- *Corresponding Author:
- Youfeng Zhou
Department of Pediatrics, Fujian Maternity and Child Health Hospital, College of Clinical Medicine for Obstetrics & Gynecology and Pediatrics, Fujian Medical University, Fuzhou, Fujian 350001, China
E-mail: qwerrtyy014725@163.com
| This article was originally published in a special issue, “New Research Outcomes in Drug and Health Sciences” |
| Indian J Pharm Sci 2023:85(6) Spl Issue “66-73” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The purpose of this study was to simulate muscular dystrophy disease by constructing a laminin-alpha 2 zebrafish model, and to study the changes of muscle transcriptome using transcriptomic methods. Using clustered regularly interspaced short palindromic repeats associated protein 9 gene editing technology; the zebrafish laminin-alpha 2 gene was knocked out or mutated. First, the appropriate guide RNA sequence was designed, and then the guide RNA and clustered regularly interspaced short palindromic repeats associated protein 9 proteins were synthesized and injected into zebrafish embryos. The lamininalpha 2 model fish is screened and identified by detecting the mutation of DNA or messenger RNA in the progeny fish genome. The muscle phenotype identification results of zebrafish model 4 showed that the muscle phenotypes of normal and unable swimming individuals in laminin-alpha 2 knockout group were abnormal, while the muscle phenotypes of clustered regularly interspaced short palindromic repeats associated protein 9 control group were not. The muscular dystrophy characteristics of laminin-alpha 2 model fish were confirmed. Functional enrichment and pathway analysis of differentially expressed genes were performed to further reveal the biological processes and regulatory pathways associated with muscular dystrophy in laminin-alpha 2 model fish. Transcriptome sequencing analysis revealed the difference in transcriptome expression between the muscle tissue of laminin-alpha 2 zebrafish model and wild-type zebrafish. These differentially expressed genes are involved in biological processes such as muscle development, metabolism and signal transduction, providing important clues for further understanding of the pathogenesis of muscular dystrophy. Through functional enrichment and pathway analysis, the key signaling pathways and molecular mechanisms involved in the pathogenesis of muscular dystrophy were identified. These findings provide theoretical basis for the discovery of new therapeutic targets and formulation of corresponding therapeutic strategies, and provide new ideas and methods for the treatment of muscle diseases.
Keywords
Laminin-alpha 2 zebrafish model, muscular dystrophy, transcriptomics, clustered regularly interspaced short palindromic repeats associated protein 9, metabolism
Muscle disease is a kind of disease that seriously affects human health and quality of life, its etiology and pathogenesis have not been fully clarified. Muscular dystrophy is a common muscle disease characterized by muscle weakness, loss of muscle mass, and abnormal muscle metabolism. Understanding the pathogenesis of muscle diseases is of great significance for diagnosis and treatment [1-3].
Muscle is one of the most important tissues in the human body and plays a key role in maintaining posture, movement ability and metabolic balance[4,5]. The occurrence of muscle diseases is related to a variety of factors, including genetic factors, environmental factors, metabolic abnormalities, neuromuscular connection disorders, etc.,[6-9]. Muscular dystrophy is one of the common types. Patients with muscular dystrophy often show symptoms such as decreased muscle strength, muscle atrophy, fatigue and uneven muscle hypertrophy, which seriously affect patients' quality of life[10-13].
However, the etiology and pathogenesis of muscle diseases are still not fully understood, which limits progress in their diagnosis and treatment. Traditional research methods are often restricted by problems such as difficulty in obtaining samples, limited technical means and limited research objects. Therefore, the development of new research methods and the establishment of suitable animal models become the key to the study of muscle diseases.
In recent years, transcriptomics has been widely applied in life science research, providing a powerful tool for studying the molecular mechanism of diseases[14]. Transcriptomics can comprehensively reveal all the transcript information in cells or tissues, so as to help us understand the changes in gene expression, the regulation of signaling pathways and the occurrence and development mechanism of related diseases. For muscle diseases, transcriptomic studies can reveal changes in gene expression related to muscle development, metabolism and signal transduction, providing important clues for disease prevention and treatment[15-17].
