- *Corresponding Author:
- Zhengbiao Liu
Department of Orthopedics, Suzhou Industrial Park Xinghu Hospital, Suzhou, Jiangsu Province 215000, China
E-mail: xhyylzb2020@163.com
| Date of Received | 14 November 2021 |
| Date of Revision | 07 August 2022 |
| Date of Acceptance | 27 June 2023 |
| Indian J Pharm Sci 2023;85(4):912-918 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To explore the influence of the long-non coding RNA-H19/microRNA-140-5p/Wnt1 axis on the osteogenic differentiation in bone marrow mesenchymal stem cells. First, we used dual-luciferase reporter gene detection to verify long-non coding-H19/miRNA-140-5p/Wnt1 regulatory axis targeting relationship. Then long-non coding-H19 was overexpressed or silenced in bone marrow mesenchymal stem cells and then long-non coding-H19, microRNA-140-5p and Wnt1 expression were assessed using quantitative real-time polymerase chain reaction and cell activity was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide. Then the osteogenic differentiation culture was performed, formation of calcium nodules were assessed via Alizarin red staining, while the expression of osteocalcin and runt-related transcription factor 2 detected via Western blot. Wild type long-non coding RNA-H19 group and wild type-Wnt1 group showed significantly decreased luciferase activity in dual-luciferase reporter assays (p<0.05). After overexpressed long-non coding RNA-H19, the microRNA-140-5p expression decreased and Wnt1 expression increased (p<0.05), and while silenced the long-non coding RNA-H19 expression, the opposite result was found (p<0.05). In terms of cell viability, there were no significant differences between groups after long-non coding RNA-H19 was overexpressed or silenced (p>0.05). After overexpressed long-non coding RNA-H19, calcium nodules formation, mineralized bone matrix level, the expression of osteocalcin and runt-related transcription factor 2 protein significantly increased (p<0.05), and while silenced the long-non coding RNA-H19 expression, the opposite result was found (p<0.05). Long-non coding RNA-H19 overexpression promotes the bone marrow mesenchymal stem cells osteogenic differentiation via targeting microRNA-140-5p/Wnt1 signaling pathway.
Keywords
Long-non coding RNA-H19, microRNA-140-5p, Wnt1, osteogenic differentiation, bone marrow mesenchymal stem cells
As bone tissue engineering technology has advanced, the use of tissue engineering to construct tissue engineering bone has become one of the most promising methods of bone repair over the past few years[1,2]. Currently, Mesenchymal Stem Cells (MSCs) are used as seed cells for bone tissue engineering. These MSCs come from Bone Marrow Mesenchymal Stem Cells (BMSCs), adipose tissue, umbilical cord and umbilical cord blood[3]. BMSCs have low immunogenicity, multidirectional differentiation ability and good transplant ability[4]. Under specific induction conditions, it can differentiate into chondrocytes, osteoblasts, adipocytes, etc. Directionally inducing BMSCs to differentiate into osteoblasts can provide treatment methods for bone remodeling, fracture repair and osteoporosis. Therefore, it is an important "seed cell" in bone tissue engineering.
Long non-coding RNAs (lncRNAs), as endogenous transcription molecules are more than 200 nucleotides in length. They usually participate in the biological process of cells at the transcription level, post-transcription level and epigenetic level. Among them, as a microRNA (miRNA) molecular sponge, regulation of miRNA to influence expression of related genes is one of the most important biological functions of lncRNAs[5,6]. Our previous study has found miRNA-140-5p expression significantly decreased during BMSCs’ osteogenic differentiation process and which is promoted via miRNA-140- 5p silence[7]. In order to analyze the upstream lncRNAs targeting miRNA-140-5p, we used Star base Database (http://starbase.sysu.edu.cn), and the result showed that lncRNA-H19 was one of potential upstream lncRNAs of miRNA-140-5p. Previous studies, lncRNA-H19 has been shown to participate in dentin differentiation in dental pulp stem cells[8], calcification of articular cartilage[9], ovarian cancer[10], pneumonia[11] and hypoxic-ischemic brain damage[12] by acting on miRNA-140-5p. However, whether lncRNA-H19 is related to osteogenic differentiation process of BMSCs is still unclear. The study was designed to investigate influence of lncRNA-H19 on osteogenic differentiation of BMSCs and possible mechanisms of action were explored.
