- *Corresponding Author:
- Min Tang
Medical College Department, Hunan Polytechnic of Environment and Biology, Hunan 421000, China
E-mail: tangmin33934@163.com
| This article was originally published in a special issue,“Trends in Therapeutic Management of Various Clinical Conditions II” |
| Indian J Pharm Sci 2021:83(2) spl issue 2;76-80 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In order to study the mechanism of abnormal deoxyribonucleic acid methylation in the process of lung cancer in rats, Wistar rats are used as the research object. The lung cancer model of rats is established by bronchial perfusion of carcinogens. Hematoxylin-eosin staining is used to observe the pathological changes of lung tissue in rats. The overall methylation level of genome is analyzed by immunohistochemistry, and the methylation status is detected by methylation specific polymerase chain reaction. The results show that the nuclei of normal bronchial epithelial cells are brown and yellow uder optical microscope. With the development of carcinogenesis, the staining gradually fades to light yellow in the infiltrating nuclei of cancer cells. Compared with normal bronchial epithelial cells, the optical density and integrated optical density values of 5-mC immunohistochemistry decrease. The methylation frequency of p16 gene in atypical hyperplasia, carcinoma in situ and invasive cancer tissues is significantly higher than that in normal and proliferative tissues. The methylation frequency of p57 gene is significantly higher in carcinoma in situ and invasive tissues than in normal and proliferative tissues. Abnormal methylation-mediated down-regulation of p16 and p57 gene transcription may be an important molecular biological event in carcinogenesis of lung cancer rats induced by carcinogens. There are still some shortcomings in the research process, but the results of the study still provide some reference and guidance for exploring more precise mechanism in the future. Therefore, this study is a significant research topic.
Keywords
Abnormal deoxyribonucleic acid methylation, lung cancer, Hematoxylin-eosin staining, immunohistochemistry, methylation specific polymerase chain reaction
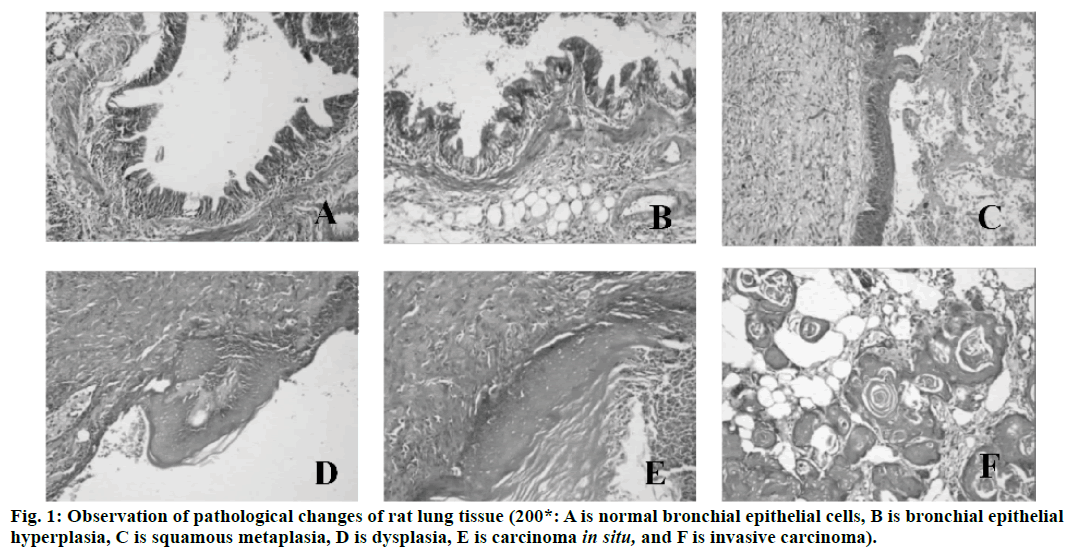
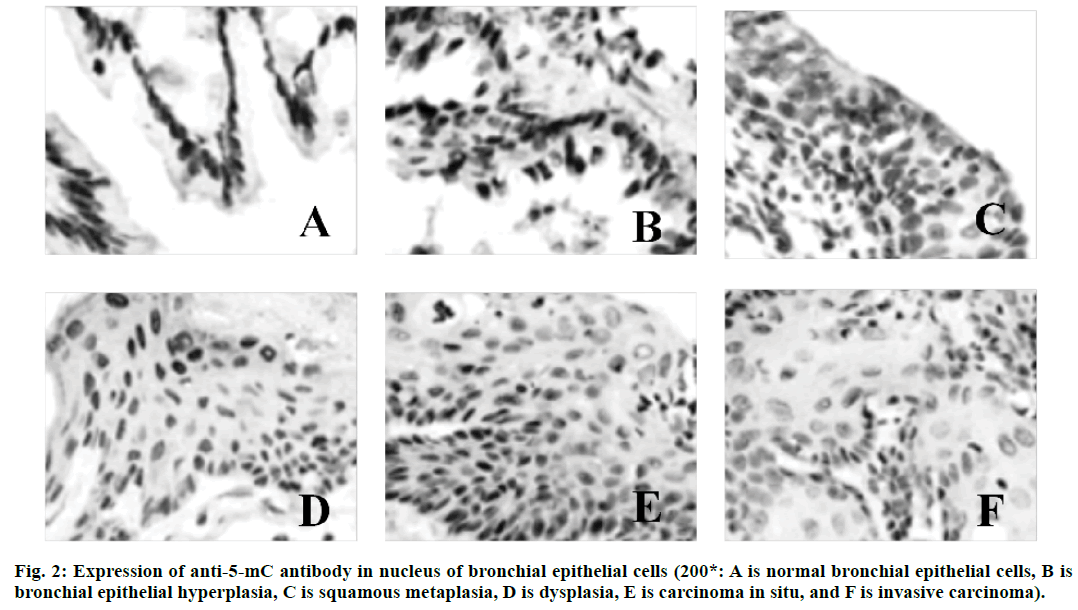
Lung cancer is a malignant lung cancer, which results from uncontrolled growth of lung tissue cells[1]. As one of the most dangerous malignant tumors to human health and life, its morbidity and mortality rate are also growing fastest. Without treatment, cancer cells will metastasize to adjacent tissues or other parts of the body[2]. High risk factors of lung cancer include smoking, ionizing radiation, chronic lung infection, air pollution, occupational and environmental exposure, heredity, etc[3]. According to the severity of the disease, cough, hemoptysis or hemoptysis in sputum, chest pain, fever, emaciation, cachexia or other extrapulmonary symptoms may occur[4]. Lung cancer can be found on chest radiography or tomography, and it can be diagnosed by bronchoscopy or CT-guided biopsy[5]. Deoxyribonucleic acid (DNA) methylation is a form of chemical modification of DNA, which can alter genetic performance without altering DNA sequence[6]. Essentially, it refers to the chemical modification of a methyl group by covalent bonding of a specific base group on DNA sequence with S-adenosyl methionine as a methyl donor under the catalysis of DNA methyltransferase[7]. DNA methylation can cause changes in chromatin structure, DNA conformation, DNA stability and the way DNA interact with proteins, thus