- *Corresponding Author:
- Zhi Hong. Xu
Department of Neurology, Suzhou Ninth People’s Hospital, No. 2666 Ludang Road, Suzhou, Jiangsu 215200, China
E-mail: feihei5893662@163.com
| This article was originally published in a special issue, “Biomedical Research in Clinical and Preclinical Pharmaceutics” |
| Indian J Pharm Sci 2020:82(5)Spl issue7;94-101 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Ischemic stroke was a brain disease with high mortality and disability rate. Previous studies have shown that the expression of Micro Ribonucleic acid-376a in the serum of stroke patients was increased, suggesting that Micro Ribonucleic acid-376a was closely related to the occurrence and development of stroke. However, its mechanism has not been reported. The purpose of this study was to explore the regulatory mechanism of Long non-coding ribonucleic acid small nucleolar host gene 1 on Ischemic stroke through targeting the Micro Ribonucleic acid-376a-mediated cystathionine β synthase hydrogen sulfide axis pathway. The Ischemic stroke model of Human Cerebral Microvascular Endothelial Cell Line/D3 cells was established by the induce of Oxygen-glucose deprivation. The Micro Ribonucleic acid concentrations of small nucleolar host gene 1, Micro Ribonucleic acid-376a, and inflammatory cytokines were detected by Reverse transcription- polymerase chain reaction. Western blotting was used to detect the expression of cystathionine β synthase and apoptosis-related proteins. The apoptosis was analyzed by flow cytometry and the concentration of hydrogen sulfide was detected by the kit. The interaction between long non-coding ribonucleic acid, micro ribonucleic acid, and target genes was confirmed by the luciferase test. Our study showed that under the condition of Oxygen-glucose deprivation induction, when Micro Ribonucleic acid-376a was up-regulated, the Micro Ribonucleic acid concentration of small nucleolar host gene 1 and cystathionine β synthase in Human Cerebral Microvascular Endothelial Cell Line/D3 cells was down-regulated. Further results showed that the overexpression of Micro Ribonucleic acid-376a promoted apoptosis and inflammation, while overexpression of small nucleolar host gene 1 alleviated these processes. In terms of mechanism, Micro Ribonucleic acid-376a was the target of small nucleolar host gene 1, and cystathionine β synthase was the target of Micro Ribonucleic acid-376a. In addition, small nucleolar host gene 1 regulates the expression of cystathionine β synthase by inhibiting Micro Ribonucleic acid-376a, thus exerting its function. Long non-coding ribonucleic acid small nucleolar host gene 1 inhibits the apoptosis and inflammation of Ischemic stroke cells by inhibiting Micro Ribonucleic acid-376a and up-regulating cystathionine β synthase/hydrogen sulfide signal. These results reveal the potential mechanism of Ischemic stroke and provide a potential target for the treatment of Ischemic stroke.
Keywords
Ischemic stroke, miRNA, LncRNA SNHG1, Inflammation
Stroke has become a major public health burden and a major cause of disability and death (81.0 % and 75.2 %)[1]. About 87 % of the stroke patients have ischemic characteristics[2]. Ischemic stroke (IS) was usually observed in the core area surrounded by the electrically silent tissue, and IS accompanied by the increased vascular cell injury, decreased vascular reactivity, inflammation, and apoptosis[3]. The damage of IS may exceed the core area and cause neuronal damage in the surrounding brain tissue by releasing inflammatory factors. IS was caused by a variety of causes and mechanisms, and at least 100 pathologies were considered to be related to IS[1]. In addition, the etiological classification of IS was still incomplete, which makes IS more difficult to treat than the other brain diseases such as coronary artery disease. Therefore, understanding the mechanism of IS is of great significance for future research and the development of treatment.
Hydrogen sulfide (H2S) was an active gas transmitter associated with stroke regulation. It was reported that H2S plays an important role in maintaining the stability of the cerebrovascular environment. The decrease of H2S level leads to the endothelial dysfunction and promotes the development of IS[4]. In the mouse model, it was also observed that H2S supplementation promoted the vascular remodeling and regeneration, which confirmed the brain protective function of H2S[5]. However, the protective effect of H2S seems to depend on its concentration. Low concentration of H2S has a protective effect, while high dose of H2S may be toxic[6]. Cystathionine β-synthase (CBS) was the main synthase of H2S in the brain. The concentration of H2S largely depends on the protein level and catalytic ability of CBS. In the mouse IS model, the overexpression of CBS increased the concentration of H2S, reduced the degree of IS, and played an anti-inflammatory effect[7]. In addition to inhibiting inflammation, H2S can also protect the brain by inhibiting the oxidants and apoptosis, although its regulatory mechanism was not well understood[8].
