- *Corresponding Author:
- Jun Li
Department of Ophthalmology
Lanzhou first people's Hospital
Lanzhou 730050, China
E-mail: 13919176578@163.com
| This article was originally published in a special issue, “Therapeutic Perspectives in Biomedical Research and Pharmaceutical Sciences and their Nursing Methods” |
| Indian J Pharm Sci 2021:83(4)Spl issue “108-114” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The present study was designed to the effect of metastasis associated lung adenocarcinoma transcript 1 on apoptosis of 661W cells and to explore its role in glaucoma. A stable and metastasis associated lung adenocarcinoma transcript 1-inhibition vector and microRNA-200a-3p overexpression vector were established and transfected into 661W cells. Cell counting kit 8 and flow cytometry were employed to evaluate the proliferation and apoptosis of transfected cells and western blot was applied to measure the changes of apoptosis markers represented by B-cell lymphoma 2 associated X-protein, caspase 3 and B-cell lymphoma 2. The metastasis associated lung adenocarcinoma transcript 1 target gene was predicted and its relationship with the target gene was determined using dual luciferase reporter assay. Inhibited metastasis associated lung adenocarcinoma transcript 1 or overexpressed microRNA-200a-3p brought inhibited proliferation, increased apoptosis rate, enhanced B-cell lymphoma 2 associated X-protein and caspase-3 protein and declined B-cell lymphoma 2 protein of 661W cells. Dual luciferase reporter assay revealed the targeted binding of metastasis associated lung adenocarcinoma transcript 1 and microRNA-205. The rescue experiment found that inhibited microRNA-200a-3p expression prevented cell proliferation, apoptosis and changes in apoptosis markers caused by the inhibition of metastasis associated lung adenocarcinoma transcript 1. Metastasis associated lung adenocarcinoma transcript 1 inhibits the apoptosis of 661W cells by targeting microRNA-200a-3p, thereby preventing the progression of glaucoma. Metastasis associated lung adenocarcinoma transcript 1 is expected to be the treatment direction of this disease.
Keywords
Metastasis associated lung adenocarcinoma transcript 1, microRNA-200a-3p, glaucoma, retinal ganglion cells
Glaucoma is a neurodegenerative disease that causes irreversible vision loss[1]. It is estimated that more than 60 million people worldwide suffer from glaucoma and this number is expected to rise to 80 million by 2020[2]. Excessive death of retinal ganglion cells (RGC) is a major cause of vision loss in glaucoma patients[3]. As is known to all, the loss or decline of vision causes the patients' independence and quality of life to plummet[4]. Therefore, preventing excessive RGC death is critical to improve the outcome of glaucoma.
Long non-coding RNA (LncRNA) with a length of 200 nucleotide (nt) beyond, which plays an important role in various life activities such as embryonic development, cell growth and disease occurrence[5,6]. Among various LncRNAs, Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) is one that located on the human 11q13 chromosome, which is highly conserved among species and is widely expressed in human tissue[7]. Previous research on MALAT1 has extensively focused on its involvement in the development of various cancers, such as breast cancer[8], ovarian cancer[9] and non-small cell lung cancer (NSCLS)[10], while rarely have done on the role of MALAT1 in eye diseases. Recent studies have found that MALAT1 is lowly expressed in glaucoma and its knocking down can lead to more severe retinal neurodegeneration[11], which is indicative of the promising value of MALAT1 in treating glaucoma. Whereas, there are few reports about its role and mechanism in glaucoma at present. In order to further explore the mechanism of MALAT1 in the development of glaucoma, we predicted the presence of target binding sites between MALAT1 and microRNA- 200a-3p (miR-200a-3p) through starBase v2.0. As to the miR-200a-3p, it belongs to miR-200 family, which has been indicated in some studies that it increases with the rise of glaucoma injury degree and is considered to be an important regulatory factor implicated in the development of glaucoma[12].
From the above research, we hypothesized that MALAT1 promoted the development of glaucoma by targeting miR-200a-3p. Based on this hypothesis, the following studies were conducted.
