
| This article was originally published in a special issue, “Recent Developments in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(4) Spl Issue “290-297” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Cisplatin represents an important strategy for gastric cancer treatment. However, cisplatin resistance often leads to failure of therapy as well as local reoccurrence. Recently, long non-coding ribonucleic acid was demonstrated to exert important roles in gastric cancer. Expression of long non-coding ribonucleic acid was measured by real time polymerase chain reaction assay in cells from different groups. Cell apoptosis was checked with Hoechst methods and cell counting kit-8 assay was used to determine cell proliferation. Bioinformatics analysis was applied to predict the target of long non-coding ribonucleic acid, which was verified in luciferase assay. We uncovered that long non-coding ribonucleic acid leukemia inhibitory factor receptor antisense RNA1 was significantly correlated to the patient survival of gastric cancer. Knockdown or overexpression of leukemia inhibitory factor receptor antisense RNA1 resulted in delayed or increased cell proliferation in gastric cancer cell lines, respectively. Moreover, in response to cisplatin treatments, cells with leukemia inhibitory factor receptor antisense RNA1 knockdown resulted in more cell apoptosis as well as less colony survival. Leukemia inhibitory factor receptor antisense RNA1 was further confirmed and bound with micro ribonucleic acid-138-5p and then regulates pyruvate dehydrogenase kinase 1 to promote gastric cancer growth. Our findings provided novel mechanism and novel targets for overcoming the cisplatin resistance of gastric cancer.
Keywords
Long non-coding ribonucleic acid leukemia inhibitory factor receptor antisense RNA1, gastric cancer, micro ribonucleic acid-138-5p, cisplatin resistance
Gastric cancer is one of the most common malignant tumors with high mortality in the whole world[1,2]. Besides surgery, chemotherapy is an effective strategy for the treatments of gastric cancer, in which the cisplatin is widely used[3,4]. However, cisplatin resistance often occurs as long as the therapy was applied[5]. Moreover, some types of cancers are with the property of cisplatin resistance[6]. To uncover the mechanism of cisplatin resistance and find new target for overcoming this resistance, there are critical questions in the treatment of gastric cancer.
Recently, long non-coding Ribonucleic Acid (lncRNA) is drawing more and more attention for their key roles in multiple biological processes, such as tissue development, cancer biology, as well as cell fate determination[7,8]. Several lncRNAs were also proved to be critical for gastric cancer regulation. For example, LncRNA in Non-Homologous End Joining Pathway 1 (LINP1) promotes proliferation and inhibits apoptosis of gastric cancer cells through repressing RNA Binding Motif protein 5 (RBM5) [9]. LncRNA Prostate Cancer Associated Transcript 18 (PCAT18) regulates gastric cancer cell viability, migration and invasion, through interaction with microRNA (miR)-301a[10]. Circulating H19 is also a potential novel diagnostic and prognostic biomarker for gastric cancer[11,12]. However, the role of most lncRNA gastric cancer remains unknown.
In the present study, we identified lncRNA, Leukemia Inhibitory Factor Receptor Antisense RNA 1 (LIFRAS1) is significantly correlated with patient survival in gastric cancer. LIFR-AS1 was also found to promote cancer cell proliferation and cisplatin resistance. Mechanical study revealed that LIFRAS1 sponged with miR-138-5p and then upregulated Pyruvate Dehydrogenase Kinase 1 (PDK1), the axis of which suggest a novel target for gastric cancer therapy.
Materials and Methods
Bioinformatical data mining:
The expression and clinical correlated data was analyzed from The Cancer Genome Atlas Stomach Adenocarcinoma (TCGA-STAD) dataset by using an online Gene Expression Profiling Interactive Analysis (GEPIA) tool (http://gepia2.cancer-pku.cn/#general) [13]. Expression of LIFR-AS1 and the correlation with patient overall survival as well as progression free survival was also analyzed. The potential target miRNA of LIFR-AS1 was predicted by using an online data base StarBase v2.0 (http://starbase.sysu.edu.cn)[14].
Cell culture and transfection:
Human gastric cancer cell lines MGC803, MKN-28, SGC-7901 and BGC-823 were purchased from the American Type Culture Collection (ATCC). All cells were maintained at 37° in 5 % Carbon dioxide (CO2) humid chamber, in Roswell Park Memorial Institute (RPMI) 1640 medium (PAA laboratories) supplemented with 10 % Fetal Bovine Serum (FBS, Gibco), streptomycin and penicillin (10 mg/ml).