Transcriptomic studies of muscle diseases have made remarkable progress in the last few years. However, due to the difficulty in acquiring muscle samples and the heterogeneity of human muscle tissue, it is limited to study muscle transcriptomics directly in human body. Therefore, finding suitable animal models to replace human studies has become an effective strategy[18-20]. As a commonly used experimental model organism, zebrafish Danio rerio has advantages such as strong fecundity, short reproductive cycle, and clear genetic background, making it an ideal model for studying muscle diseases[21].
In this study, we will use zebrafish as a model organism to construct a zebrafish model of Laminin- Alpha 2 (LAMA2) muscular dystrophy, and use transcriptomic methods to conduct an in-depth study of its muscle transcriptome. Zebrafish have a highly conserved genome, and their muscle development and physiological processes are very similar to those of humans, so they are highly comparable and reliable.
In order to construct the LAMA2 zebrafish model, we will use Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) gene editing technology to knockout or mutate the LAMA2 gene of zebrafish. CRISPR/ Cas9 technology is an important breakthrough in genome editing in recent years because it is efficient, accurate and economical, and can realize the targeted editing of specific genes. Through the establishment of this model, we will simulate muscular dystrophy disease and deeply study the changes of transcriptome in its muscle tissue, so as to reveal the pathogenesis of muscular dystrophy[22-25].
In transcriptomics studies, we will apply highthroughput transcriptome sequencing techniques, such as RNA Sequencing (RNA-Seq), to sequence the collected muscle tissue samples of zebrafish. The RNA-Seq technique enables the quantification and identification of transcripts on a genome-wide scale, providing a comprehensive understanding of transcriptome changes. By analyzing the sequencing data, we will identify and compare the transcriptome expression differences in muscle tissue between LAMA2 model fish and wild-type zebrafish, and identify differentially expressed genes related to muscle development, metabolism and signal transduction. Through the construction of LAMA2 zebrafish model and transcriptomic studies, we can deeply understand the development process of muscle diseases, discover new therapeutic targets and formulate corresponding therapeutic strategies, and make contributions to improving the quality of life of patients. We will further promote the application of bioinformatics technology in the biomedical field, and open up new ways for the prevention and treatment of muscle diseases.
Materials and Methods
RNA-Seq data source:
Muscle tissue sample collection: Muscle tissue samples were respectively collected from LAMA2 model fish and wild-type zebrafish. During the collection process, we tried to select muscle tissues with the same anatomical position and similar size, and through rapid freezing and preservation, to maintain the integrity of RNA.
RNA extraction and library preparation: Total RNA was extracted from the collected muscle tissue, and the RNA was purified and quality controlled using appropriate reagents and methods. We then used the RNA-Seq library preparation kit to construct the RNA-Seq library according to the manufacturer's recommended procedures, including RNA reverse transcription, fragmentation, and sequencing splicing.
RNA-Seq: Constructed RNA-Seq libraries will be sent to high-throughput sequencing platforms, such as Illumina HiSeq or NextSeq, for sequencing. Through high throughput sequencing technology, we will obtain sequencing readings of all transcripts in muscle tissue.
Experimental methods:
Construction of the LAMA2 zebrafish model: Using CRISPR/Cas9 gene editing technology, we designed appropriate primers and probes to trigger LAMA2 gene knockout or mutation in zebrafish fertilized eggs by introducing Cas9 protein and the single guide RNA (sgRNA) of the LAMA2 gene. Through screening and identification, the zebrafish individuals with LAMA2 gene mutation were obtained, and the LAMA2 muscular dystrophy zebrafish model was established.
Collecting muscle tissue samples of zebrafish: Muscle tissue samples were collected from LAMA2 model fish and wild-type zebrafish. Using appropriate techniques and tools, we remove the muscle tissue from the fish body and maintain the integrity and purity of the sample as much as possible. The collected samples will be used for subsequent transcriptomic analysis.