Materials and Methods
Cell source and treatment:
P661W, which was a RGC precursor-like cell line with the characteristics of retinal ganglia and photoreceptor cells[13], was purchased from American Type Culture Collection (ATCC) and used as the cell material in this study. The repurchased cells were placed in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, USA) containing 10 % fetal bovine serum (FBS, Gibco, USA) and 1 % penicillin-streptomycin (100X, Solarbio, USA) in advance and then cultured at 37° and 5 % CO2 before transfection. Cell transfection: miR-200a- 3p overexpression plasmid (miR-200a-3p-mimics), MALAT1 inhibitory plasmid (si-MALAT1) and blank controls (si-NC, miR-NC) were established and the established cell lines were transferred to 24-well plates. After 48 h, the cell plasmid was transfected into cells with lipofectamine 2000 (Invitrogen, USA) kit in strict accordance with the kit instructions.
Real-time quantitative reverse transcription polymerase chain reaction (RT-qPCR):
The total RNA was extracted from the target cells by TRIzol reagent (Invitrogen, USA). The total RNA was extracted by ultraviolet (UV) spectrophotometer (Shanghai yuangue instrument co., LTD.) and agarose gel electrophoresis to detect the purity, concentration and integrity of the extracted total RNA. (Metash Instruments Co., Ltd., Shanghai, China) and agarose gel electrophoresis. Then, 5 μg of total RNA was processed for reverse transcription with the help of TaqManTM reverse transcription reagents (Invitrogen, USA) in strict accordance with the given instructions. Then the amplification, where SYBR Premix ExTaq II (Takara, Dalian, China) and Applied Biosystems® 7500 PCR instrument (Applied Biosystems, USA) were employed in this part. The amplification system: SYBR Premix Ex Taq II (2X):10 μl, complementary DNA (cDNA) 2 μl, upstream and downstream primers. 0.8 μl each and sterile purified water was completed to 20 μl. PCR reaction conditions: pre-denaturation at 95° for 30 s, denaturation at 95° for 5 s, annealing extension at 60° for 30 s, totaling for 40 cycles. Beta-actin was set as the internal reference of MALAT1 and U6 as that of miR-200a-3p. Shanghai GenePharma Co., Ltd. was in charge of the design and synthesis of all primers. The data were analyzed by 2-△△ct[14].
Western blot (WB):
After culture, the total protein of cells was extracted by Radioimmunoprecipitation assay (RIPA) lysis (Thermo Scientific, USA) and the protein concentration was measured by Bicinchoninic acid (BCA) (Thermo Scientific, USA). Then, the protein concentration was arranged to 4 μg/μl and then 12 % SDS-PAGE (sodium dodecyl sulphate–polyacrylamide gel electrophoresis) was applied for separation before it was transferred to oolyvinylidene fluoride (PVDF) membrane. Next, the membrane was soaked in phosphate-buffered saline (PBS) for 5 min and blocked with skim milk powder (5 %) for 2 h. Followed by the addition of B-cell lymphoma 2 associated X-protein (BAX) (1:1000), B-cell lymphoma 2 (Bcl-2) (1:1000), Caspase 3 (1:1000) and β-catenin primary antibody (1:1000) (Abcam, USA) and sealing for one night at the temperature of 4°. After that, the membrane was rinsed to discard the primary antibody, added with horseradish peroxidase-labeled goat anti-rabbit secondary antibody (1:2000), incubated at 37° for 1 h and then rinsed 3 times with PBS, 5 min each. Through blotting the excess liquid on the membrane with a filter paper first and then illuminated by Electrochemiluminescence (ECL) to develop, the development was finally carried out in a dark room. The protein bands were scanned to calculate the relative expression level of the target protein.
Cell counting kit 8 (CCK-8) assay for cell proliferation:
The transfected cells were gathered and digested with 0.25 % trypsin and the cell concentration was adjusted by routine inoculation of 100 μl/ml in 96-well plates. At the set time points, which were 24, 48, 72 and 96 h respectively, 200 μl CCK8 (Beyotime Biotechnology Co., Ltd., Beijing, China) mixture (10:1) was added and the culture was resumed for 3 h. Then the absorbance of each well was measured with a microplate at 490 nm and the corresponding growth curve was drawn.
Apoptosis detection:
The transfected cells were collected and digested with 0.25 % trypsin. After digestion, they were configured into 1×106/ml suspension. Followed by the successive addition of AnnexinV-FITC and PI (Yeasen Biotechnology, Co., Ltd., Shanghai, China) and then incubated at room temperature for 5 min shielded from light. Finally, apoptosis was measured by CytoFLEX flow cytometer (Beckman, USA).