Real time Polymerase Chain Reaction (PCR) assay:
Total RNA was extracted from different cells with TRIzol reagent and transcribed into complementary Deoxyribonucleic Acid (cDNA) with a Takara cDNA synthesis kit. Then real time quantitative PCR (qPCR) was performed with a SYBR qPCR kit (Takara), according to the instructions. The data was analyzed using Roche software. The primers used in this study are as follows: LIFR-AS1 Forward: TGGCCTCGGTTTTCTAGCAG; LIFR-AS1 Reverse: TGTGCCGAAGAGAACACCAT; Glyceraldehyde- 3-Phosphate Dehydrogenase (GAPDH) Forward: AAGGTGAAGGTCGGAGTCAAC; GAPDH Reserve: GGGTCATTGATGGCAACAATA; miR- 138-5p Forward: TGTTGTGAATCAGGCCGTTGC, Reverse: CTGTAGTGTGGTGTGGCCCTG; U6 Forward: CTCGCTTCGGCAGCACA and Reserve: AACGCTTCACGAATTTGCGT.
Cell Counting Kit-8 (CCK-8) assay:
Cell proliferation assay was performed by using a CCK-8 kit. Briefly, transfected cells were seeded in 96 well plates at the concentration of 2E3 per well. At 24, 48 and 72 h, 10 μl CCK-8 solution was added into each well and the absorbance at Optical Density 450 (OD450) was measured.
Western blot assay:
After transfected with miR-138-5p, cells were lysed with Radioimmunoprecipitation Assay (RIPA) lysis buffer (Thermo Fisher) to extract total proteins. After that, the protein was separated by using a Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) and transferred to Polyvinylidene Difluoride (PVDF) membrane. After that, the membrane was blocked with PDK1 primary antibody (Cell Signaling Technology (CST), 1:1000) at 4° overnight. An Enhanced Chemiluminescence (ECL) kit (Thermal Fisher) was used to detect the chemiluminescence intensity, according to the manufacture’s instruction.
Cell survival assay:
For cell apoptosis determination, we performed a Hoechst 33342 staining method using an apoptosis kit. The high blue cells were considered as apoptotic cells, according to the manufacturer’s instructions.
For cell survival assay, cells were plated into 60 mm plates at different number for each group. 24 h later, cisplatin (10 μM) was used to treat cells for 72 h. After that, cells were washed with Phosphate Buffered Saline (PBS) and returned to the incubator. After 10 d growth, cells were fixed with ethanol and then stained with crystal violet. The colony formation efficacy was calculated to measure the cell survival rate.
Dual luciferase reporter assay:
The potential target, miRNA of LIFR-AS1 was predicted by using an online database StarBase v2.0 (http://starbase.sysu.edu.cn), from which miR-138-5p was selected for further validation. The sequence of LIFR-AS1 and LIFR-AS1 with mutant in miR-138- 5p binding sites was inserted into a Promega pGL3 luciferase reporter vector. The wild type or mutant LIFR-AS1 together with miR-138-5p mimics were cotransfected using a dual luciferase reporter assay kit (Promega). The Renilla luciferase activities were used to normalize the firefly luciferase activities, according the manufacturer’s instructions.
Statistical analysis:
Data were expressed as mean±Standard Error of the Mean (SEM). The difference between two groups was analyzed with student’s t-test and difference among more than three groups was analyzed with analysis of variance, p<0.05 was regarded as statistically significant. All the experiments were repeated at least three independent times. All the experiments were approved by the Ethics Committee of the Dahua hospital and followed the instructions for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH) (Publication No. 96-01).