Differential expression analysis: Differential expression analysis of two conditions/groups (two biological replicates per condition) was performed using the DESeq2 R package (1.16.1). DESeq2 provide statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting p values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted p<0.05 found by DESeq2 were assigned a differentially expressed gene[26].
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis: GO enrichment analysis of differentially expressed genes was implemented by the top GO R package, in which gene length bias was corrected[27]. GO terms with corrected p<0.05 were considered significantly enriched by differential expressed genes.
Protein-Protein Interaction (PPI) analysis of differentially expressed genes: PPI analysis of differentially expressed genes was based on the Search Tool for the Retrieval of Interacting Genes (STRING) database, which known and predicted PPIs[28].
Results and Discussion
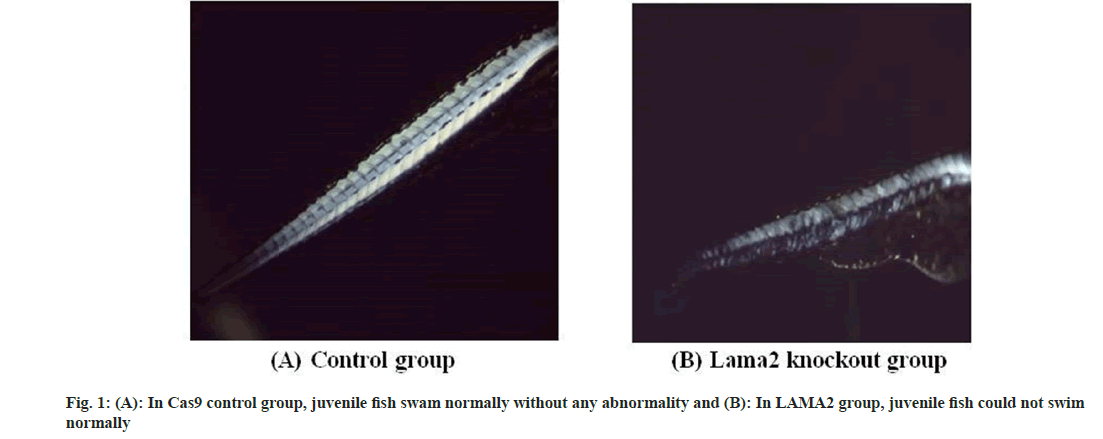
The establishment of the LAMA2 zebrafish model was successfully verified through molecular biology and genetic analysis. The experiment was divided into two groups; LAMA2 knockdown group and Cas9 control group. In LAMA2 knockdown group, muscle phenotypes of juvenile fish in normal swimming state and unable to swim state were selected for identification. Juvenile fish in Cas9 control group had no abnormal state and could swim normally, so individuals in Cas9 group were randomly selected as muscle phenotype control. Detailed phenotypic analysis of the model was performed, including assessment of muscle structure and function. The differences between the model and wild-type zebrafish were compared. The muscle phenotype identification results of zebrafish model 4 showed that the muscle phenotypes of normal and unable swimming individuals in LAMA2 knockout group were abnormal, while the muscle phenotypes of Cas9 control group were not. Muscular dystrophy was identified in LAMA2 model fish (fig. 1A and fig. 1B).
Using the transcriptome sequencing technology to LAMA2 model fish and wild type zebrafish muscle tissue samples were analyzed, and won the 3054 DEGs, 1201 DEGs of up and 1853 DEGs of down (fig. 2). In hierarchal clustering analysis, the regions with different colors represent different clustering information and have similar gene expression patterns in the same group. They may have similar functions or participate in the same biological processes (fig. 3).