MicroRNA (miRNA) was a regulatory non-coding RNA which usually contains 20-24 nt. They can interact with the specific base-paired target molecules RNA through the RNA-induced silencing complex (RISC) and mediate their degradation. MiRNA was involved in many biological processes, including IS. For example, in stroke samples from 82 patients, at least 8 miRNA were associated with the differential gene expression and miRNAs were considered to be the biomarkers[9]. miR-152 can protect the neurons in the IS mouse model and in vitro cell model by regulating the Nuclear factor erythroid 2-related factor 2 (NRF2) signal[10]. In addition, miR-130 plays a neuroprotective role by inhibiting Phosphatase and tensin homolog (PTEN)/phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT) signal pathway[11]. It was reported that the risk factors of IS, such as inflammation, apoptosis, oxidative stress, and excitotoxicity were all regulated by miRNAs[12]. Some studies have reported that the expression of miR-376a was increased in the serum of stroke patients[13], suggesting that miR-376a was closely related to the occurrence and development of stroke, while with unclear mechanism. Bioinformatics prediction reveals the direct interaction between miR- 376a and CBS, but it has not been reported in the literature. Therefore, it is worth exploring whether miR-376a can regulate the inflammation and apoptosis of stroke through CBS/H2S pathway.
Long non-coding RNA (lncRNA) was a newly emerged regulatory RNA, with a length of about 200 nm. Through the conserved sequences, lncRNA interrupts its RNA degradation ability, and protects and fixes the interaction between its target gene and miRNA[14,15]. The abnormal expression of lncRNA was related to the occurrence of many diseases. Studies have shown that the lncRNA SNHG1 can alleviate the injury of cerebral microvascular endothelial cells induced by the Oxygenglucose deprivation (OGD) through microRNA -338/HIF-1α, suggesting that SNHG1 also plays an important role in the pathogenesis of stroke[3]. Bioinformatics analysis further showed the targeting effect of SNHG1 on miR-376a. Therefore, we speculate that the lncRNA SNHG1 alleviates apoptosis and inflammation of IS through the CBS/H2S pathway.
In this study, we revealed that the lncRNA SNHG1 can reduce the OGD-induced apoptosis and inflammation in the IS cell model, which may be regulated by the CBS/ H2S pathway targeting miR-376a. This study expands our understanding of the regulatory mechanism of IS, and provides theoretical support for future research, and expands the treatment of IS by providing valuable targets.
Materials and Methods
Cell culture and transfection:
Human cerebrum microvascular endothelial cells (HCMIEC/D3) were purchased from Cell Bank of the typical Culture Preservation Committee of the Chinese Academy of Sciences (Shang Hai). Lipofectamine 2000 was used for the cell transfection, and the RNA in this study was as follows, miR-376 mimics and miR- 376 inhibitor, sh-CBS and sh-NC, pcDNA3.1-SNHG1 (OE-SNHG) and pcDNA3.1-NC. The transfected cells were induced by the OGD for 2.5 h, and the culture medium was replaced by OGD buffer (37π, 95 % N2, and 5 % CO2).
RNA extraction and Reverse transcriptionpolymerase chain reaction (RT-PCR):
The cells were collected according to the manufacturer’s instructions, and the RNA was extracted by TRIzol reagent, which was then used for the cDNA synthesis by the reverse transcription kit. The cDNA was used in the RT-PCR in the Applied biosystems QuantStudio 7 Flex (ABI Q7) system by the Ex Taq II kit (SYBR premix). The reaction conditions were as follows (40 cycles), 94π 5 s, 60π 34 s, 72π 30 s. The results were analyzed by 2−ΔΔCt.