Materials and Methods
Cell line:
This study was approved via Ethics Committee of the Suzhou Industrial Park Xinghu Hospital. Chinese company Beina Chuanglian Biotechnology Co., Ltd provided the human BMSCs. The BMSCs were recovered and passaged after the growth density reached about 85 %. The Dulbecco's Modified Eagle Medium (DMEM) (Hyclone, United States of America (USA)) was removed and 1 ml 0.25 % trypsin was used for digestion. A microscope was used to observe the cells and when cell’s morphology became round, a FBS and 1 ml of DMEM medium was applied to stop the digestion of the cells. Cells were suspended and transferred to an EP tube, then cells were centrifuged and suspended, and then the cells were passaged.
Cell transfection:
Three generations of BMSCs were cultured in 6-well culture plates (3×105/ml). After the cells adhered, they were transfected with plasmid cloning Deoxyribonucleic Acid (pcDNA)-lncRNA-H19 (RiboBio, China), pcDNA-Negative Control (NC) (RiboBio, China), small interfering (si)-lncRNA-H19 (RiboBio, China), si-NC (RiboBio, China) according to lipofectamine 2000 (ThemoFisher, USA) transfection instructions and blank control groups were set up. The cells were incubated for 5 h in 5 % Carbon dioxide (CO2) at 37° incubator. Then, DMEM medium was applied to replace the transfection medium, and the cultivation was continued for 48 h and then cells were collected for the next experiment.
3-(4,5-Dimethylthiazol-2-yl)-2,5 Diphenyl Tetrazolium Bromide (MTT) assay for cell viability:
After transfection for 48 h, 100 μl MTT (0.5 mg/ml) solution (Beyotime, China) was applied to the well of each group, cells were incubated at 37°, 5 % CO2 incubator for 5 h, then wells were added another 100 μl 20 % Sodium Dodecyl Sulfate (SDS) (containing 50 % dimethylformamide) solution and then continuously incubated at 37° for 24 h. A 570 nm wavelength was used to detect the Optical Density (OD) values of each well.
The expression levels of lncRNA-H19, miRNA- 140-5p and Wnt1 were detected by quantitative Real-Time Polymerase Chain Reaction (qRT-PCR). TaqMan miRNA extraction kit (Applied Biosystems, USA) was used to extract total Ribonucleic Acid (RNA) from the BMSCs according to instructions. For lncRNA-H19 and Wnt1 mRNA, the TransScript® First-Strand complementary DNA (cDNA), Synthesis Super Mix (TransGen, China) was used to synthesize the cDNA. For miRNA-140-5p, the cDNA was synthesized by TransScript® miRNA First-Strand cDNA Synthesis Super Mix (Trans gen, China). The internal references were Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH) and U6. The relative primers were as follows; lncRNA-H19 forward 5'-GCACCTTGGACATCTGGAGT-3' and reverse 5'-TTCTTTCCAGCCCTAGCTCA-3'; miRNA-140-5p forward 5'-ACACTCCAGCTG GGAGGCGGGGCGCCGCGGGA-3' and reverse 5'-CTCAACTGGTGTCGTGGA-3'; Wnt1 forward 5'- TGGTTTGCAAAGACCACCTCCA-3' and reverse 5'-TGATTCCAGGAGGCAAACGCAT-3'; GAPDH forward 5'-AGAAGGCTGGGGCTCATTTG-3' and reverse 5'- AGGGGCCATCCACAGTCTTC-3'; U6 forward 5'-TGCGGGTGCTCGCTTCGGCAGC-3' and reverse 5'-CCAGTGCAG GGTCCGAGGT-3'. Using the 2-ΔΔCt method to analyze genes relative expression level.
Osteogenic differentiation culture:
The transfected or transfected BMSCs were digested into single cell, cultured in 24-well culture plates containing DMEM medium, at a density of 5×105/ ml in a 37°, 5 % CO2 incubator under saturated humidity. When cell growth fusion reached about 50 %, induction solution for osteogenic differentiation (DMEM containing 10 μmol/l dexamethasone, 10 mmol/l Beta (β)-glycerol sodium phosphate, 50 μmol/l vitamin C and 10 % Fetal Bovine Serum (FBS)) was used instead of culture medium, and cultured for 14 d. They were divided into control, si- NC, pcDNA-NC, si-lncRNA-H19 and pcDNA-H19 groups.
Alizarin red staining:
14 d after cells differentiation, culture solution was removed and 4 % paraformaldehyde was added for 30 min. Following three rinses with 0.01 mol/l Phosphate Buffer Solution (PBS), 2 % Alizarin red staining solution (Beyotime, China) were added, and incubated the sample for 10 min. Under a microscope, parts of the cells in each group were examined. Other remaining cells in each group extracted Alizarin red with 100 mm cetylpyridinium chloride solution (Sigma, USA) to quantify the cell mineralization and 570 nm wavelength was used to determine the OD.