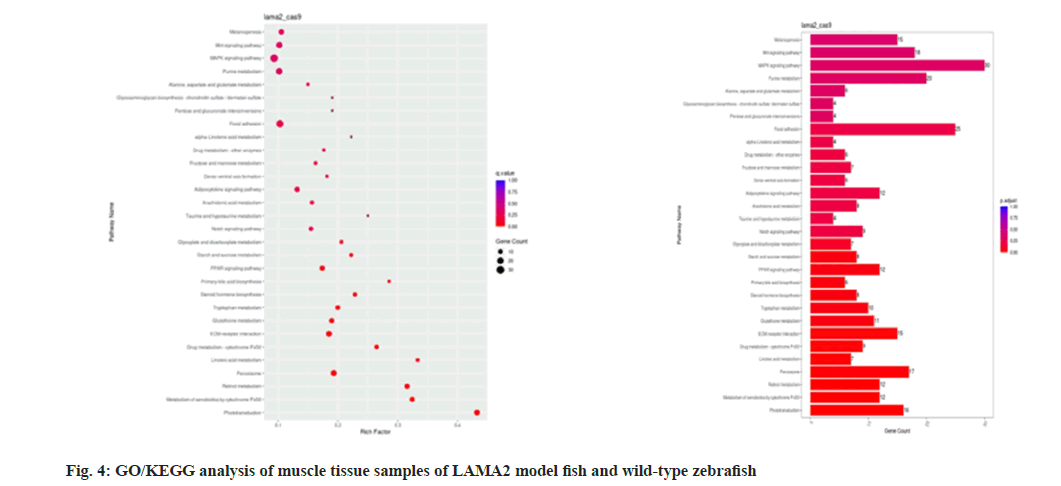
Functional enrichment and pathway analysis of differentially expressed genes were performed to further reveal the biological processes and regulatory pathways associated with muscular dystrophy in LAMA2 model fish. Identify key signaling pathways and molecular mechanisms that may be involved in the pathogenesis of muscular dystrophy (fig. 4). Through differential expression gene analysis, MYH1, MYH2, MYH3, MYH4, ACTA1, TNNT1, TNNT3, TNNI1, MYOD1, MYOG, MYF5, MYF6, PAX3, PAX7, MRF4, MYH1, MYH2, MYH3, MYH4, Acta1, TNnt1, TNnt3, TNNI1, myod1, myog, myF5, myF6,IGF1, IGF2, FGF2, FGF6, related to metabolism are AMPK, PGC1α, PPARγ, SIRT1, GLUT1, GLUT4, G6PD, HK2, FASN, ACACA, PPARG, LPL, MAFbx, MuRF1, AKT1, mTOR and signal transduction related genes are WNT1, WNT3, CTNNB1, AXIN2, TGFB1, TGFBR1, SMAD2, SMAD3, MAPK1, MAPK3, MAPK8, MAPK14,PIK3CA, AKT1, FOXO1 and GSK3B (Table 1).
| Category | Gene |
|---|---|
| Genes related to muscle development | MYH1, MYH2, MYH3, MYH4, ACTA1, TNNT1, TNNT3, TNNI1, MYOD1, MYOG, MYF5, MYF6, PAX3, PAX7, MRF4, IGF1, IGF2, FGF2 and FGF6 |