Western blotting:
The collected cells were lysed by the Radioimmunoprecipitation assay (RIPA) buffer for 20 min (0π), and then centrifuged at 4π, 12 000 rpm for 15 min. The supernatant was collected for protein quantification by the Bicinchoninic Acid (BCA) kit. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used for the separation of 20 μg protein, then transformed to the nitrocellulose membrane (NC membrane), which was blocked by the 5 % skimmed milk for 1 h at room temperature. The NC membrane was incubated with the primary antibody at 4π overnight. After washed 3 times (5 min/time), the NC membrane was incubated with the secondary antibody (labeled with horse radish peroxidase) at room temperature for 1 h. The NC membrane was washed again and the protein signals were detected by the enhanced chemiluminescence (ECL). Protein intensity was analyzed by Image J software, and β-actin was used as the internal reference. Antibodies to CBS, Bcl-2, Bax, caspase-3, and β-actin were purchased from CST.
Detection of H2S:
The cells were cultured in the medium without phenol red and treated under OGD or other necessary treatment/ transfection conditions. A total of 500 μl medium was collected and fully mixed with 25 μl 1 % zinc acetate. The mixture was then mixed with N,N-Dimethyl-pphenylenediamine sulfate salt (20 μM; 133 μl) dissolved in 7.2 M hydrochloric acid and ferric trichloride (30 μM; 133 μl) dissolved in 1.2 M hydrochloric acid. Then, 250 ul 10 % trichloroacetic acid was added to the mixture, and centrifuged at 4π, 24 000 rpm for 5 min. The absorbance of the supernatant was detected by an infinite 2000 at 670 nm wavelength.
Flow cytometry analysis:
According to the manufacturer’s instructions, cell apoptosis was detected by annexin V-FITC apoptosis kit (Sigma). In short, the cells were collected and washed once with Phosphate-buffered saline (PBS). The cells were then re-suspended using the binding buffer (100 μl) and mixed with annexin V-FITC (5 μl). Then, 5 μl propidium iodide was added to the mixture and incubate in the dark for 10 min. The binding buffer (400 μl) was mixed with the mixture and analyzed by FACS Calibur.
Dual-luciferase assay:
According to the manufacturer’s instructions, luciferase reporter gene assay was performed by the co-transfection of luciferase reporter plasmid and luciferase control reporter vector into HCMIEC/D3 cells. Luciferase was detected by the double luciferase reporter assay system 48 h after transfection. In short, the cells were lysed by the lysate buffer at room temperature for 20 min. The 100 μl supernatant was transferred to the photometer tube, mixed with 20 μl luciferase detection reagent, and the signal was detected on the GloMax20/20 photometer.
Statistical analysis:
All data were processed by SPSS18.0 software, t-test was used in the comparison between two groups, and variance was used in the comparison between three groups. *p<0.05, **p<0.01, ***p<0.001 means the difference was statistically significant. All experiments were repeated at least 3 times. The data were expressed as average±standard deviation (SD).
Results and Discussion
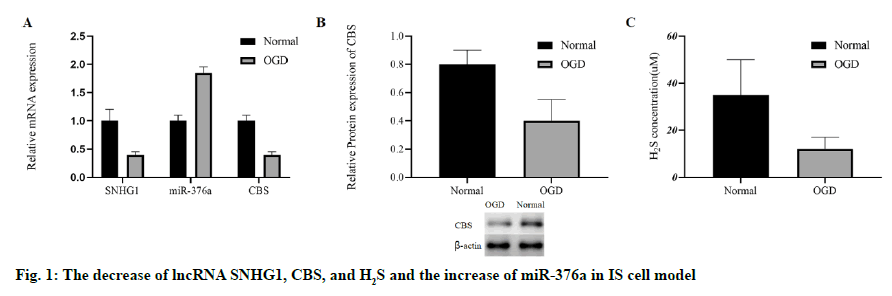
To evaluate whether IS can change the levels of lncRNA SNHG1, miR-376a, CBS and H2S, we stimulated the HCMEC/D3 cells with OGD to establish an in vitro IS cell model, as mentioned in the previous study[3]. After OGD stimulation, the miR-376a was up-regulated, and the RNA levels of SNHG1 and CBS in HCMEC/ D3 cells were down-regulated (fig. 1A). After OGD stimulation, the protein concentration of CBS was also inhibited (fig. 1B). In addition, OGD significantly inhibited the level of H2S (fig. 1C). To sum up, the in vitro IS model was successfully established by OGD. OGD cannot only increase the level of miR-376a, but also inhibit SNHG1, CBS and H2S.