Double luciferase reporter assay:
MALAT1 was cloned into pmirGLO double luciferase target expression vector using LipofectamineTM 2000 kit and the established MALAT1-3'UTR wild type (Wt), MALAT1-3'UTR mutant (Mut), miR-200a-3p-inhibition and miR-NC were successfully transfected into 661W cells. The change of luciferase activity was detected 48 h after culture.
Statistical analysis:
In this study, the collected data was statistically analyzed using statistical package for the social sciences (SPSS) 18.0 and the required pictures were plotted by GraphPad 7. The Kolmogorov–Smirnov (KS) test was employed to analyze the distribution of the dose data, among which the data of normal distribution was expressed in the form of mean±standard deviation (mean±SD). An independent sample t-test was adopted for inter-group comparison and one-way analysis of variance (ANOVA) was employed for multi-group comparison. The post hoc pairwise comparison was performed by least significant difference (LSD)-t test, the multi-time expressions were analyzed by repeated measurement analysis of variance and the post hoc test was conducted by Bonferroni. p<0.05 indicated a statistically significant difference.
Results and Discussion
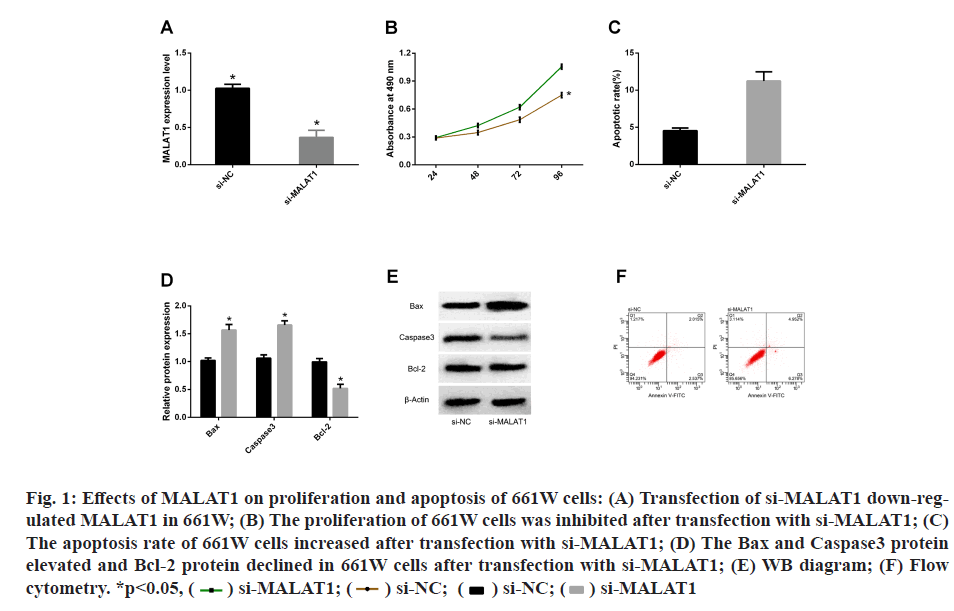
In order to determine the role of MALAT1 in glaucoma, we established a model of low expression of MALAT1 in 661W cells by transfection with si-MALAT1 and si- NC. QPC detection showed that the transfection of si- MALAT1 successfully down-regulated the MALAT1 in 661W cells, indicating that the model was established successfully. Subsequently, we employed CCK-8 and flow cytometry to evaluate cell proliferation and apoptosis after transfection. It was found that 661W was inhibited from proliferating after transfection with si-MALAT1 and the apoptosis rate increased. WB detection of apoptotic markers revealed that Bax, Caspase3 protein increased and Bcl-2 protein decreased in 661W cells after transfection with si-MALAT1 (fig. 1).