controlling gene expression[8]. DNA methylation reaction can be divided into two types, which are ab initio methylation and retention methylation[9]. As a stable modification state, DNA methylation can be inherited to newborn offspring DNA along with the process of DNA replication, which is an important epigenetic mechanism[10]. In conclusion, there are few studies on abnormal DNA methylation in the carcinogenesis of lung cancer. Therefore, in this study, the rat model of lung cancer is established by carcinogens, and the mechanism of DNA abnormal methylation in the process of lung cancer in rats is explored, so as to provide new ideas for the treatment of lung cancer in the future. Ninety healthy Wistar rats (Nanjing Junke Bioengineering Co., Ltd.,China) are selected, half male and half female, aged 6-8 w, weighing about 200 g. They are randomly divided into experimental group (80 rats) and control group (10 rats). The operation of animal experiments conforms to the International Regulations on the Protection and Management of Animals and is approved by the ethical committee of The Second Affiliated Hospital, University of South China. From 3 d before perfusion to 5 d after perfusion, each rat is injected with 20,000 units of penicillin (Shanghai Hengfei Biotechnology Co., Ltd., China) and 50 mg of streptomycin (Shanghai Hengfei Biotechnology Co., Ltd., China) to prevent infection. Rats are not fed on the day of perfusion. Ketamine hydrochloride is injected intraperitoneally (0.15 ml, Zhejiang Jiuxu Pharmaceutical Co., Ltd., China). After waiting for about 5 min, the rats in the early stage of anesthesia are anesthetized by inhalation of ether (Shanghai Malu Pharmaceutical Co., Ltd., China). After complete anesthesia, rats in the experimental group are given bronchial perfusion of carcinogens (0.1 ml lipiodol suspension containing 10 mg 3-methylcholane (Shanghai Chengshao Biological Technology Co., Ltd., China) and diethylnitrosamine (Shanghai McLean Biochemical Technology Co., Ltd., China)). Rats in the control group receive bronchial iodized oil infusion (0.1 ml, Shandong Xiya Chemical Industry Co., Ltd., China). At 20, 40, 60, 80 and 100 d after perfusion, 16 rats in the experimental group and 2 rats in the control group are killed at random. The perfused tissue (left lung) is removed and cut into two parts on average. One is placed in liquid nitrogen (-80°) for methylation specific polymerase chain reaction(PCR) (MSP) test, and the other is placed in 4 % polyformaldehyde stationary solution (Shanghai Bogu Biotechnology Co., Ltd., China). After routine dehydration, the paraffin is embedded and sliced continuously at a thickness of 3 μm. Xylene (Shanghai Yubo Biotechnology Co., Ltd., China) is used for dewaxing. Gradient alcohol is used for dehydration. After 10 min of dyeing with hematoxylin (Shanghai Baoman Biotechnology Co., Ltd., China), deionized water (Shanghai Yaji Biotechnology Co., Ltd., China) is used to rinse for 3 min, and then ethanol hydrochloride is used to differentiate. Deionized water (Shanghai Yaji Biotechnology Co., Ltd., China) is used to rinse for 3 min. Then, it is dyed with Yihong solution (Beijing Dingguo Changsheng Biotechnology Co., Ltd., China) for 3 min. Deionized water (Shanghai Yaji Biotechnology Co., Ltd., China) is used to rinse for 3 min. Gradient alcohol is used for dehydration. After decolorization and transparency of xylene, neutral resin (Shanghai Yantuo Biotechnology Co., Ltd., China) is used to seal it. The pathological changes of lung tissue are observed under light microscopy (leica, Germany). Tissue slices are baked in a thermostat (65°, Beijing Lambokons Technology Co., Ltd., China). 