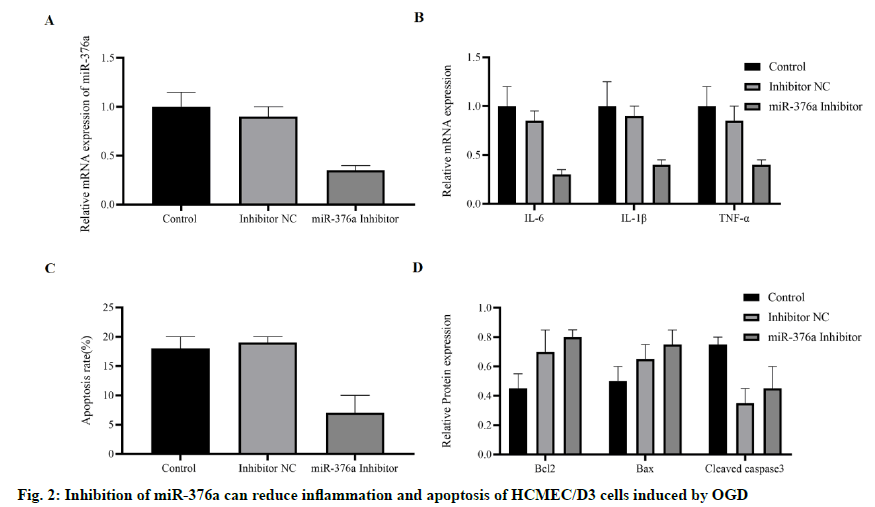
To explore whether miR-376a was involved in IS-related biological processes, especially inflammation and apoptosis. miR-376a inhibitor was transfected into HCMEC/D3 cells, and then induced by OGD. The results showed that the expression of miR-376a was inhibited (fig. 2A). The mRNA concentrations of inflammatory cytokines Interleukin-6, Interleukin-1 β, and Tumor Necrosis Factor-α induced by OGD were detected by RT-PCR. It was found that they were all inhibited by miR-376a inhibitor (fig. 2B). The apoptosis of these cells was detected, and compared with the control group or normal control group; the miR-376a inhibitor significantly inhibited the OGD-induced apoptosis (fig. 2C). To verify this finding, apoptosis-related proteins were detected. The expression of anti-apoptotic protein B-cell lymphoma 2 (Bcl-2) was significantly increased, while the expression of pro-apoptotic protein Bax and caspase-3 were down-regulated (fig. 2D). Based on these findings, we concluded that the miR- 376a promotes apoptosis and inflammation of HCMEC/ D3 cells induced by OGD.
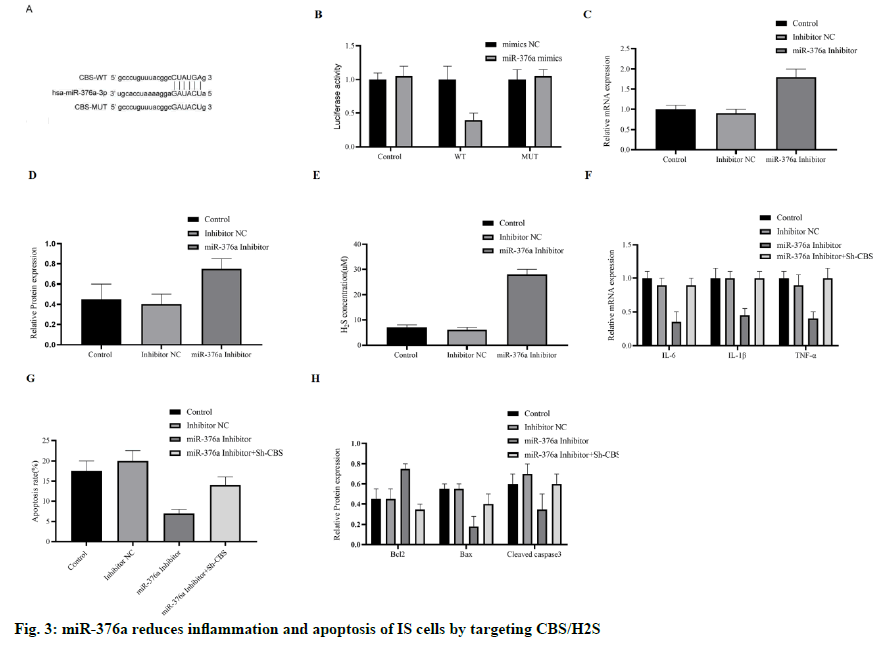
After OGD induction, the increase of miR-376a and the decrease of CBS were detected in HCMEC/D3 cells. To explore the relationship between miR-376a and CBS, we carried out bioinformatics analysis. Finally, the direct interaction region between miR-376a and CBS mRNA was determined (fig. 3A), which shows that the miR-376a targets CBS directly. To further verify this finding, we constructed a luciferase reporting system with wild type (WT) or mutant (MUT) CBS mRNA binding sites. As we expected, miR-376a inhibited luciferase activity in WT group, but did not inhibit luciferase activity in MUT group (fig. 3B). Next, we examined the mRNA level and protein