Fig. 1: Effects of MALAT1 on proliferation and apoptosis of 661W cells: (A) Transfection of si-MALAT1 down-regulated
MALAT1 in 661W; (B) The proliferation of 661W cells was inhibited after transfection with si-MALAT1; (C)
The apoptosis rate of 661W cells increased after transfection with si-MALAT1; (D) The Bax and Caspase3 protein
elevated and Bcl-2 protein declined in 661W cells after transfection with si-MALAT1; (E) WB diagram; (F) Flow
cytometry. *p<0.05, si-MALAT1
si-MALAT1
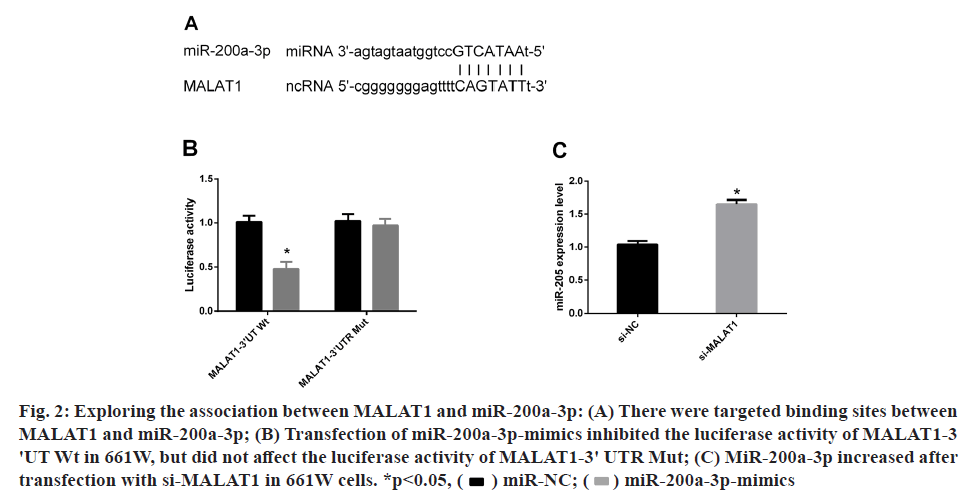
For the sake of understanding the molecular mechanism through which MALAT1 promoted 661W apoptosis, we predicted through starBase2.0 website that miR- 200a-3p could be used as a downstream target gene of MALAT1. Therefore, a dual luciferase activity test was performed to prove the connection between them. It was observed that transfection of miR-200a-3p-mimics inhibited the luciferase activity of MALAT1-3'UT Wt in 661W, but had no effect on the luciferase activity of MALAT1- 3'UTR Mut. Subsequently, miR-200a-3p was noticed to be increased in 661W after transfection with si-MALAT1 by PCR-qRT. All these indicated that miR-200a-3p was indeed the downstream target gene of MALAT1 (fig. 2).
Fig. 2: Exploring the association between MALAT1 and miR-200a-3p: (A) There were targeted binding sites between
MALAT1 and miR-200a-3p; (B) Transfection of miR-200a-3p-mimics inhibited the luciferase activity of MALAT1-3
'UT Wt in 661W, but did not affect the luciferase activity of MALAT1-3' UTR Mut; (C) MiR-200a-3p increased after
transfection with si-MALAT1 in 661W cells. *p<0.05, miR-200a-3p-mimics
miR-200a-3p-mimics
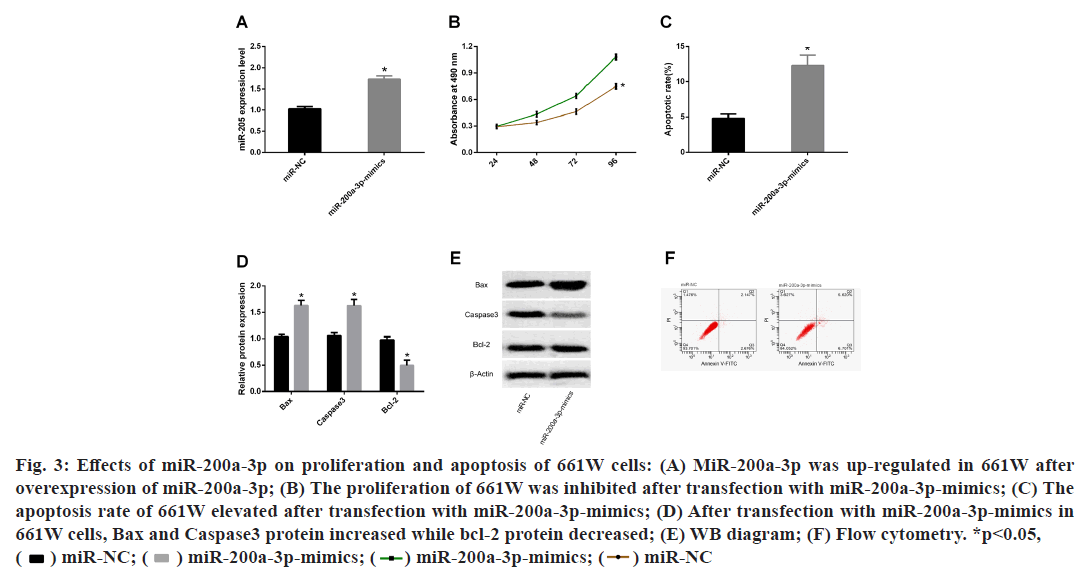