| Metabolism-related genes | AMPK, PGC1α, PPARγ, SIRT1, GLUT1, GLUT4, G6PD, HK2, FASN, ACACA, PPARG, LPL, MAFbx, MuRF1, AKT1 and mTOR |
| Genes involved in signal transduction | WNT1, WNT3, CTNNB1, AXIN2, TGFB1, TGFBR1, SMAD2, SMAD3, MAPK1, MAPK3, MAPK8, MAPK14, PIK3CA, AKT1, FOXO1 and GSK3B |
Table 1: Functional Enrichment and Pathway Analysis Through Differential Expression Genes
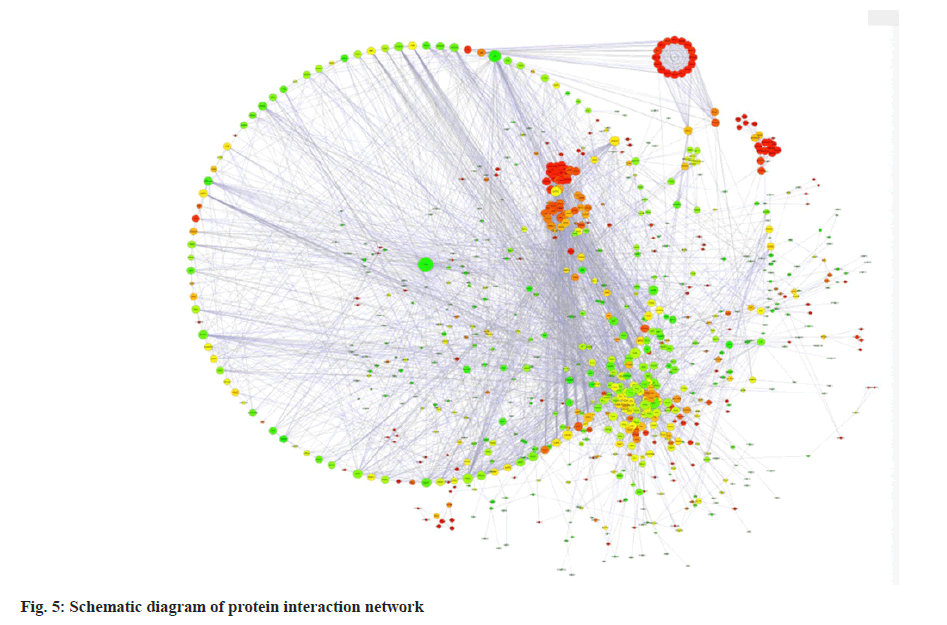
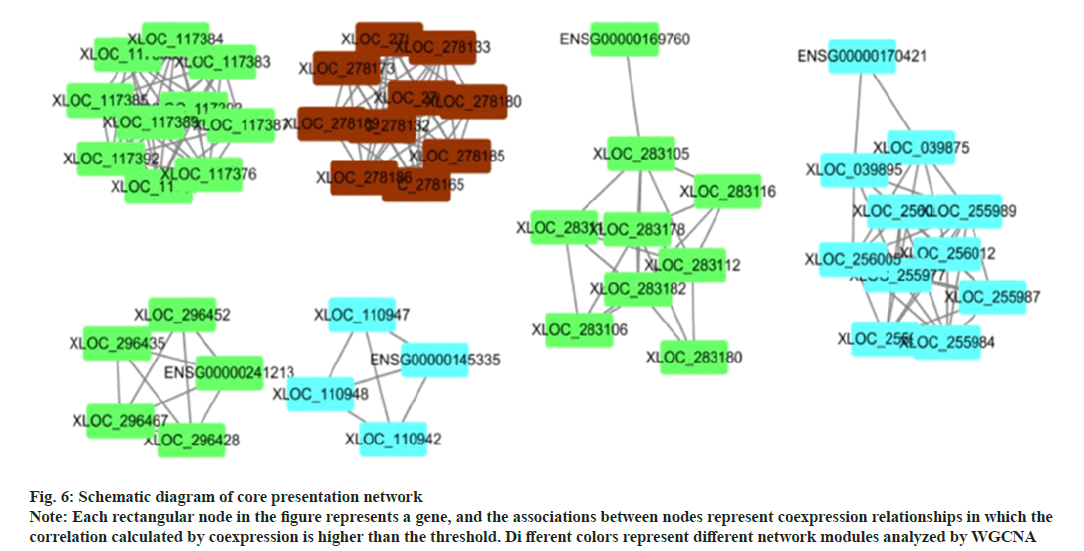
The interaction relationship in the STRING protein interaction database (http://string-db.org/) was applied. For the species contained in the database, the interaction relationship of target gene set (such as differential gene list) was directly extracted from the database to construct the network (fig. 5). The filter was conducted according to the expression levels of all genes, and the Standard Deviation (SD) values of all samples were sorted from largest to smallest. The top 25% genes were selected for co-expression analysis (fig. 6).