concentration of CBS in OGD-HCMEC/D3 cells treated with miR- 376a inhibitor, and their expression was up-regulated (fig. 3C and 3D). H2S was also significantly upregulated by miR-376a inhibitor (fig. 3E). These results suggest that miR-376a directly targets and inhibits the expression of CBS.
We speculate whether miR-376a regulates the apoptosis and inflammation through CBS, and then constructed the anti-CBS Short Hairpin RNA (shRNA), and cotransfects HCMEC/D3 cells with miR-376a inhibitor. When miR-376a inhibitor inhibits the inflammatory cytokines (IL-6, IL-1 β, and TNF-α), sh-CBS reversed these effects (fig. 3F), apoptosis has the same trend. Inhibition of CBS reduces the anti-apoptotic ability of miR-376a inhibitor (fig. 3G); Western blotting also supports these findings. This effect was blocked by the sh-CBS, when miR-376a inhibitor up-regulation of Bcl-2, and down-regulation of Bax and caspase-3 (fig. 3H). In short, miR-376a promotes the inflammation and apoptosis of HCMEC/D3 cells induced by OGD by directly targeting CBS.
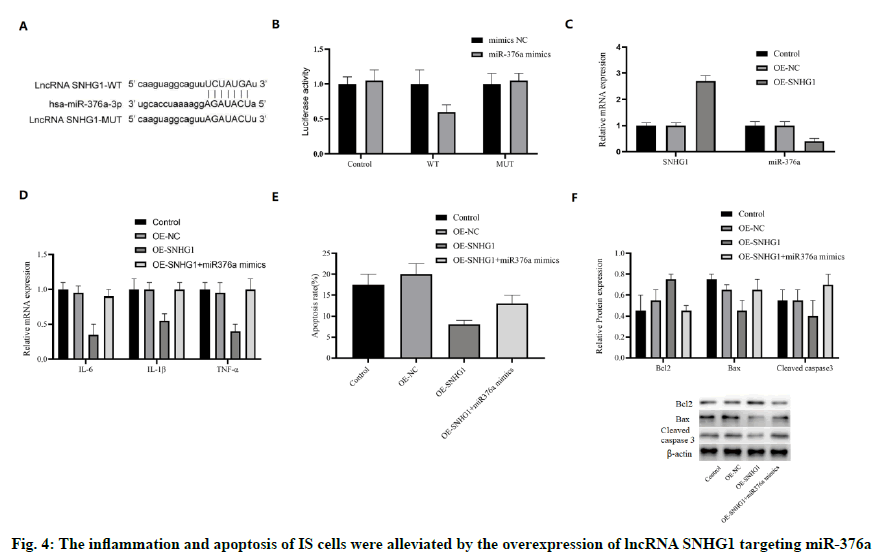
Another bioinformatics analysis was conducted to reveal whether SNHG1 could interact with miR-376a. In fact, SNHG1 and miR-376a shared an associative sequence (fig. 4A). A luciferase reporter system containing WT and MUT binding sequences of SNHG1 was constructed and introduced into HCMEC/D3 cells with miR-376a. Luciferase activity was inhibited by miR-376a in WT group, but not in MUT group (fig. 4B), which shows that there was a direct interaction between SNHG1 and miR-376a. To verify this finding, SNHG1 was overexpressed by OE-SNHG1 in OGD-HCMEC/ D3 cells. As expected, the mRNA concentration of miR-376a was inhibited by SNHG1 (fig. 4C). These results show that miR-376a was the direct target of lncRNA SNHG1.