Results from finding downstream target genes of MALAT1, had confirmed that was the downstream target gene of MALAT1, but it was not clear whether MALAT1 could promote the apoptosis of 661W through miR-200a-3p. Therefore, we observed the changes in proliferation and apoptosis of 661W cells after transfection with miR-200a-3p-mimics. The results displayed that after miR-200a-3p overexpression treatment, miR-200a-3p was up-regulated in 661W cells, the proliferation ability was inhibited and the apoptosis rate was increased. WB detection of apoptosis markers revealed that after transfecting miR-200a-3pmimics, Bax and Caspase3 protein increased while Bcl- 2 protein declined in 661W cells (fig. 3).
Fig. 3: Effects of miR-200a-3p on proliferation and apoptosis of 661W cells: (A) MiR-200a-3p was up-regulated in 661W after
overexpression of miR-200a-3p; (B) The proliferation of 661W was inhibited after transfection with miR-200a-3p-mimics; (C) The
apoptosis rate of 661W elevated after transfection with miR-200a-3p-mimics; (D) After transfection with miR-200a-3p-mimics in
661W cells, Bax and Caspase3 protein increased while bcl-2 protein decreased; (E) WB diagram; (F) Flow cytometry. *p<0.05, 
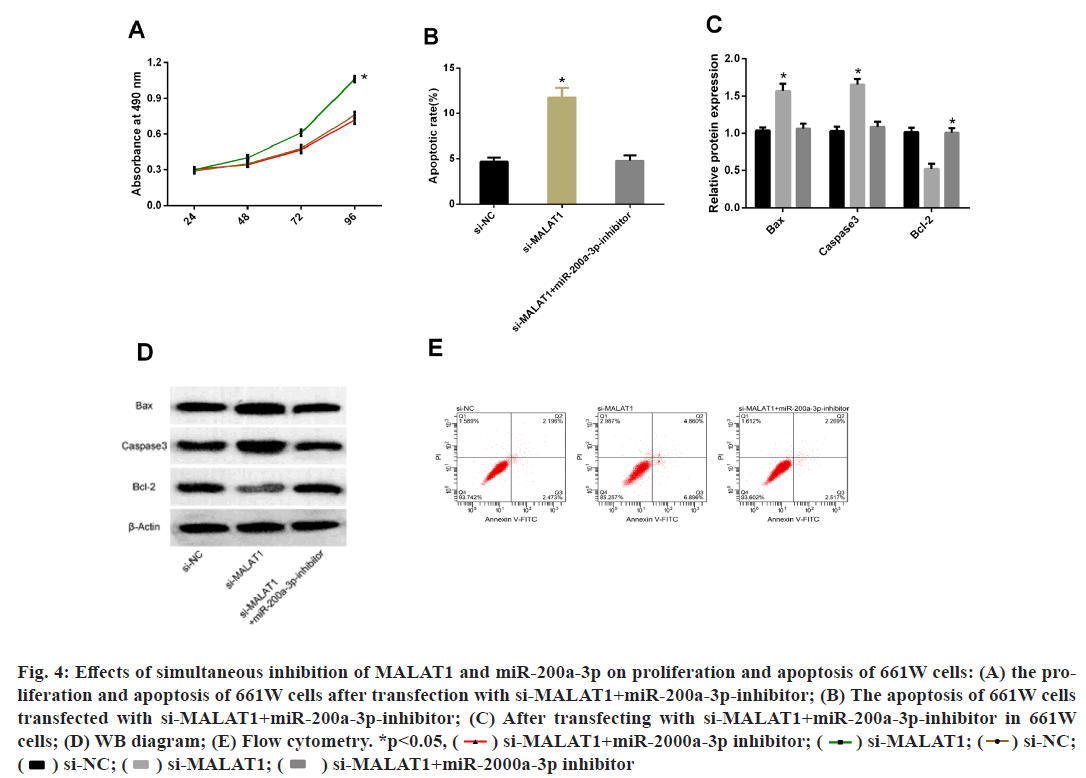
From Result miR-200a-3p promoted apoptosis of 661W cells, it was known that MALAT1 could bind to miR-200a-3p in a targeted manner. Subsequently, to further understand the relationship between the two, we simultaneously performed MALAT1 and miR-200a-3p inhibition treatment on 661W and then observed their biological function changes. The results showed that the proliferation and apoptosis of 661W cells after transfecting si-MALAT1+miR-200a-3pinhibitor were not statistically different from those transfected with miR-NC, but the proliferation ability increased and the apoptosis rate decreased compared with those transfected with si-MALAT1. WB detection of apoptosis markers