2 h later, xylene is used for dewaxing and gradient alcohol for dehydration. After washing with distilled water, tissue slices are immersed in phosphate buffer (Shanghai Shifeng Biotechnology Co., Ltd., China). 5 min later, the 0.01 M citrate buffer solution (pH 6.0, Xiamen Research Biotechnology Co., Ltd., China) is heated to 95° by electric furnace (Dongguan Spectrum Standard Experimental Equipment Technology Co., Ltd., China), and the tissue slices are heated for 10 min. Hydrochloric acid (3.5 mol/l, Xi’an Chuangyuan Chemical Co., Ltd., China) is incubated at room temperature for 15 min, and phosphate buffer is rinsed for 3×3 min. At room temperature, it is incubated with 3 % hydrogen peroxide (Suzhou Binshun Chemical Co., Ltd., China) for 10 min and washed with phosphate buffer for 3×3 min. At room temperature, goat serum is added (Wuhan Purity Biotechnology Co., Ltd., China) and sealed normally for 20 min to remove excess liquid. Anti -5-mC antibody (1:200, Shanghai Kemin Biotechnology Co., Ltd., China) is added. Phosphate buffer solution is used instead of anti-5-mC antibody as negative control and is placed overnight at 4°. It is taken out, placed at 37° for 1 h, and rinsed with phosphate buffer for 3×3 min. The secondary antibody (Beijing Taizer Ruida Technology Co., Ltd., China) is added dropwise, and incubated at 37° for 15 min. Phosphate buffer is used to wash for 3×3 min. Horseradish enzyme-labeled chain enzyme avidin working solution (Beijing Jiamei Qianyuan Biotechnology Co., Ltd., China) is added dropwise, incubated at 37° for 15 min, and washed with phosphate buffer for 3×3 min. DAB (Tianjin Taize Xingye Biotechnology Co., Ltd., China) is used to stain for 2 min and rinsed with tap water for 10 min. After staining with hematoxylin for 2 min, the ethanolic acid is differentiated and rinsed with tap water for 10 min. The expression of anti-5-mC antibody in bronchial epithelial cells is observed under light microscope. The average optical density (OD) and integrated optical density (IOD) of 5-mC immunohistochemistry are determined semi-quantitatively by computer image analysis. Genome-wide methylation of DNA from tumor tissues, pre-cancerous tissues and normal lung tissues is carried out. The reaction systems are as follows: 7.0 μl template DNA (10 ng/μL), 2.5 μl 10×PCR Buffer (Bao Bioengineering (Dalian) Co., Ltd., China), 1.5 ml MgCl2 (2.5 mmol/l, Qingyuan Shengli Biotechnology Co., Ltd., China), 0.5 ml deoxyribonucleotide triphosphate (dNTPs) Mix (10 mmol/l, Shanghai Hengfei Biotechnology Co., Ltd., China), 0.4 μl Primer (s) (25 μmol/l, Shanghai Kemin Biotechnology Co., Ltd.), 0.4 μl Taq enzyme (5 U/μl), RNase Free H2O (Beijing Baiaosentai Biotechnology Co., Ltd., China) are used to supplement the total volume to 25.0 μl. The conditions of PCR reaction are as follows: 5 min at 95°, 40 s at 95°, 60 s at 40°, and 60 s at 72°. 40 cycles are repeated, and the reaction is carried out at 72° for 10 min. 10 μl of PCR product is taken and electrophoresed on a 3 % agarose gel (Wuhan Purity Biotechnology Co., Ltd., China), and the results are directly interpreted under UV light. Data processing and analysis are performed using SPSS21.0 statistical software. Quantitative data consistent with normal distribution are described by mean±standard deviation. Comparison between the two groups is performed by two independent samples test or rank sum test. The comparison of different group rates is performed by chi-square