In addition, we also studied whether SNHG1 can regulate apoptosis and inflammation of IS through miR-376a. Both SNHG1 and miR-376a mimics were cotransfected into OGD-HCMEC/D3 cells. The mRNA expression of IL-6, IL-1 β, and TNF-α was inhibited by the overexpression of SNHG1, which could be restored to the normal level by miR-376a mimics (fig. 4D). In addition, miR-376a mimics also blocked the antiapoptotic effect of SNHG1 (fig. 4E). SNHG1 upregulated the anti-apoptotic protein Bcl-2, while down-regulated the pro-apoptotic protein Bax and caspase-3, which were also reversed by miR-376a (fig. 4F). Therefore, we infer that the inflammation and apoptosis of IS cells can be reduced by the overexpression of lncRNA SNHG1 targeting miR-376a.
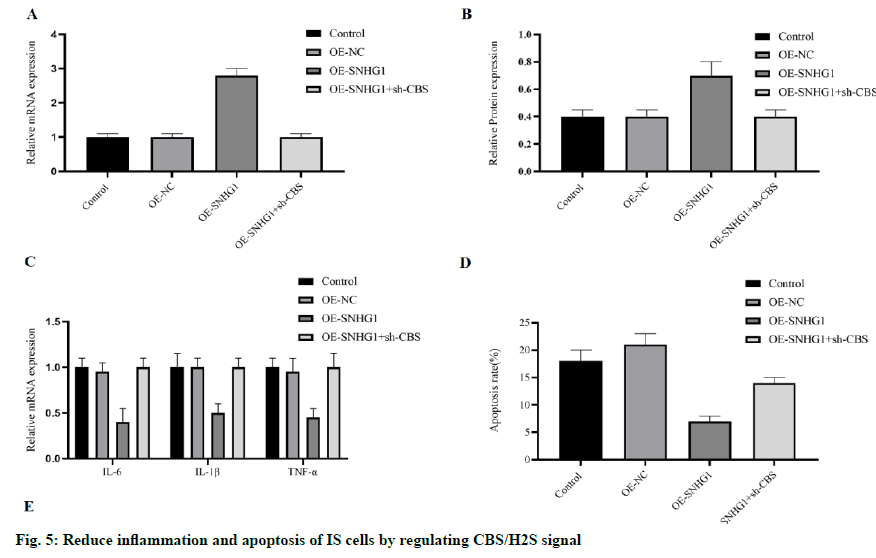
Based on the previous findings, the existence of a lncRNA SNHG1-miR-376a- CBS/H2S signal axis can be predicted. Then we try to reveal whether this signal axis exists in IS cells. Ectopic SNHG1 increased the concentration of mRNA and protein of CBS in OGDHCMEC/ D3 (fig. 5A and 5B), both of which were antagonized by sh-CBS. The inflammatory cytokines IL-6, IL-1 β, and TNF-α were all inhibited by SNHG1 in OGD-HCMEC/D3 and as expected, counteracted by sh-CBS (fig. 5C). We also detected the cell apoptosis, and sh-CBS counteracted the anti-apoptotic effect of SNHG1 on OGD-HCMEC/D3 cells (fig. 5D). In addition, when SNHG1 upregulated Bcl-2 and inhibited Bax and caspase-3, these effects were blocked by sh-CBS (fig. 5E). All these results lead to the conclusion that lncRNA SNHG1 can reduce the inflammation and apoptosis of IS cells by inhibiting miR-376a and upregulating CBS/H2S signal.
CBS/H2S signal plays an important role in maintaining the balance of brain tissue, which was a regulator in stroke[4,5]. Study found that miRNA and lncRNA influence the regulation of stroke-related processes through PI3K or Hypoxia-Inducible Factor (HIF)- 1 pathways[3,11]. However, the regulation mechanism between these factors and CBS/H2S signal was still unclear.