Results and Discussion
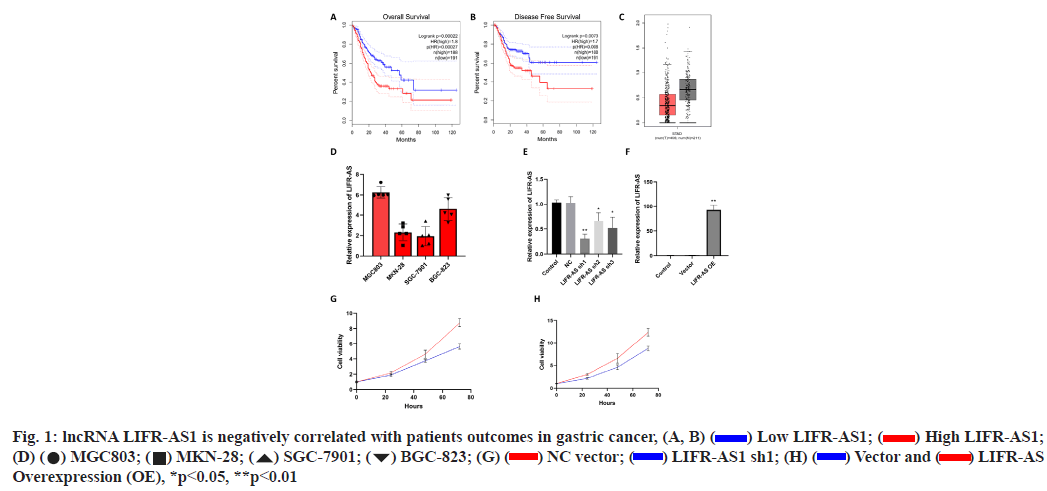
Elevation of lncRNA LIFR-AS1 is negatively correlated with patient’s outcomes in gastric cancer was shown here (fig. 1A- fig. 1H). By using an online GEPIA analysis database (http://gepia.cancer-pku.cn/), the correlation of lncRNA LIFR-AS with patient survival was analyzed in gastric cancer. From the analysis, the expression of lncRNA LIFR-AS1 was found to be negatively correlated with the overall survival as well as disease free survival in gastric cancer patients (fig. 1A and fig. 1B). Significant difference was also observed in the expression of LIFR-AS1 in gastric cancer tissues derived from cancer patients (fig. 1C). Then we checked the expression level of LIFR-AS1 in several gastric cancer cell lines, including MGC803, MKN-28, SGC- 7901 and BGC-823. Relative expression of LIFR-AS1 in gastric cancer cells was measured through real time PCR assay and it was found that the expression of LIFRAS1 in MGC803 and BGC-823 is higher than that in MKN-12 and SGC-7901 cells (fig. 1D). To determine the influence of LIFR-AS1 on cell proliferation, specific short hairpin RNAs (shRNAs) interfering LIFR-AS1 were designed and used to knockdown the expression level of LIFR-AS1 in MGC803 cells. Our data showed that the sequence of shRNA1 inhibited the LIFR-AS1 expression to 62.1 % reduction (fig. 1E). LIFR-AS1 also significantly inhibited cell proliferation (fig. 1G). After then, LIFR-AS1 was elevated in low expressing cells, SGC-7901. Real time PCR was used to detect the effect of overexpression. Our results showed that the vector group had no influence on the level of LIFR-AS1. Surprisingly, overexpressing group induced dramatically up-regulation of LIFR-AS1 (fig. 1F). From CCK-8 assay, we observed that LIFR-AS1 overexpression significantly increased cell proliferation (fig. 1H).
Knockdown of LIFR-AS1 increased cellular sensitivity to cisplatin (fig. 2A-fig. 2F). Then we determined the sensitivity of cells to cisplatin, a main strategy of chemotherapy. Firstly, we performed survival fraction assay in LIFR-AS1 knockdown cells and overexpression cells. It was found that LIFR-AS1 knockdown resulted in less colony survival after incubation with cisplatin (fig. 2A). In contrast, LIFR-AS1 overexpression increased cellular resistance to cisplatin (fig. 2D). After that, we further detected the cell apoptosis by using Hoechst 33342 staining assay. Our data showed that in LIFR-AS1 knockdown cells, more apoptotic cells were observed after cisplatin treatments (fig. 2B and fig. 2C). Cells apoptosis was determined with Hoechst assay in vector or LIFR-AS1 expressing cells. In response to cisplatin, less apoptosis cells were observed in the LIFR-AS1 overexpressing cells (fig. 2E and fig. 2F).
LIFR-AS1 knockdown augmented apoptosis signaling pathway when combined with cisplatin (fig. 3A-fig. 3E). The data above showed that LIFR-AS1 knockdown induced more apoptosis in cisplatin treated cells. Then we detected the expression of apoptosis pathway and found that in LIFR-AS1 knockdown cells, cisplatin induced the elevation of apoptosis pathway. In detail, in LIFR-AS1 knockdown cells, cleaved caspase 3 increased after cisplatin treatment, which is an executor of cell apoptosis. Raw density was also analyzed with Image J software and beta-actin was used as an internal control (fig. 3A and fig. 3B). Knockdown of LIFR-AS1 also increased the level of Bcl-2 Associated X-protein (Bax) (fig. 3A and fig. 3C) and p53 (fig. 3A and fig. 3E), which promotes the process of apoptosis. However, no significant difference was found in the expression level of anti-apoptosis factor B-cell lymphoma 2 (Bcl- 2) between Negative Control (NC) and LIFR-AS1 knockdown cells (fig. 3A and fig. 3D).