test. There are no obvious hyperplasia and neoplastic lesions in the control group. The rats in the experimental group are observed by hematoxylin-eosin staining (HE staining), and the lesions at various stages of lung cancer are observed under light microscope, including bronchial epithelial hyperplasia, squamous metaplasia, atypical hyperplasia, carcinoma in situ, and invasive carcinoma. The results are shown in fig. 1. A total of 49 rats in 80 experimental groups develop lung squamous cell carcinoma. There are 26 cases of bronchial epithelial hyperplasia, 13 cases of squamous metaplasia, 17 cases of dysplasia, 15 cases of carcinoma in situ and 21 cases of invasive carcinoma. Since a plurality of lesion stages are usually found in a diseased tissue, they are each counted separately. The expression of anti-5-mC antibody in the nucleus of bronchial epithelial cells is observed under a light microscope, and the results are shown in fig. 2. The nucleus of normal bronchial epithelial cells is brownish yellow, and the staining gradually fades with the development of the carcinogenesis process, and the infiltrating cancer cell nucleus is pale yellow. The results of OD and IOD values of 5-mC immunohistochemistry are shown in fig. 1. It can be concluded that the OD and IOD values of 5-mC immunohistochemistry of cancerous cells are reduced compared with normal bronchial epithelial cells. There is statistical significance (p<0.01). The results are shown in Tables 1 and 2. The frequency of methylation of p16 gene in dysplasia, carcinoma in situ and invasive carcinoma is significantly higher than that in normal and proliferative tissues, and the difference is statistically significant (p<0.05). The p57 gene is not detected in the proliferative phase, but the frequency of methylation in the carcinoma in situ and invasive carcinoma is significantly higher than that in the normal and proliferative phase, and the difference is statistically significant (p<0.05). In this study, the mechanism of DNA aberrant methylation in rat lung cancer is studied. Wistar rats are used as research object to establish a lung cancer rat model by bronchial perfusion carcinogen. HE staining is used to observe the pathological changes of rat lung tissue. Immunohistochemistry is used to analyze the overall methylation level of the genome, and MSP is used to detect the methylation of the gene. The results show that under normal light microscope, the nucleus of normal bronchial epithelial cells is brownish yellow, and the staining gradually fades with the development of carcinogenesis, and the infiltrating cancer cells are light yellow. Compared with normal bronchial epithelial cells, the OD and IOD values of 5-mC immunohistochemistry are decreased. The frequency of p16 gene methylation in dysplasia, carcinoma in situ and invasive carcinoma is significantly higher than that in normal and proliferative tissues. The p57 gene has a significantly higher methylation frequency in the carcinoma in situ and invasive carcinoma than in the normal and proliferative phase. Therefore, in this study, it is found that abnormal methylation-mediated down-regulation of p16 and p57 gene transcription may be important molecular biological events during carcinogenesis in carcinogeninduced lung cancer rats. However, there are some shortcomings in the research process. For example, the sample data collection is small and the result is biased to a certain extent. Therefore, the data capacity will be further increased in the later research process, so that the obtained results are more valuable.