In this study, we found that in OGD-induced IS cell model, the expression of lncRNA SNHG1, CBS, and H2S were all down-regulated, while the expression of miR-376a was up-regulated (fig. 1). It was previously reported that the SNHG1 expression was up-regulated in IS and attenuated the IS-related damage by regulating miR-338/HIF-1α axis[3], miR-18/HIF-1α/vascular endothelial growth factor axis[16] or miR-199[17]. Our findings were consistent with these reports, although our results showed that miR-376 and CBS/H2S signals were affected. It was worth noting that CBS/ H2S signaling has previously been reported to reduce IS by inhibiting inflammation and apoptosis[7,8]. These evidences suggest that SNHG1/miR-376a/CBS/H2S may be a new anti-IS signal axis. We have shown that miR-376a promotes inflammation and apoptosis of IS by targeting CBS/H2S (fig. 2 and fig. 3). Recent studies have shown that miRNA can indeed participate in IS-related regulation, such as miR-152, miR-130, and other miRNAs[11,12]. However, the specific mechanisms of these miRNAs vary greatly. miR-152 regulates the Nrf2 signal, while miR-130 inhibits PTEN/PI3K/AKT signal pathway[10,11]. Some miRNAs were only observed to be dysfunctional, and their targets were not found, such as miR-376a[12,13]. We did observe that the trend of miR-376 in IS cells was similar to that previously reported. In addition, in this study, we first revealed that CBS, as a direct target of miR-376a, regulates the level of H2S, and finally affects the inflammation and apoptosis of IS cell model. The link between miR- 376a and apoptosis has also been reported in many other diseases, with different target genes, such as c-myc in lung cancer, neurociliary protein-1 NR in breast cancer, and Forkhead box protein P2 (FOXP 2) in lymphoma[18-20]. This difference may be caused by the complex regulatory networks between different tissues, cell types, and diseases. In addition, Su et al. reported that miR-376a was up-regulated in porcine coronary microembolization (CME) model, and bioinformatics analysis showed that it was associated with inflammation and apoptosis[21]. Our results may partly explain that miR-376a regulates inflammation and apoptosis in IS.
This study found that the inflammatory markers include C reactive protein, erythrocyte sedimentation rate and related apoptosis of IS cells can be reduced by inhibiting the expression of miR-376a, and overexpression of lncRNA SNHG1. It is cIearIy understood that apoptotic cell death plays an important role in inflammatory processes. This was consistent with the previous reports that SNHG1 has an anti-IS effect[3]. However, this mechanism was different from what we have found. It has been reported that SNHG1 reduces IS by regulating microRNA-338/HIF-1α rather than CBS/ H2S. This difference may be related to the different heredity and expression profiles of different cell lines. We have proved the direct interaction and regulation between SNHG1 and miR-376a, which expands the understanding of IS regulation mechanism.
In summary, we revealed that in IS, SNHG1 can inhibit apoptosis and inflammation, and play a neuroprotective function. These functions were mainly achieved by the inhibition of miR-376a. In conclusion, in IS cells, the increase of SNHG1 can inhibit the expression of miR- 376a, up-regulate CBS/H2S signal and reduce IS. This study broadens the scope and knowledge of IS-related regulatory networks, reveals the target of miR-376a in IS, and reveals a new regulatory signal axis lncRNA SNHG1/miR-376a/CBS/H2S in IS. These findings will support future studies on IS regulation and provide potential therapeutic targets for the clinical application.
Acknowledgement
This work was supported by Suzhou Ninth People’s Hospital.
Declaration of interest
The authors report no conflicts of interest.
References
- Radu RA, Terecoasă EO, Băjenaru OA, Tiu C. Etiologic classification of ischemic stroke: Where do we stand? Clin Neurol Neurosurg 2017;159:93-106.
- Del Brutto VJ, Chaturvedi S, Diener HC, Romano JG, Sacco RL. Antithrombotic therapy to prevent recurrent strokes in ischemic cerebrovascular disease: JACC Scientific Expert Panel. J Am Coll Cardiol 2019;74:786-803.