exhibited that after transfection of si-MALAT1+miR-200a-3p-inhibitor with 661W cells, there was no statistical changes in Bax, Caspase3 or Bcl-2 compared with those transfected with miR-NC, while compared with the cells transfected with si- MALAT1, Bax and Caspase 3 protein were decreased, while Bcl-2 protein was increased (fig. 4)
Fig. 4: Effects of simultaneous inhibition of MALAT1 and miR-200a-3p on proliferation and apoptosis of 661W cells: (A) the proliferation
and apoptosis of 661W cells after transfection with si-MALAT1+miR-200a-3p-inhibitor; (B) The apoptosis of 661W cells
transfected with si-MALAT1+miR-200a-3p-inhibitor; (C) After transfecting with si-MALAT1+miR-200a-3p-inhibitor in 661W
cells; (D) WB diagram; (E) Flow cytometry. *p<0.05, si-NC;
si-NC; si-MALAT1+miR-2000a-3p inhibitor
si-MALAT1+miR-2000a-3p inhibitor
Glaucoma is the second leading cause of blindness, which can be divided into the open-angle and angle-closure[15]. Although glaucoma patients can be treated by surgery and drugs at present, there is often a progressive vision loss[16]. Therefore, it is high time to elucidate the mechanisms of the onset and progression of glaucoma to improve patient outcomes. With the exploration of LncRNA in medicine, it has been found that LncRNA is closely related to the occurrence and progression of various neurodegenerative diseases including glaucoma[17-19]. In this paper, we found through CCK- 8 and flow cytometry that 661W cells were inhibited from proliferating after transfection with si-MALAT1, accompanied by elevated apoptosis, increased Bax and Caspase3 and reduced Bcl-2 protein, indicating that MALAT1 could slow the progression of glaucoma by preventing apoptosis of 661W cells. However, it is still unclear how MALAT1 prevents the apoptosis of 661W cells.