Western blot analysis:
We extracted the total protein using Radioimmunoprecipitation Assay Buffer (RIPA) containing phenylmethylsulphonyl fluoride (Beyotime, China) as a protease inhibitor. Briefly, following transfer of protein to Polyvinylidene Difluoride (PVDF) membrane, PVDF membrane were successively incubated with primary and secondary antibodies. Antibodies were as follows; 1:400 diluted rabbit anti-Wnt1 polyclonal antibody (Abcam, UK), 1:400 diluted rabbit anti-Runt-Related Transcription Factor 2 (RUNX2) polyclonal antibody (Abcam, UK), 1:1000 diluted mouse anti-β-actin monoclonal antibody (Abcam, UK), 1:1000 diluted Horseradish Peroxidase (HRP)-labeled goat anti-rabbit Immunoglobulin G (IgG) antibody (Abcam, United Kingdom (UK)) or 1:1000 diluted HRP-labeled goat anti-mouse IgG antibody (Abcam, UK). The images and quantity of protein band was assessed with Molecular Imager ChemiDoc XRS System (BioRad, USA).
Luciferase reporter gene experiment:
Wild Type (WT)-lncRNA-H19 sequence, Mutant Type (MT)-lncRNA-H19 sequence, WT-Wnt1 3′ Untranslated Regions (3′ UTRs) sequence and MTWnt1 3′UTR sequence was separately cloned to the luciferase reporter vector psiCHECK-2 (Synbio, China). Lipofectamine 2000 (Themo Fisher, USA) was used to co-transfect HEK293 cells with psiCHECK-2 and miRNA-140-5p mimic (Themo Fisher, USA ). We incubated cells for 48 h in a saturated humidity, 37°, 5 % CO2 incubator, lysed them, and analyzed them with the dual-luciferase reporter system (Promega, USA).
Statistical analysis:
To analyze the data obtained in this study, Statistical Package for the Social Sciences (SPSS) 21.0 software was used. We presented the data as mean+standard deviation. One-way Analysis of Variance (ANOVA) or independent t tests were used to compare groups. The significance level was set at p<0.05.
Results and Discussion
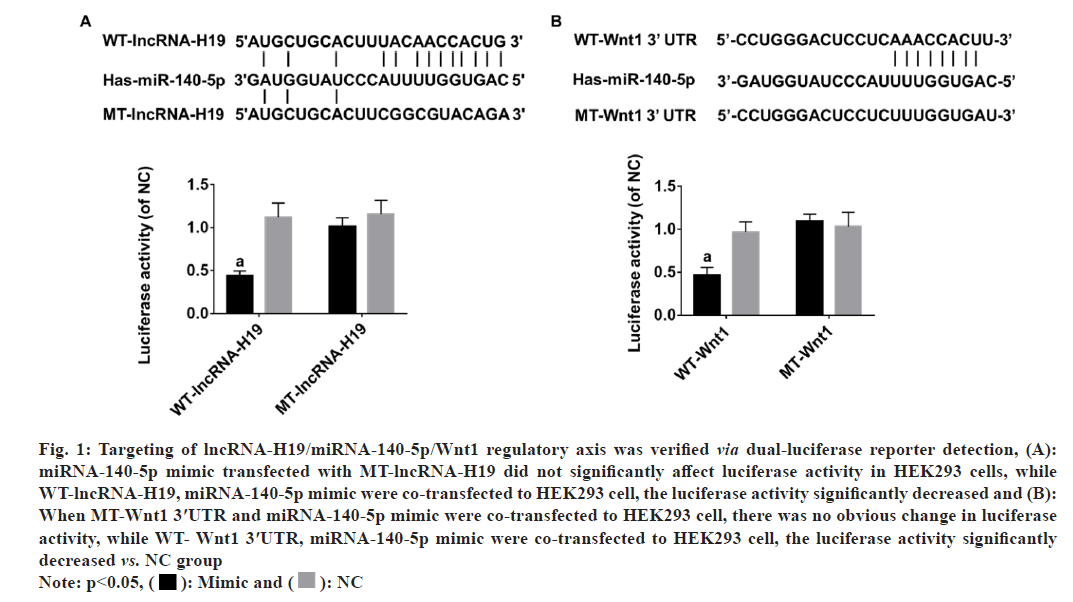
When MT-lncRNA-H19, miRNA-140-5p mimic were co-transfected to HEK293 cells, result of dualluciferase reporter assay showed no obvious change in luciferase activity, while WT-lncRNA-H19 and miRNA-140-5p mimic were co-transfected into HEK293 cells, luciferase activity markedly decreased (p<0.05) (fig. 1A). When MT-Wnt1 3′UTR, miRNA- 140-5p mimic were co-transfected to HEK293 cell, result of dual-luciferase reporter assay showed there was no obvious change in luciferase activity, while WT-Wnt1 3′UTR, miRNA-140-5p mimic were co-transfected to HEK293 cell, luciferase activity markedly decreased (p<0.05) (fig.1B).