LncRNA LIFR-AS1 sponges with miR-138-5p and regulated cell response to cisplatin (fig. 4A-fig. 4D). By using an online prediction tool, miR-138-5p was predicted to bind with LIFR-AS1 with two sets of seed sequences (fig. 4A). Then we measured the expression level of miR-138-5p in different gastric cancer cells and found significantly high miR-138-5p expression in MKN-28 cells and SGC-7901 cells (fig. 4B), where both exhibited low expression in LIFR-AS1. The negative relationship promoted us to investigate the direct interaction of these two non-coding RNAs. Through a luciferase analysis, we observed that miR-138-5p mimics significantly reduced the luciferase activity in LIFR-AS1 expressing cells, but not in LIFR-AS1 mutant group (fig. 4C). These data proved that LIFR-AS1 directly bound with miR-138-5p with a sequence specific manner. Then we measured the influence of miR-138-5p on cell survival in LIFR-AS1 knockdown cells. It was found that cells with LIFR-AS1 knockdown are sensitive to cisplatin treatment, which was significantly rescued by miR-138-5p inhibitor transfection (fig. 4D). These data showed that lncRNA LIFR-AS1 regulates cellular sensitivity through miR- 138-5p.
miR-138-5p target PDK1 contributes to the effects of lncRNA LIFR-AS1 (fig. 5A-fig. 5D). The interaction between miR-138-5p and LFIR-AS1 provided novel mechanism of lncRNA on cisplatin resistance of gastric cancer. We also aimed to determine the downstream mechanism. It was reported that miR-138-5p inhibited PDK1 in colorectal cancer, so we determined whether miR-138-5p could regulate PDK1 in gastric cancer cells. Our data proved that miR-138-5p mimics reduced the messenger RNA (mRNA) level and protein level expression of PDK1 in MGC803 cells (fig. 5A and fig. 5B). Then we conducted luciferase activity assay and found that miR-138-5p mimics significantly inhibited the luciferase activity of PDK1 3'-Untranslated Region (3' UTR) expressing cells (fig. 5C). Finally, we measured the effects of PDK1 expression on cell survival. We found that PDK1 overexpression promoted cell survival in LIFR-AS1 knockdown cells (fig. 5D).
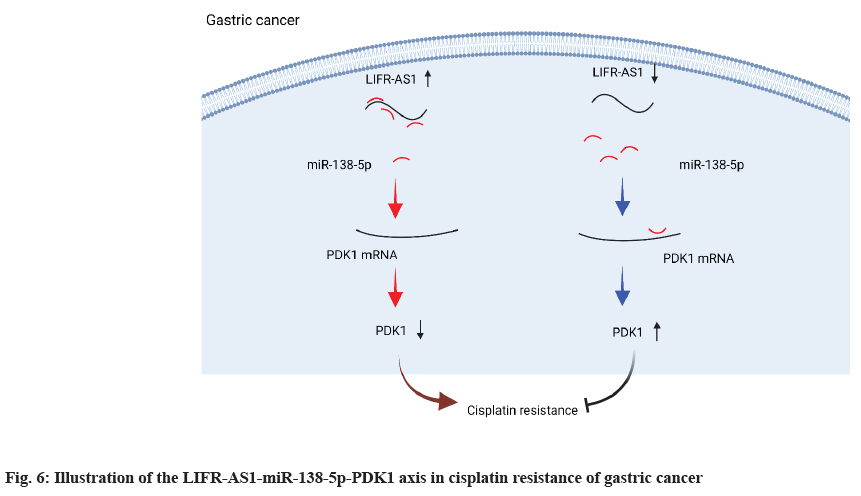
In the present study, we identified a novel lncRNA, LIFR-AS1 in gastric cancer and investigated its role in gastric cancer cell proliferation and its sensitivity to cisplatin treatment (fig. 6). Our data showed that LIFR-AS1 is significantly correlated to patient survival of gastric cancer. By using gastric cancer cell lines, it was observed that knockdown of LIFR-AS1 or overexpression regulated cell proliferation and the cellular sensitivity of gastric cancer cells. Mechanically, we identified that LIFR-AS1 directly bound with miR-138-5p through a sequence dependent manner. Further investigation proved that miR-138-5p related regulation on PDK1 accounts for the role of LIFR-AS1 in cellular resistance to cisplatin. Our findings provide novel targets and mechanisms for the therapy of gastric cancer.
Firstly, we identified that lncRNA LIFR-AS1 could be regarded as a biomarker for gastric cancer. Recently, non-coding RNA was emerging as a novel biomarkers and regulators in multiple biological processes[15-17]. Several studies have investigated the role of lncRNAs, microRNAs as well as exosomal components for its potential as biomarkers[7]. Here, we identified that lncRNA LIFR-AS1 was significantly related to patient survival. Our study provided novel potential for LIFRAS1 as a biomarker to predict the outcomes of gastric cancer patients. Previously, several lncRNAs had been identified as prognostic biomarker for gastric cancer. Serum exosomal lncRNA H19 levels were significantly upregulated in patients with gastric cancer both before and after surgery compared with healthy controls[11]. The Gastric Cancer-associated long noncoding RNA1 (lncRNA-GC1) was also proved as a novel early detection and detection marker for gastric cancer[18]. Thus, in our present study LIFR-AS1, also exhibited the potential as a novel biomarker in gastric cancer. The dysregulation of LIFR-AS1 was a possible clinical application alone or in combination with other biomarkers.