| Carcinogenesis stage | OD value | IOD value |
|---|---|---|
| Normal bronchial epithelial cells | 0.38±0.01 | 110.24±3.13 |
| Bronchial epithelial hyperplasia | 0.35±0.01 | 104.53±4.14 |
| Squamous metaplasia | 0.33±0.01 | 97.89±5.76 |
| Atypical hyperplasia | 0.31±0.01 | 88.41±6.51 |
| Carcinoma in situ | 0.28±0.02 | 80.53±7.02 |
| Invasive cancer | 0.24±0.02 | 70.43±7.96 |
Table 1: Comparison of OD and IOD Values of 5-Mc at Various Stages of Carcinogenesis
| Carcinogenesis stage | P16 methylation frequency (%) | P57 methylation frequency (%) |
|---|---|---|
| Normal bronchial epithelial cells | 0 | 0 |
| Bronchial epithelial hyperplasia | 7.69 | 0 |
| Squamous metaplasia | 23.08 | 15.38 |
| Atypical hyperplasia | 35.29 | 23.53 |
| Carcinoma in situ | 46.67 | 40.0 |
| Invasive cancer | 57.14 | 47.62 |
Table 2: Methylation Analysis of P16 and P57 Genes in Various Stages of Lung Cancer Carcinogenesis in Rats
References
- Ji M, Wang X, Wu W. ART manipulation after controlled ovarian stimulation may not increase the risk of abnormal expression and DNA methylation at some CpG sites of H19, IGF2 and SNRPN in foetuses: a pilot study. Reprod Biol Endocrin 2018;16(1):63.
- Meyer K F, Kraussetschmann S, Kooistra W. Prenatal exposure to tobacco smoke sex dependently influences methylation and mRNA levels of the Igf axis in lungs of mouse offspring. Am J Physiol Lung Cell Mol Physiol 2017;312(4): L542.
- Willeumier J J, Nm V D H, Bollen L. Epidermal growth factor receptor mutations should be considered as a prognostic factor for survival of patients with pathological fractures or painful bone metastases from non-small cell lung cancer. Bone Joint J 2017;99-B(4):516.
- Garde N J, Jantus-Lewintre E, Gil-Raga M. Role of RAS mutation status as a prognostic factor for patients with advanced colorectal cancer treated with first-line chemotherapy based on fluoropyrimidines and oxaliplatin, with or without bevavizumab: A retrospective analysis. Mol Clin Oncol 2017;6(3):403.
- Italiya K S, Sharma S, Kothari I. Simultaneous estimation of lisofylline and pentoxifylline in rat plasma by high performance liquid chromatography-photodiode array detector and its application to pharmacokinetics in rat. J Chromatogr B Analyt Technol Biomed Life Sci 2017;1061:49-56.
- Renaud S, Schaeffer M, Falcoz P E. Perioperative bevacizumab improves survival following lung metastasectomy for colorectal cancer in patients harbouring v-Ki-ras2 Kirsten rat sarcoma viral oncogene homologue exon 2 codon 12 mutations. Eur J Cardiothorac Surg 2017;51(2):255.
- Chen M, Cao H, Yingying J I. Association between the ALK Gene Status and the Efficacy of First-line Pemetrexed Chemotherapy in Patients with Advanced Lung Adenocarcinoma. Chin J Lung Cancer 2017;20(11):732.
- Wen J, Bao S, Cai Y. A reliable and stable method for determination of brigatinib in rat plasma by UPLC–MS/MS: Application to a pharmacokinetic study. J Chromatogr B Analyt Technol Biomed Life Sci 2017;1068-1069:84-89.
- Guerrera F, Renaud S, Tabbó F. Epidermal growth factor receptor mutations are linked to skip N2 lymph node metastasis in resected non-small-cell lung cancer adenocarcinomas. Eur J Cardiothorac Surg 2017;23(suppl 1):i10.4-i11.
- Kovi RC, Johnson CS, Balbo S, Hecht SS, O’Sullivan MG. Metastasis to the F344 Rat Pancreas from Lung Cancer Induced by 4-(Methylnitrosamino)- 1-(3-pyridyl)-1-butanone and Enantiomers of Its Metabolite 4-(Methylnitrosamino)-1-(3-pyridyl)- 1-butanol, Constituents of Tobacco Products. Toxicol Pathol 2018;46(2):184-92.