Fig. 1: Targeting of lncRNA-H19/miRNA-140-5p/Wnt1 regulatory axis was verified via dual-luciferase reporter detection, (A):
miRNA-140-5p mimic transfected with MT-lncRNA-H19 did not significantly affect luciferase activity in HEK293 cells, while
WT-lncRNA-H19, miRNA-140-5p mimic were co-transfected to HEK293 cell, the luciferase activity significantly decreased and (B):
When MT-Wnt1 3′UTR and miRNA-140-5p mimic were co-transfected to HEK293 cell, there was no obvious change in luciferase
activity, while WT- Wnt1 3′UTR, miRNA-140-5p mimic were co-transfected to HEK293 cell, the luciferase activity significantly
decreased vs. NC group
Note: p<0.05, ( ): Mimic and (
): Mimic and ( ): NC
): NC
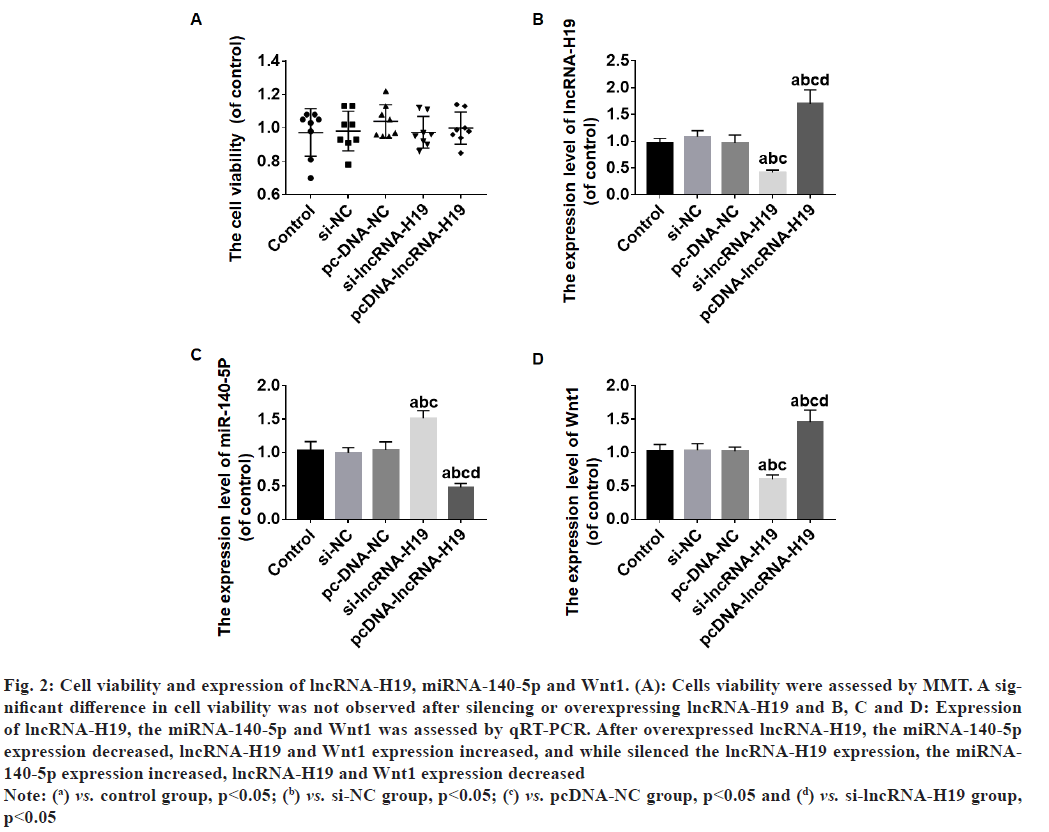
The viability of the cells was not significantly different between the groups after lncRNA-H19 was overexpressed or silenced (p>0.05) (fig.2A). After overexpressed lncRNA-H19, the miRNA-140- 5p expression decreased, lncRNA-H19 and Wnt1 expression increased (p<0.05) (fig. 2B-fig. 2D), and while silenced the lncRNA-H19 expression, the miRNA-140-5p expression increased, lncRNA-H19 and Wnt1 expression decreased (p<0.05) (fig. 2Bfig. 2D).