- Yang X, Zi XH. LncRNA SNHG1 alleviates OGD induced injury in BMEC via miR-338/HIF-1alpha axis. Brain Res 2019;1714:174-81.
- Narne P, Pandey V, Phanithi PB. Role of nitric oxide and hydrogen sulfide in ischemic stroke and the emergent epigenetic underpinnings. Mol Neurobiol 2019;56:1749-69.
- Nath N, Prasad HK, Kumar M. Cerebroprotective effects of hydrogen sulfide in homocysteine-induced neurovascular permeability: Involvement of oxidative stress, arginase, and matrix metalloproteinase-9. J Cell Physiol 2019;234:3007-19.
- Dou Y, Wang Z, Chen G. The role of hydrogen sulfide in stroke. Med Gas Res 2016;6:79-84.
- Zhang M, Wu X, Xu Y, He M, Yang J, Li J, et al. The cystathionine beta-synthase/hydrogen sulfide pathway contributes to microglia-mediated neuroinflammation following cerebral ischemia. Brain Behav Immun 2017;66:332-46.
- Wang JF, Li Y, Song JN, Pang HG. Role of hydrogen sulfide in secondary neuronal injury. Neurochem Int 2014;64:37-47.
- Theofilatos K, Korfiati A, Mavroudi S, Cowperthwaite MC, Shpak M. Discovery of stroke-related blood biomarkers from gene expression network models. BMC Med Genomics 2019;12:118.
- Zhang A, Qian Y, Qian J. MicroRNA-152-3p protects neurons from oxygen-glucose-deprivation/reoxygenation-induced injury through upregulation of Nrf2/ARE antioxidant signaling by targeting PSD-93. Biochem Biophys Res Commun 2019;517:69-76.
- Zheng T, Shi Y, Zhang J, Peng J, Zhang X, Chen K, et al. MiR-130a exerts neuroprotective effects against ischemic stroke through PTEN/PI3K/AKT pathway. Biomed Pharmacother 2019;117:109117.
- Li G, Morris-Blanco KC, Lopez MS, Yang T, Zhao H, Vemuganti R, et al. Impact of microRNAs on ischemic stroke: From pre- to post-disease. Prog Neurobiol 2018;163:59-78.
- van Kralingen JC, McFall A, Ord EN, Coyle TF, Bissett M, McClure JD, et al. Altered extracellular vesicle microRNA expression in ischemic stroke and small vessel disease. Transl Stroke Res 2019;10:495-508.
- Qi X, Zhang DH, Wu N, Xiao JH, Wang X, Ma W. ceRNA in cancer: possible functions and clinical implications. J Med Genet 2015;52:710-8.
- Zhang Y, Xu Y, Feng L, Li F, Sun Z, Wu T, et al. Comprehensive characterization of lncRNA-mRNA related ceRNA network cross 12 major cancers. Oncotarget 2016;7:64148.
- Zhang L, Luo X, Chen F, Yuan W, Xiao X, Zhang X, et al. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1alpha/VEGF signaling in ischemic stroke. J Cell Biochem 2018;119:5460-72.
- Wang Z, Wang R, Wang K, Liu X. Upregulated long noncoding RNA SNHG1 promotes the angiogenesis of brain microvascular endothelial cells after oxygen-glucose deprivation treatment by targeting miR-199a. Can J Physiol Pharmacol 2018;96:909-15.
- Wang Y, Cong W, Wu G, Ju X, Li Z, Duan X, et al. MiR-376a suppresses the proliferation and invasion of non-small-cell lung cancer by targeting c-Myc. Cell Biol Int 2018;42:25-33.
- Zhang L, Chen Y, Wang H, Zheng X, Li C, Han Z. miR-376a inhibits breast cancer cell progression by targeting neuropilin-1 NR. Onco Targets Ther 2018;11:5293-302.
- Yao S, Liu Y, Yao Z, Zhao Y, Wang H, Xu Y, et al. MicroRNA-376a regulates cell proliferation and apoptosis by targeting forkhead box protein P2 in lymphoma. Oncol Lett 2018;16:3169-76.
- Su Q, Li L, Zhao J, Sun Y, Yang H. MiRNA expression profile of the myocardial tissue of pigs with coronary microembolization. Cell Physiol Biochem 2017;43:1012-24.