LcnRNA regulates the downstream target of miRNA through sponge action on miRNA[20]. In order to understand how MALAT1 prevents the apoptosis of 661W cells, this paper predicted the downstream target gene of MALAT1 through starBase2.0 website. It turned out that there were target binding sites between MALAT1 and miR-200a-3p. miR is a class of single-stranded non-coding RNA, containing 20- 24 nucleotides, which can promote the degradation of transcripts by binding to the 3'UTR of its target gene mRNA sequence, thus participating in biological events[21]. Misexpression or mutation of miR has been identified as the cause of the occurrence and development of many human diseases[22]. Previous studies have found that multiple miRs can affect the development of glaucoma, such as miR-93-59, miR- 187, and miR-17-5p[23-25], showing potential therapeutic value. Like the transfection of si-MALAT1, we found that transfection of miR-200a-3p-mimics shared the same function in this study, that is, the transfection could inhibit the proliferation of 661W cells, increase the apoptosis rate, enhance Bax and Caspase 3 protein and decrease Bcl-2 protein, indicating that miR-200a- 3p could induce the apoptosis of 661W cells, thereby accelerating the progress of glaucoma. There is evidence showing that miR-200a-3p can be regulated by a variety of lcnRNA, thus affecting the development of the disease. For example, it is regulated by lcnRNA SNHG16 to promote the development of colorectal cancer[26] and in liver cancer, it can be regulated by lncRNA highly upregulated in liver cancer (HULC) to facilitate the occurrence and development of the disease[27]. A recent study demonstrates that miR-200a- 3p can also be regulated by MALAT1 to promote the development of non-small cell lung cancer (NSCLC) [28]. These results indicate that lcnRNA can participate in tumor development by regulating miR-200a-3p. The findings in present study indicated that transfection of miR-200a-3p-mimics could inhibit MALAT1-3'UT Wt luciferase activity in 661W, but it did not affect MALAT1-3'UTR Mut luciferase activity and that miR- 200a-3p increased after transfection of si-MALAT1 in 661W cells, indicating that MALAT1 could be targeted and regulated by miR-200a-3p. At the end of the paper, rescue experiments were carried out to further explore the correlation between MALAT1 and miR-200a-3p. The results demonstrated that the transfection of si- MALAT1+miR-200a-3p-inhibitor with 661W reversed the changes in proliferation, apoptosis and apoptosisrelated proteins induced by the transfection of si- MALAT1. According to the above results, MALAT1 can at least partially prevent the apoptosis of 661w cells by regulating miR-200a-3p.
This paper discusses the mechanism of MALAT1 in glaucoma at the molecular level and proves that MALAT1 can prevent the apoptosis of 661W cells by regulating miR-200a-3p, thus slowing the development of glaucoma, reflecting the potential therapeutic value of MALAT1/miR-200a-3p in glaucoma. Studies have found that miR-200a-3p can contribute to β-amyloid-induced neuronal death through Sirtuin 1 (SIRT1) in Alzheimer's disease[29]. Therefore, it is speculated that MALAT1 may influence the development of glaucoma through the miR-200a-3p/SIRT1 axis, which will be discussed in future experimental designs. In addition, this paper only discussed the cytological effects of MALAT1/miR-200a-3p axis, but failed to explore the clinical value of the two. In follow-up studies, relevant clinical trials can be regulated to supplement the value of both.
Taken together, MALAT1 can prevent the apoptosis of 661W cells by regulating miR-200a-3p, thereby slowing the progression of glaucoma.
Funding:
The work was supported by Identification and optimization of new targets for glaucoma treatment based on medical big data (self-pooled funds).
Conflict of interests:
The authors declared no conflicts of interest.
References
- Guo R, Shen W, Su C, Jiang S, Wang J. Relationship between the pathogenesis of glaucoma and miRNA. Ophthalmic Res 2017;57(3):194-9.
- Tham YC, Li X, Wong TY, Quigley HA, Aung T, Cheng CY. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology 2014;121(11):2081-90.
- Davis BM, Crawley L, Pahlitzsch M, Javaid F, Cordeiro MF. Glaucoma: the retina and beyond. Acta Neuropathol 2016;132(6):807-26.
- Hindle AG, Thoonen R, Jasien JV, Grange RM, Amin K, Wise J, et al. Identification of candidate miRNA biomarkers for glaucoma. Invest Ophthalmol Vis Sci 2019;60(1):134-46.
- Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer cell 2016;29(4):452-63.
- Hong Q, Li O, Zheng W, Xiao WZ, Zhang L, Wu D, et al. LncRNA HOTAIR regulates HIF-1 α/AXL signaling through inhibition of miR-217 in renal cell carcinoma. Cell Death Dis 2017;8(5):e2772.
- Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA Malat1: Its physiological and pathophysiological functions. RNA Biol 2017;14(12):1705-14.
- Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang Y, et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat Genet 2018;50(12):1705-15.
- Gordon MA, Babbs B, Cochrane DR, Bitler BG, Richer JK. The long non‐coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2‐mediated alternative splicing. Mol Carcinog 2019;58(2):196-205.
- Cui Y, Li G, Zhang X, Dai F, Zhang R. Increased MALAT1 expression contributes to cisplatin resistance in non-small cell lung cancer. Oncol Lett 2018;16(4):4821-8.