Fig. 2: Cell viability and expression of lncRNA-H19, miRNA-140-5p and Wnt1. (A): Cells viability were assessed by MMT. A significant
difference in cell viability was not observed after silencing or overexpressing lncRNA-H19 and B, C and D: Expression
of lncRNA-H19, the miRNA-140-5p and Wnt1 was assessed by qRT-PCR. After overexpressed lncRNA-H19, the miRNA-140-5p
expression decreased, lncRNA-H19 and Wnt1 expression increased, and while silenced the lncRNA-H19 expression, the miRNA-
140-5p expression increased, lncRNA-H19 and Wnt1 expression decreased
Note: (a) vs. control group, p<0.05; (b) vs. si-NC group, p<0.05; (c) vs. pcDNA-NC group, p<0.05 and (d) vs. si-lncRNA-H19 group,
p<0.05
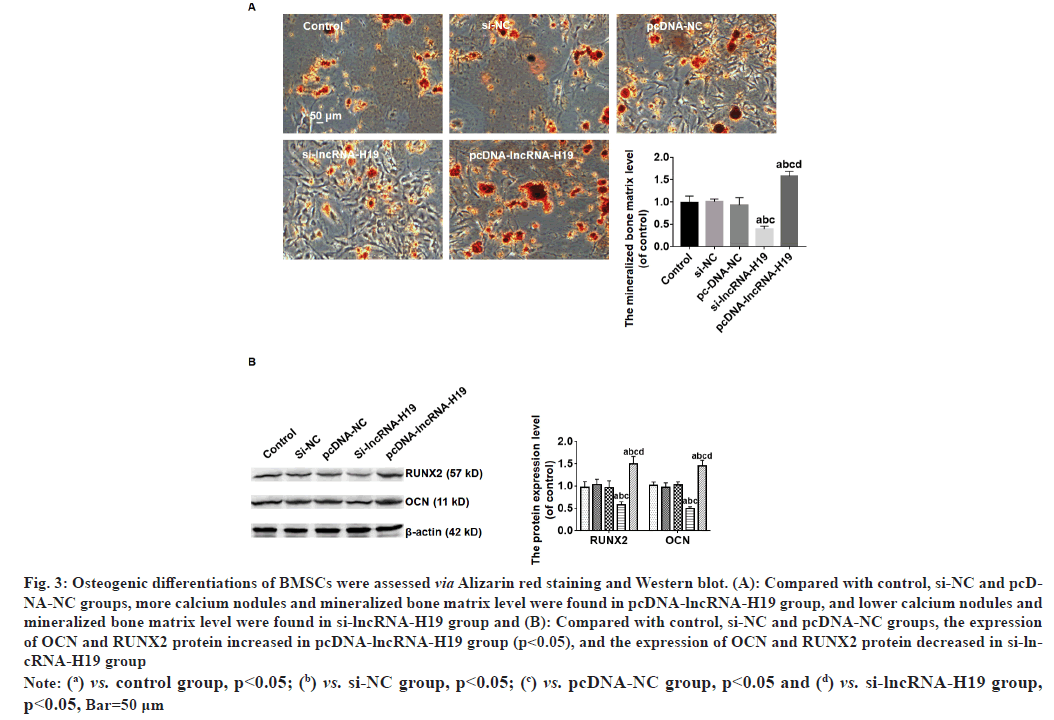
Compared with control, si-NC and pcDNA-NC groups, more calcium nodules and mineralized bone matrix level was found in pcDNA-lncRNA-H19 group, and lower calcium nodules and mineralized bone matrix level were found in si-lncRNA-H19 group (p<0.05) (fig. 3A). A marked increase in expression of Osteocalcin (OCN), RUNX2 proteins was observed in the pcDNA-lncRNA-H19 group compared with control, si-NC and pcDNA-NC groups (p<0.05), and the expression of OCN and RUNX2 protein significantly decreased in si-lncRNA-H19 group (p<0.05) as shown in fig. 3B.