Fig. 6: Schematic diagram of core presentation network Note: Each rectangular node in the figure represents a gene, and the associations between nodes represent coexpression relationships in which the correlation calculated by coexpression is higher than the threshold. Di fferent colors represent different network modules analyzed by WGCNA
The aim of this study was to construct a zebrafish model of LAMA2 muscular dystrophy and to reveal its underlying molecular mechanism through muscular transcriptomics studies[29-32]. Through the analysis and discussion of the experimental results, we can deeply understand the characteristics of muscle development and function abnormalities in the LAMA2 model, further elucidate the pathophysiological process of the disease, and provide new clues for the development of therapeutic strategies[33-36].
In this study, we successfully constructed a LAMA2 zebrafish model to simulate the clinical phenotype of muscular dystrophy. Using single-cell sequencing and RNA-Seq techniques, we obtained and comprehensively analyzed the transcriptome data of muscle tissue in the LAMA2 model. In single-cell sequencing data, we identified and classified multiple muscle cell subtypes and identified changes in gene expression associated with muscle development and function. RNASeq data analysis revealed a large number of differentially expressed genes and pathways in the LAMA2 model, which are involved in biological processes such as muscle structure, metabolic regulation, and cellular stress. Through differential expression gene analysis, MYH1, MYH2, MYH3, MYH4, ACTA1, TNNT1, TNNT3, TNNI1, MYOD1, MYOG, MYF5, MYF6, PAX3, PAX7, MRF4, MyH1, MyH2, MyH3, MyH4, Acta1, TNnt1, TNnt3, TNNI1, myod1, myog, myF5, myF6, IGF1, IGF2, FGF2, FGF6, related to metabolism are AMPK, PGC1α, PPARγ, SIRT1, GLUT1, GLUT4, G6PD, HK2, FASN, ACACA, PPARG, LPL, MAFbx, MuRF1, AKT1, mTOR and signal transduction related genes are WNT1, WNT3, CTNNB1, AXIN2, TGFB1, TGFBR1, SMAD2, SMAD3, MAPK1, MAPK3, MAPK8, MAPK14,PIK3CA, AKT1, FOXO1 and GSK3B[37-42].
Further discussing the results of the zebrafish model of LAMA2 muscular dystrophy, we can compare the study results with existing clinical and molecular genetic data. Although zebrafish models cannot fully simulate human diseases, we can find some common pathological features and molecular mechanisms through comparison. For example, we observed muscle degeneration and atrophy in the LAMA2 model, which is consistent with clinical manifestations of muscular dystrophy in humans[29-32]. In addition, by analyzing differentially expressed genes and pathways, we can identify some potential therapeutic targets and drug candidates. For example, targeting the differentially expressed gene family of muscle structural proteins[33-35]. We can explore the efficacy of related drugs to promote muscle regeneration and functional recovery.
Although we achieved a series of meaningful results in this study, there are some research limitations and future directions for improvement. First of all, this study focuses on transcriptomic analysis, which can be combined with proteomics and metabolomics to obtain more comprehensive molecular information in the future. Second, although we have verified the changes of some differentially expressed genes, the function and regulatory mechanism of these genes still need to be further studied. In addition, further functional experiments and gene knockout can be used to confirm the importance of differentially expressed genes in muscle development and function.In conclusion, this study provides an in-depth understanding of LAMA2 muscular dystrophy zebrafish through the construction of the model and muscle transcriptomics studies. We reveal differentially expressed genes and pathways associated with abnormal muscle development and function in the LAMA2 model, providing important clues for further study of the pathophysiological mechanisms and potential therapeutic targets of this disease. This study provides new ideas and directions for the treatment and clinical management of muscular dystrophy and is expected to make important contributions to the health and well-being of patients.
Successful construction of the LAMA2 zebrafish model; successfully simulated the clinical phenotype of LAMA2 muscular dystrophy, including muscle degeneration and atrophy. This provides a reliable model system for studying the pathophysiological process of the disease and seeking treatment strategies.
Muscle transcriptomic studies reveal differentially expressed genes and pathways. Using single-cell sequencing and RNA-Seq techniques, we identified changes in gene expression in muscle cells in the LAMA2 model. We identified multiple muscle cell subtypes and identified differentially expressed genes and pathways related to muscle development and function.