For the first time, our data proved that LIFR-AS1 is related to cisplatin resistance in gastric cancer. Chemotherapy is one of the most important strategies in gastric cancer treatments. Among all, cisplatin resistance was a main reason for treatment failure as well as reoccurrence[19]. Previously, multiple protein factors as well as cytokines were found to be related to cisplatin resistance[20,21]. Recently, the role of lncRNA was drawing more and more attention. For instance, lncRNA Differentiation Antagonizing Non- Protein Coding RNA (DANCR) plays a role in the cellular sensitivity of colon cancer cells to cisplatin treatments[22]. Increased expression of ZXF1 promotes cisplatin resistance in lung cancer cell via Mitogen- Activated Protein Kinase (MAPK) axis[23]. LncRNA Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) promotes Epithelial-Mesenchymal Transition (EMT) process and cisplatin resistance of oral squamous cell carcinoma, the mechanism of which was related to Phosphatidylinositol-3-Kinase (PI3K)/Protein kinase B (AKT)/mammalian Target of Rapamycin (mTOR) signaling pathway. Here, we found that a novel lncRNA, LIFR-AS1 was related to cisplatin resistance and knockdown of LIFR-AS1 increased cellular sensitivity of gastric cancer to cisplatin treatments.
One of the most important interacting targets of lncRNA is the species of microRNAs. Through a bioinformatics tool, we predicted that miR-138-5p could be a potential target of lncRNA LIFR-AS1, which was also validated through experiments. A negative correlation was also found in the expression of miR- 138-5p and lncRNA LIFR-AS1. Importantly, miR-138- 5p play critical roles in the proliferation and metastasis of gastric cancer cells[24] and several critical targets were proved to be potentially targeted by miR-138-5p[25-27]. Among all, in colon cancer as well as non-small cell lung cancer, miR-138-5p significantly suppressed the expression of PDK1[27,28], however, there is no report in gastric cancer. By using gastric cancer cell lines, we found that miR-138-5p significantly bound with LIFRAS1 and targets PDK1 3’UTR sequence. Our study demonstrated a critical role of LIFR-AS1-miR-138-5p- PDK1 signaling pathway in the regulation of cisplatin sensitivity of gastric cancer.
In conclusion, our data identified a novel lncRNA LIFR-AS1 was significantly correlated with the patient survival of gastric cancer. We also found that LIFR-AS1 was related to cisplatin related cell death. Mechanically, we proved that LIFR-AS1 directly binds with miR-138- 5p and regulated miR-138-5p-PDK1 axis. Our findings provided a novel mechanism and potential target for overcoming cisplatin resistance in gastric cancer.
Ethical statement:
All the experiments were approved by the Ethics Committee of the Dahua Hospital and followed the instructions for the Care and Use of Laboratory Animals published by the NIH (Publication No. 96- 01).
Author’s contributions:
Kun Yu and Yao Xiao contributed equally to this work.
Acknowledgements:
This study was supported by the grant from Dahua hospital of Xuhui District, Shanghai, China.
Conflict of interests:
The authors have no conflicts of interest to disclaim.
References
- Kang C, Dhillon S, Deeks ED. Trifluridine/tipiracil: A review in metastatic gastric cancer. Drugs 2019;79(14):1583-90.
[Crossref] [Google scholar] [PubMed]
- Long B, Qin L, Zhang B, Li Q, Wang L, Jiang X, et al. CAR T‑cell therapy for gastric cancer: Potential and perspective. Int J Oncol 2020;56(4):889-99.
[Crossref] [Google scholar] [PubMed]
- Tan Z. Recent advances in the surgical treatment of advanced gastric cancer: A review. Med Sci Monit 2019;25:3537-41.
[Crossref] [Google scholar] [PubMed]
- Chen C, Zhang F, Zhou N, Gu YM, Zhang YT, He YD, et al. Efficacy and safety of immune checkpoint inhibitors in advanced gastric or gastroesophageal junction cancer: A systematic review and meta-analysis. Oncoimmunology 2019;8(5):e1581547.
[Crossref] [Google scholar] [PubMed]
- Zhou K, Guo H, Zhang J, Zhao D, Zhou Y, Zheng Z, et al. Potential role of TET2 in gastric cancer cisplatin resistance. Pathol Res Pract 2019;215(11):152637.