Fig. 3: Osteogenic differentiations of BMSCs were assessed via Alizarin red staining and Western blot. (A): Compared with control, si-NC and pcDNA-
NC groups, more calcium nodules and mineralized bone matrix level were found in pcDNA-lncRNA-H19 group, and lower calcium nodules and
mineralized bone matrix level were found in si-lncRNA-H19 group and (B): Compared with control, si-NC and pcDNA-NC groups, the expression
of OCN and RUNX2 protein increased in pcDNA-lncRNA-H19 group (p<0.05), and the expression of OCN and RUNX2 protein decreased in si-lncRNA-
H19 group
Note: (a) vs. control group, p<0.05; (b) vs. si-NC group, p<0.05; (c) vs. pcDNA-NC group, p<0.05 and (d) vs. si-lncRNA-H19 group,
p<0.05, Bar=50 μm
Bone defect is one of the most common diseases in clinical orthopedics. The main reasons for bone defect are trauma, bone tumor resection and infection. Despite the rapid development of related medical technologies, the repair of bone defect is still one of the major issues that need to be solved[13,14]. With the development of tissue engineering technology, it provides a very promising treatment for bone defect repair. Stem cells, absorbable biological scaffolds, and regulatory factors are the three major elements of tissue engineering[15]. However, the directional regulation of stem cell’s osteogenic differentiation is still a hot and difficult point of study and it is also one of the key points restricting its clinical application[16]. Osteogenic differentiation is a dynamic process with time series, and it is also a complex and highly regulated process. It can be roughly divided into four consecutive stages; lineage orientation, cell proliferation, extracellular matrix maturation and matrix mineralization. The osteogenic differentiation process is affected by many factors which regulate multiple signaling pathways of osteoblast differentiation and ultimately promote the expression of osteoblast differentiation genes[17].
miRNAs have many important regulatory functions in cell through inhibiting the target mRNAs translation process and thereby inhibiting protein synthesis[18-20]. Researchers are becoming increasingly concerned about how it influences stem cell differentiation[21-23]. Our previous study has found that the miRNA- 140-5p expression decreased during osteogenic differentiation process of BMSCs, and BMSCs osteogenic differentiation is promoted by miRNA- 140-5p down-regulation through targeting the Wnt l signal pathway[7], however, the relative mechanism about miRNA-140-5p low expression in this process is still unclear. LncRNAs, as a miRNA molecular sponge, LncRNAs have increasingly been studied in relation to osteogenic differentiation[24-28]. The predicted results of Star base Database indicated that lncRNA-H19 may target the miRNA-140- 5p. Our result of dual-luciferase reporter detection showed WT-lncRNA-H19, miRNA-140-5p mimic were co-transfected into HEK293 cell, the luciferase activity significantly decreased and result of qRTPCR showed miRNA-140-5p expression decreased in BSMCs after overexpressed lncRNA-H19. The abovementioned results indicated LncRNAs act as the miRNA-140-5p molecular sponge in osteogenic differentiation process of BSMCs. In the complex process of osteoblast differentiation, there are three most important signaling pathways; BMP, Wnt and Notch signaling pathways[29-31]. In order to determine target gene of miRNA-140-5p, we used Target Scan software. The results indicated that Wnt1 is likely one of the miRNA-140-5p target genes. Our result of dual-luciferase reporter assay showed WT-Wnt1 3′UTR, miRNA-140-5p mimic were co-transfected to HEK293 cell, the luciferase activity significantly decreased, and result of qRT-PCR showed Wnt1 expression increased in BSMCs while miRNA-140- 5p decreased after overexpressed lncRNA-H19. The abovementioned results verified Wnt1 is miRNA- 140-5p’s target gene.
To investigate how lncRNA-H19 affects osteogenic differentiation of BMSCs, lncRNA-H19 expression were upregulated or silenced in this research. The result of MTT detection showed the OD values of all groups were not significantly different, which suggests that the lncRNA-H19 expression change has no obvious effect on the cell viability of BMSCs. After overexpressed lncRNA-H19, calcium nodules formation, mineralized bone matrix level and expression of OCN and RUNX2 proteins (osteogenic differentiation marker protein) significantly increased, and while silenced the lncRNA-H19 expression, the opposite result was found. The above mentioned result indicated lncRNA-H19 overexpression enhanced osteogenic differentiation of BMSCs and lncRNA-H19 low expression inhibited the osteogenic differentiation.
In summary, our results indicated lncRNA-H19 overexpression promote osteogenic differentiation of BMSCs via targeting miRNA-140-5p/Wnt1 signaling pathway. It provides potential therapeutic targets for bone defects treatment via regulating osteogenic differentiation of BMSCs.
Ethics approval
Ethics Committee of Suzhou Industrial Park Xinghu Hospital has approved this study.