The molecular mechanisms underlying abnormal muscle development and function are revealed. Our results indicate abnormalities in the LAMA2 model involved in biological processes such as cytoskeletal recombination, mitochondrial function impairment, and inflammatory responses. These changes may be closely related to pathological features such as muscle degeneration and atrophy.
Providing clues to therapeutic targets and new drug development through analysis of differentially expressed genes and pathways, we identified a number of potential therapeutic targets and drug candidates. In particular, the study of differentially expressed gene families of muscle structural proteins is expected to provide new directions for therapeutic strategies for muscle regeneration and functional recovery.
Funding:
This article was supported by the scientific innovation start-up foundation of Fujian Maternity and Child Health Hospital (YCXM20-23).
Conflicts of interests:
The authors declare no conflict of interests.
References
- Katoch S, Patial V. Zebrafish: An emerging model system to study liver diseases and related drug discovery. J Appl Toxicol 2021;41(1):33-51.
[Crossref] [Google Scholar] [PubMed]
- Lai KP, Gong Z, Tse WK. Zebrafish as the toxicant screening model: Transgenic and omics approaches. Aquat Toxicol 2021;234:1-9.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Chen M, Chen C. Using the zebrafish as a genetic model to study erythropoiesis. Int J Mol Sci 2021;22(19):1-18.
[Crossref] [Google Scholar] [PubMed]
- Bashirzade AA, Zabegalov KN, Volgin AD, Belova AS, Demin KA, de Abreu MS, et al. Modeling neurodegenerative disorders in zebrafish. Neurosci Biobehav Rev 2022;138:104679.
[Crossref] [Google Scholar] [PubMed]
- Xie H, Li M, Kang Y, Zhang J, Zhao C. Zebrafish: An important model for understanding scoliosis. Cell Mol Life Sci 2022;79(9):1-16.
[Crossref] [Google Scholar] [PubMed]
- Xia H, Chen H, Cheng X, Yin M, Yao X, Ma J, et al. Zebrafish: An efficient vertebrate model for understanding role of gut microbiota. Mol Med 2022;28(1):1-20.
[Crossref] [Google Scholar] [PubMed]
- Wang J, Cao H. Zebrafish and medaka: Important animal models for human neurodegenerative diseases. Int J Mol Sci 2021;22(19):1-17.
[Crossref] [Google Scholar] [PubMed]
- Rosa JG, Lima C, Lopes-Ferreira M. Zebrafish larvae behavior models as a tool for drug screenings and pre-clinical trials: A review. Int J Mol Sci 2022;23(12):1-12.
[Crossref] [Google Scholar] [PubMed]
- Vasyutina M, Alieva A, Reutova O, Bakaleiko V, Murashova L, Dyachuk V, et al. The zebrafish model system for dyslipidemia and atherosclerosis research: Focus on environmental/exposome factors and genetic mechanisms. Metabolism 2022;129:155138.
[Crossref] [Google Scholar] [PubMed]
- Angelini C, Marozzo R, Pegoraro V. Current and emerging therapies in Becker Muscular Dystrophy (BMD). Acta Myol 2019;38(3):172-79.
[Google Scholar] [PubMed]
- Taheri F, Taghizadeh E, Pour MJ, Rostami D, Renani PG, Rastgar-Moghadam A, et al. Limb-girdle muscular dystrophy and therapy: Insights into cell and gene-based approaches. Curr Gene Ther 2019;19(6):386-94.
[Crossref] [Google Scholar] [PubMed]
- Datta N, Ghosh PS. Update on muscular dystrophies with focus on novel treatments and biomarkers. Curr Neurol Neurosci Rep 2020;20(6):1-14.
[Crossref] [Google Scholar] [PubMed]
- Kanagawa M, Toda T. Muscular dystrophy with ribitol-phosphate deficiency: A novel post-translational mechanism in dystroglycanopathy. J Neuromuscul Dis 2017;4(4):259-67.