- Yao J, Wang XQ, Li YJ, Shan K, Yang H, Wang YN, et al. Long non‐coding RNA MALAT 1 regulates retinal neurodegeneration through CREB signaling. EMBO Mol Med 2016;8(4):346-62.
- Liu Y, Chen Y, Wang Y, Zhang X, Gao K, Chen S, et al. microRNA profiling in glaucoma eyes with varying degrees of optic neuropathy by using next-generation sequencing. Invest Ophthalmol Vis Sci 2018;59(7):2955-66.
- Sayyad Z, Sirohi K, Radha V, Swarup G. 661W is a retinal ganglion precursor-like cell line in which glaucoma-associated optineurin mutants induce cell death selectively. Sci Rep 2017;7(1):1-3.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001;25(4):402-8.
- Wang X, Li Z, Bai J, Song W, Zhang F. miR-17-5p regulates the proliferation and apoptosis of human trabecular meshwork cells by targeting phosphatase and tensin homolog. Mol Med Rep 2019;19(4):3132-8.
- Wang JJ, Liu C, Shan K, Liu BH, Li XM, Zhang SJ, et al. Circular RNA-ZNF609 regulates retinal neurodegeneration by acting as miR-615 sponge. Theranostics 2018;8(12):3408.
- Johnson WM, Finnegan LK, Hauser MA, Stamer WD. lncRNAs, DNA methylation, and the pathobiology of exfoliation glaucoma. J Glaucoma 2018;27(3):202-9.
- Yue D, Guanqun G, Jingxin L, Sen S, Shuang L, Yan S, et al. Silencing of long noncoding RNA XIST attenuated Alzheimer's disease‐related BACE1 alteration through miR‐124. Cell Biol Int 2020;44(2):630-6.
- Yan W, Chen ZY, Chen JQ, Chen HM. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson's disease through stabilizing PINK1 protein. Biochem Biophys Res Commun 2018;496(4):1019-24.
- He B, Bai Y, Kang W, Zhang X, Jiang X. LncRNA SNHG5 regulates imatinib resistance in chronic myeloid leukemia via acting as a CeRNA against MiR-205-5p. Am J Cancer Res 2017;7(8):1704.
- Gareev IF, Novicova LB, Beylerli OA. Circulating microrPas as new potential biomarkers for the diagnosis of high-grade gliomas. Zh Nevrol Psikhiatr im SS Korsakova 2019;119(5):86-90.
- Li G, Luo J, Xiao Q, Liang C, Ding P. Predicting microRNA-disease associations using label propagation based on linear neighborhood similarity. J Biomed Inform 2018;82:169-77.
- Li R, Jin Y, Li Q, Sun X, Zhu H, Cui H. MiR-93-5p targeting PTEN regulates the NMDA-induced autophagy of retinal ganglion cells via AKT/mTOR pathway in glaucoma. Biomed Pharmacother 2018;100:1-7.
- Zhang QL, Wang W, Li J, Tian SY, Zhang TZ. Decreased miR-187 induces retinal ganglion cell apoptosis through upregulating SMAD7 in glaucoma. Biomed Pharmacother 2015;75:19-25.
- Wang X, Li Z, Bai J, Song W, Zhang F. miR-17-5p regulates the proliferation and apoptosis of human trabecular meshwork cells by targeting phosphatase and tensin homolog. Mol Med Rep 2019;19(4):3132-8.
- Li Y, Lu Y, Chen Y. Long non-coding RNA SNHG16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer via sponging miR-200a-3p. Biosci Rep 2019;39(5):BSR20182498.
- Li SP, Xu HX, Yu Y, He JD, Wang Z, Xu YJ, et al. LncRNA HULC enhances epithelial-mesenchymal transition to promote tumorigenesis and metastasis of hepatocellular carcinoma via the miR-200a-3p/ZEB1 signaling pathway. Oncotarget 2016;7(27):42431.
- Wei S, Wang K, Huang X, Zhao Z, Zhao Z. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int J Immunopathol Pharmacol 2019;33:2058738419859699.
- Zhang QS, Liu W, Lu GX. miR-200a-3p promotes β-Amyloid-induced neuronal apoptosis through down-regulation of SIRT1 in Alzheimer’s disease. J Biosci 2017;42(3):397-404.