Funding:
This work was supported by Suzhou Science and Technology Plan Project (SYS2020072).
Authors' contributions:
The experiments were designed by Zhengbiao Liu. And were carried out by Renwei Wang, Hong Zhang, Zaiqing Zhang and Wei Liu. The data were analyzed by Reyuan Zang and Chenyu Wang. The paper was written by Zhengbiao Liu. All authors read and approved the final version of the paper.
Conflict of interests:
The authors declared no conflict of interests.
References
- Liu F, Sun T, An Y, Ming L, Li Y, Zhou Z, et al. The potential therapeutic role of extracellular vesicles in critical-size bone defects: Spring of cell-free regenerative medicine is coming. Front Bioeng Biotechnol 2023;11:1050916.
[Crossref] [Google Scholar] [PubMed]
- Zhang R, Hou Y, Sun L, Liu X, Zhao Y, Zhang Q, et al. Recent advances in carbon dots: Synthesis and applications in bone tissue engineering. Nanoscale 2023;15(7):3106-19.
- Minervini G, Del Mondo D, Russo D, Cervino G, D’Amico C, Fiorillo L. Stem cells in temporomandibular joint engineering: State of art and future persectives. J Craniofac Surg 2022;33(7):2181-7.
[Crossref] [Google Scholar] [PubMed]
- Xu S, Xu Y. Recent progress of BMSCs acting as seeding cell for tissue engineered cartilage. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi 2008;22(2):163-6.
[Google Scholar] [PubMed]
- Huang T, Wu Z, Zhu S. The roles and mechanisms of the lncRNA-miRNA axis in the progression of esophageal cancer: A narrative review. Jo Thorac Dis 2022;14(11):4545.
[Crossref] [Google Scholar] [PubMed]
- Lv Y, Wang Y, Zhang Z. Potentials of lncRNA–miRNA–mRNA networks as biomarkers for laryngeal squamous cell carcinoma. Hum Cell 2023;36(1):76-97.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Xu H, Zhou Z. Low expression of miRNA-140-5p promotes osteogenic differentiation of human bone marrow mesenchymal stem cells. Chin J Clin Res 2022;35(8):1041-50.
- Zhong J, Tu X, Kong Y, Guo L, Li B, Zhong W, et al. LncRNA H19 promotes odontoblastic differentiation of human dental pulp stem cells by regulating miR-140-5p and BMP-2/FGF9. Stem Cell Res Ther 2020;11(1):202.
[Crossref] [Google Scholar] [PubMed]
- Yang B, Xu L, Wang S. Regulation of lncRNA-H19/miR-140-5p in cartilage matrix degradation and calcification in osteoarthritis. Ann Palliat Med 2020;9(4):1896-904.
[Crossref] [Google Scholar] [PubMed]
- Xu H, Ding Y, Yang X. Overexpression of long noncoding RNA H19 downregulates miR-140-5p and activates PI3K/AKT signaling pathway to promote invasion, migration and epithelial-mesenchymal transition of ovarian cancer cells. Biomed Res Int 2021;2021:6619730.
[Crossref] [Google Scholar] [PubMed]
- Yang H. Silencing of long non-coding RNA H19 alleviates lipopolysaccharide (LPS)-induced apoptosis and inflammation injury by regulating miR-140-5p/TLR4 axis in cell models of pneumonia. Curr Mol Med 2023;23(3):275-84.
[Crossref] [Google Scholar] [PubMed]
- Lu Q, Hou HM, Li S, Yuan J, Liu H, Xu Y. Long non-CODING RNA H19 deteriorates hypoxic-ischemic brain damage by interacting with microRNA-140-5p and STAT3. Nanoscale Res Lett 2022;17(1):43.
[Crossref] [Google Scholar] [PubMed]
- Pan Z, Huang F, Li J, Tang X. Current concepts of diagnostic techniques and measurement methods for bone defect in patient with anterior shoulder instability. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi 2019;33(6):762-7.
[Crossref] [Google Scholar] [PubMed]
- Wang W, Yeung KW. Bone grafts and biomaterials substitutes for bone defect repair: A review. Bioact Mater 2017;2(4):224-47.
[Crossref] [Google Scholar] [PubMed]
- Roi A, Ardelean LC, Roi CI, Boia ER, Boia S, Rusu LC. Oral bone tissue engineering: Advanced biomaterials for cell adhesion, proliferation and differentiation. Materials 2019;12(14):2296.