[Crossref] [Google scholar] [PubMed]
- Zhu X, Zhang K, Wang Q, Chen S, Gou Y, Cui Y, et al. Molecular mechanism of cisplatin to enhance the ability of TRAIL in reversing multidrug resistance in gastric cancer cells. Zhonghua Zhong Liu Za Zhi 2015;37(6):404-11.
- Zhang K, Shi H, Xi H, Wu X, Cui J, Gao Y, et al. Genome-wide lncRNA microarray profiling identifies novel circulating lncRNAs for detection of gastric cancer. Theranostics 2017;7(1):213-27.
[Crossref] [Google scholar] [PubMed]
- Zhang Y, Zhang Q, Zhang M, Yuan M, Wang Z, Zhang J, et al. DC-SIGNR by influencing the lncRNA HNRNPKP2 upregulates the expression of CXCR4 in gastric cancer liver metastasis. Mol Cancer 2017;16(1):1-6.
[Crossref] [Google scholar] [PubMed]
- Lu XC, Zhou HY, Wu J, Jin Y, Yao XM, Wu XY. LncRNA LINP1 promotes proliferation and inhibits apoptosis of gastric cancer cells by repressing RBM5. Eur Rev Med Pharmacol Sci 2020;24(13):7205.
[Crossref] [Google scholar] [PubMed]
- Dou J, Tu D, Zhao H, Zhang X. LncRNA PCAT18/miR-301a/TP53INP1 axis is involved in gastric cancer cell viability, migration and invasion. J Biochem 2020;168(5):547-55.
[Crossref] [Google scholar] [PubMed]
- Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the role of circulating long non‐coding RNA H19 as a promising novel biomarker in plasma of patients with gastric cancer. J Clin Lab Anal 2016;30(6):1100-5.
[Crossref] [Google scholar] [PubMed]
- Yörüker EE, Keskin M, Kulle CB, Holdenrieder S, Gezer U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed Rep 2018;9(2):181-6.
- Li D, Xing Y, Tian T, Guo Y, Qian J. Overexpression of LRRC59 is associated with poor prognosis and promotes cell proliferation and invasion in lung adenocarcinoma. Onco Targets Ther 2020;13:6453-63.
[Crossref] [Google scholar] [PubMed]
- Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2. 0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 2014;42(D1):D92-7.
[Crossref] [Google scholar] [PubMed]
- Giulietti M, Righetti A, Principato G, Piva F. LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis 2018;39(8):1016-25.
[Crossref] [Google scholar] [PubMed]
- Guo X, Yang J, Liang B, Shen T, Yan Y, Huang S, et al. Identification of novel LncRNA biomarkers and construction of LncRNA-related networks in Han Chinese patients with ischemic stroke. Cell Physiol Biochem 2018;50(6):2157-75.
[Crossref] [Google scholar] [PubMed]
- Wu C, Wei Y, Zhu Y, Li K, Zhu Y, Zhao Y, et al. Identification of cancer‐related potential biomarkers based on lnc RNA-pseudogene-mRNA competitive networks. FEBS Lett 2018;592(6):973-86.
[Crossref] [Google scholar] [PubMed]
- Guo X, Lv X, Ru Y, Zhou F, Wang N, Xi H, et al. Circulating exosomal gastric cancer-associated long noncoding RNA1 as a biomarker for early detection and monitoring progression of gastric cancer: A multiphase study. JAMA Surg 2020;155(7):572-9.
[Crossref] [Google scholar] [PubMed]
- Wang X, Xu Z, Sun J, Lv H, Wang Y, Ni Y, et al. Cisplatin resistance in gastric cancer cells is involved with GPR30‐mediated epithelial‐mesenchymal transition. J Cell Mol Med 2020;24(6):3625-33.
[Crossref] [Google scholar] [PubMed]
- Cho HJ, Kim IK, Park SM, Baek KE, Nam IK, Park SH, et al. VEGF‐C mediates RhoGDI2‐induced gastric cancer cell metastasis and cisplatin resistance. Int J Cancer 2014;135(7):1553-63.
[Crossref] [Google scholar] [PubMed]
- Duan S, Yin J, Bai Z, Zhang Z. Effects of taxol resistance gene 1 on the cisplatin response in gastric cancer. Oncol Lett 2018;15(6):8287-94.
[Crossref] [Google scholar] [PubMed]
- Shi H, Li K, Feng J, Liu G, Feng Y, Zhang X. LncRNA-DANCR interferes with miR-125b-5p/HK2 axis to desensitize colon cancer cells to cisplatin vis activating anaerobic glycolysis. Front Oncol 2020;10:1034.