[Crossref] [Google Scholar] [PubMed]
- Salazar-Noratto GE, Luo G, Denoeud C, Padrona M, Moya A, Bensidhoum M, et al. Understanding and leveraging cell metabolism to enhance mesenchymal stem cell transplantation survival in tissue engineering and regenerative medicine applications. Stem Cells 2020;38(1):22-33.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Liu S, Li J, Zhao S, Yi Z. Roles for miRNAs in osteogenic differentiation of bone marrow mesenchymal stem cells. Stem Cell Res Ther 2019;10(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Deng H, Sun C, Sun Y, Li H, Yang L, Wu D, Gao Q, Jiang X. Lipid, protein, and microRNA composition within mesenchymal stem cell-derived exosomes. Cell Reprogram 2018;20(3):178-86.
[Crossref] [Google Scholar] [PubMed]
- Ferreira AF, Calin GA, Picanço-Castro V, Kashima S, Covas DT, de Castro FA. Hematopoietic stem cells from induced pluripotent stem cells–considering the role of microRNA as a cell differentiation regulator. J Cell Sci 2018;131(4):jcs203018.
[Crossref] [Google Scholar] [PubMed]
- Park JH, Theodoratou E, Calin GA, Shin JI. From cell biology to immunology: Controlling metastatic progression of cancer via microRNA regulatory networks. Oncoimmunology 2016;5(11):e1230579.
[Crossref] [Google Scholar] [PubMed]
- Gao Y, Qiao H, Zhong T, Lu Z, Hou Y. MicroRNA‑29a promotes the neural differentiation of rat neural stem/progenitor cells by targeting KLF4. Mol Med Rep 2020;22(2):1008-16.
[Crossref] [Google Scholar] [PubMed]
- Bame M, McInnis MG, O'Shea KS. MicroRNA alterations in induced pluripotent stem cell-derived neurons from bipolar disorder patients: Pathways involved in neuronal differentiation, axon guidance, and plasticity. Stem Cells Dev 2020;29(17):1145-59.
[Crossref] [Google Scholar] [PubMed]
- Liu Y, Liu X, Ye P, Zhang X, Schilling AF, Yonezawa T, Gao G, Cui X. MicroRNA-191 regulates differentiation and migration of mesenchymal stem cells and their paracrine effect on angiogenesis. Biotechnol Lett 2020;42(9):1777-88.
[Crossref] [Google Scholar] [PubMed]
- Qu HL, Sun LJ, Li X, Liu F, Sun HH, He XT, et al. Long non‐coding RNA AC018926. 2 regulates palmitic acid exposure‐compromised osteogenic potential of periodontal ligament stem cells via the ITGA2/FAK/AKT pathway. Cell Prolif 2023:e13411.
[Crossref] [Google Scholar] [PubMed]
- Wang H, Fan M, An Y, He D. Molecular mechanism of long noncoding RNA SNHG14 in osteogenic differentiation of bone marrow-derived mesenchymal stem cells through the NEDD4L/FOXA2/PCP4 axis. Stem Cells Int 2023;2023:7545635.
[Crossref] [Google Scholar] [PubMed]
- Zhang Q, Long Y, Jin L, Li C, Long J. Non-coding RNAs regulate the BMP/Smad pathway during osteogenic differentiation of stem cells. Acta Histochem 2023;125(1):151998.
[Crossref] [Google Scholar] [PubMed]
- Sufianov A, Beilerli A, Begliarzade S, Ilyasova T, Kudriashov V, Liang Y, et al. The role of noncoding RNAs in the osteogenic differentiation of human periodontal ligament-derived cells. Noncoding RNA Res 2022;8(1):89-95.
[Crossref] [Google Scholar] [PubMed]
- Saranya I, Akshaya RL, Selvamurugan N. Regulation of Wnt signaling by non-coding RNAs during osteoblast differentiation. Differentiation 2022;128:57-66.
- Nishimura R, Hata K, Kida J. Regulation of osteoblasts and chondrocytes by Wnt signaling. Clin Calcium 2019;29(3):299-307.
[Google Scholar] [PubMed]
- Mahendra CK, Tan LT, Lee WL, Yap WH, Pusparajah P, Low LE, et al. Angelicin—A Furocoumarin compound with vast biological potential. Front Pharmacol 2020;11:366.
[Crossref] [Google Scholar] [PubMed]
- Zieba JT, Chen YT, Lee BH, Bae Y. Notch signaling in skeletal development, homeostasis and pathogenesis. Biomolecules 2020;10(2):332.
[Crossref] [Google Scholar] [PubMed]