- Yu T, Bai W, Su Y, Wang Y, Wang M, Ling C. Enhanced expression of lncRNA ZXF1 promotes cisplatin resistance in lung cancer cell via MAPK axis. Exp Mol Pathol 2020;116:104484.
- Pang L, Li B, Zheng B, Niu L, Ge L. miR-138 inhibits gastric cancer growth by suppressing SOX4. Oncol Rep 2017;38(2):1295-302.
[Crossref] [Google scholar] [PubMed]
- Tian S, Guo X, Yu C, Sun C, Jiang J. miR-138-5p suppresses autophagy in pancreatic cancer by targeting SIRT1. Oncotarget 2017;8(7):11071-82.
[Crossref] [Google scholar] [PubMed]
- Wang X, Zhao Y, Cao W, Wang C, Sun B, Chen J, et al. miR-138-5p acts as a tumor suppressor by targeting hTERT in human colorectal cancer. Int J Clin Exp Pathol 2017;10(12):11516-25.
[Google scholar] [PubMed]
- Wang Y, Zhang D, Li Y, Fang F. MiR-138 suppresses the PDK1 expression to decrease the oxaliplatin resistance of colorectal cancer. Onco Targets Ther 2020;13:3607-18.
[Crossref] [Google scholar] [PubMed]
- Huang T, Wen Y, Peng B, Ding G, Yang L, Wang Z. Upregulated lncRNA H19 promotes non-small cell lung cancer cell proliferation through miR-138/PDK1 axis. Int J Clin Exp Pathol 2017;10(8):9012-20.
[Google scholar] [PubMed]
 High LIFR-AS1;
(D)
High LIFR-AS1;
(D)  LIFR-AS
Overexpression (OE), *p<0.05, **p<0.01
LIFR-AS
Overexpression (OE), *p<0.05, **p<0.01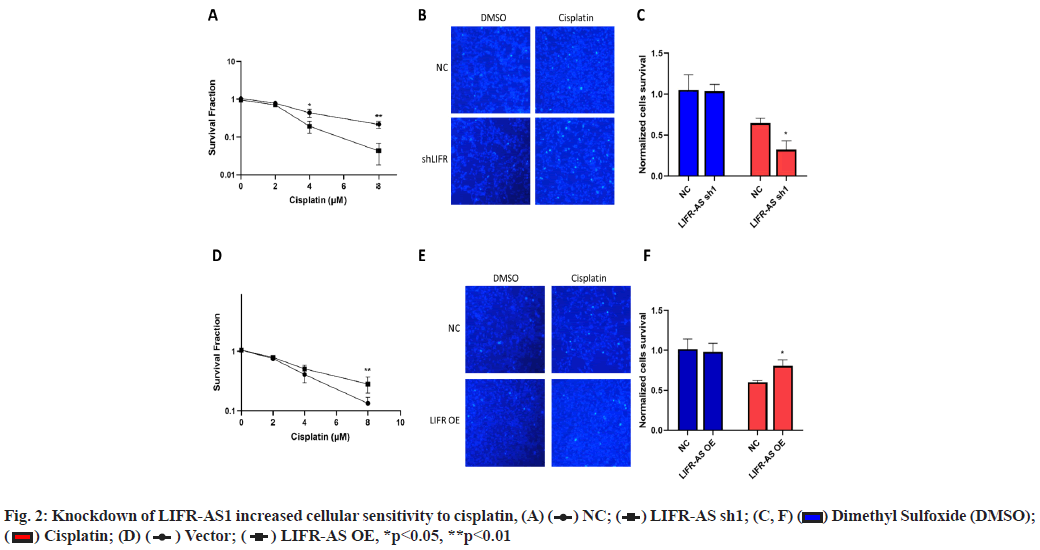
 Dimethyl Sulfoxide (DMSO);
Dimethyl Sulfoxide (DMSO);  LIFR-AS OE, *p<0.05, **p<0.01
LIFR-AS OE, *p<0.05, **p<0.01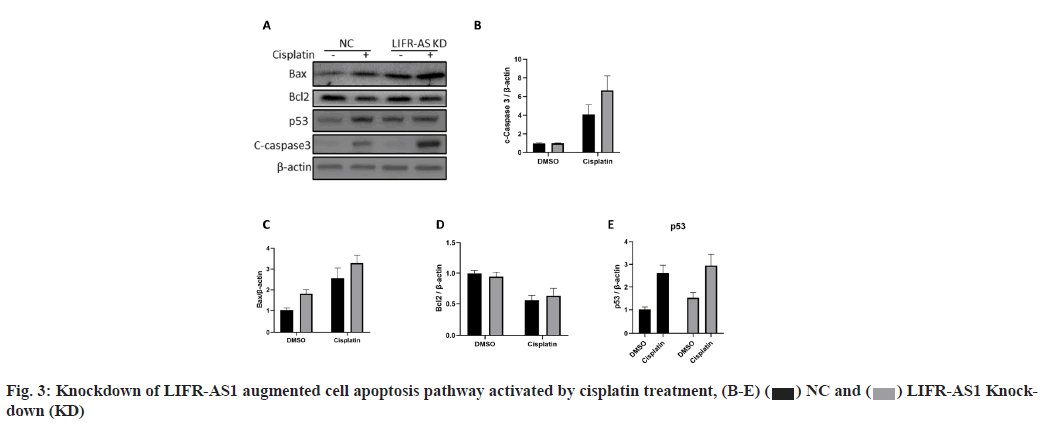
 LIFR-AS1 Knockdown (KD)
LIFR-AS1 Knockdown (KD)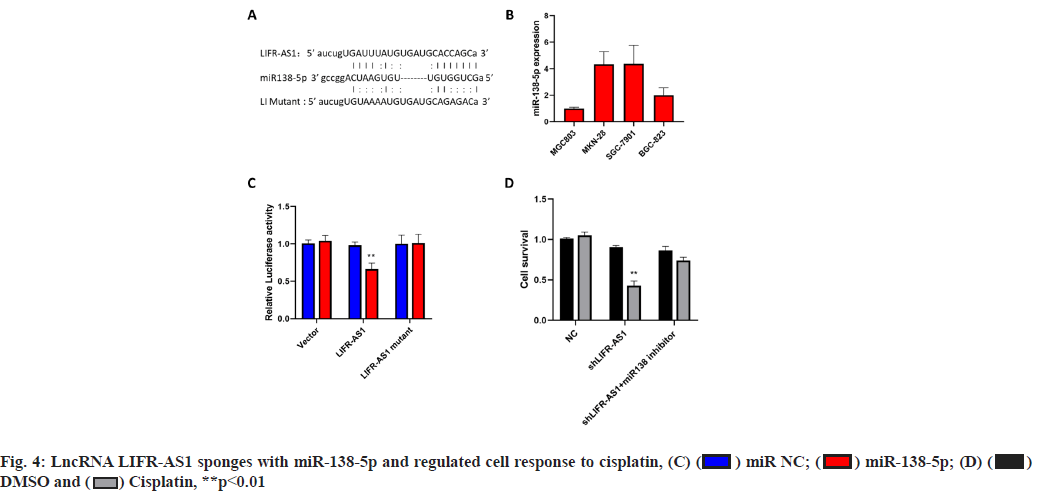
 DMSO and
DMSO and  Cisplatin, **p<0.01
Cisplatin, **p<0.01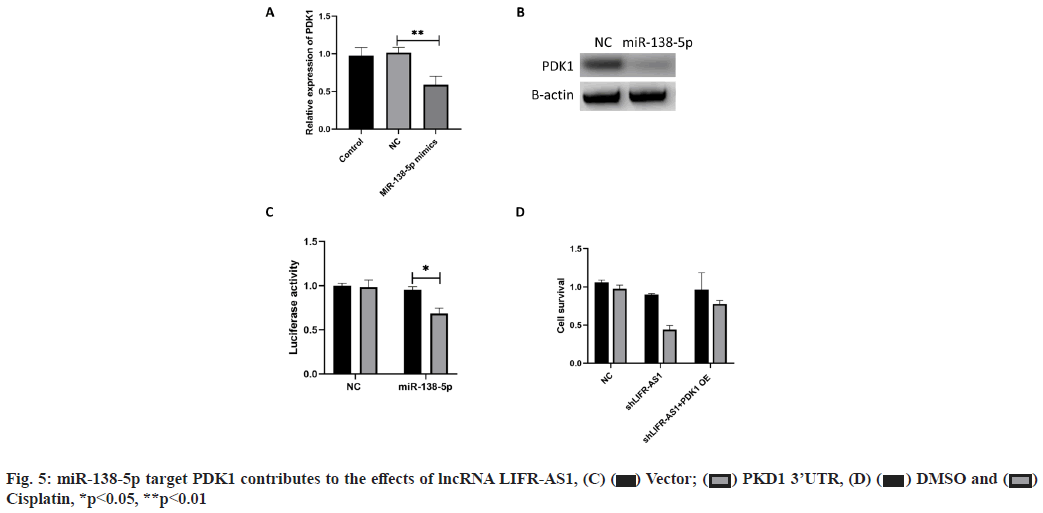
 Cisplatin, *p<0.05, **p<0.01
Cisplatin, *p<0.05, **p<0.01