[Crossref] [Google Scholar] [PubMed]
- Misaka T, Yoshihisa A, Takeishi Y. Titin in muscular dystrophy and cardiomyopathy: Urinary titin as a novel marker. Clin Chim Acta 2019;495:123-8.
[Crossref] [Google Scholar] [PubMed]
- Narasimhaiah D, Uppin MS, Ranganath P. Genetics and muscle pathology in the diagnosis of muscular dystrophies: An update. Indian J Pathol Microbiol 2022;65(5):259-70.
[Crossref] [Google Scholar] [PubMed]
- Hu P, Yuan L, Deng H. Molecular genetics of the POMT1-related muscular dystrophy-dystroglycanopathies. Mutat Res Rev Mutat Res 2018;778:45-50.
[Crossref] [Google Scholar] [PubMed]
- Wang S, Peng D. Cardiac involvement in Emery-Dreifuss muscular dystrophy and related management strategies. Int Heart J 2019;60(1):12-8.
[Crossref] [Google Scholar] [PubMed]
- Mandeville RM, Narayanaswami P. The birth of informed decisions: Pregnancy and muscular dystrophy. Muscle Nerve 2021;63(6):787-9.
[Crossref] [Google Scholar] [PubMed]
- Pozsgai E, Griffin D, Potter R, Sahenk Z, Lehman K, Rodino-Klapac LR, et al. Unmet needs and evolving treatment for limb girdle muscular dystrophies. Neurodegener Dis Manag 2021;11(5):411-29.
[Crossref] [Google Scholar] [PubMed]
- Rodrigues M, Yokota T. An overview of recent advances and clinical applications of exon skipping and splice modulation for muscular dystrophy and various genetic diseases. Methods Mol Biol 2018:31-55.
[Crossref] [Google Scholar] [PubMed]
- Chen Y, Song J, Ruan Q, Zeng X, Wu L, Cai L, et al. Single-cell sequencing methodologies: From transcriptome to multi-dimensional measurement. Small Methods 2021;5(6):2100111.
[Crossref] [Google Scholar] [PubMed]
- Liu S, Wang Z, Zhu R, Wang F, Cheng Y, Liu Y. Three differential expression analysis methods for RNA sequencing: Limma, EdgeR, DESeq2. J Vis Exp 2021;(175):e62528.
[Crossref] [Google Scholar] [PubMed]
- Honda M, Oki S, Kimura R, Harada A, Maehara K, Tanaka K, et al. High-depth spatial transcriptome analysis by photo-isolation chemistry. Nat Commun 2021;12(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Riaño-Pachón DM, Espitia-Navarro HF, Riascos JJ, Margarido GR. Modern approaches for transcriptome analyses in plants. In: Advances in plant omics and systems biology approaches. Springer Int Publishing 2022:11-50.
[Crossref] [Google Scholar] [PubMed]
- Nynca J, Słowińska M, Wiśniewska J, Jastrzębski J, Dobosz S, Ciereszko A. Ovarian transcriptome analysis of diploid and triploid rainbow trout revealed new pathways related to gonadal development and fertility. Animal 2022;16(8):1-13.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Xiao Y, Yang H, Lu L, Liu X, Lyu W. Transcriptome analysis reveals the genes involved in growth and metabolism in Muscovy ducks. Biomed Res Int 2021;2021:1-9.
[Crossref] [Google Scholar] [PubMed]
- Zhang B, Bai P, Wang D. Growth behavior and transcriptome profile analysis of strain under long-vs. short-term simulated microgravity environment. Pol J Microbiol 2022;71(2):161-71.
[Crossref] [Google Scholar] [PubMed]
- Coulter M, Entizne JC, Guo W, Bayer M, Wonneberger R, Milne L, et al. BaRTv2: A highly resolved barley reference transcriptome for accurate transcript-specific RNA-seq quantification. Plant J 2022;111(4):1183-202.
[Crossref] [Google Scholar] [PubMed]
- Saura CA, Deprada A, Capilla-López MD, Parra-Damas A. Revealing cell vulnerability in Alzheimer’s disease by single-cell transcriptomics. Semin Cell Dev Biol 2023;139:73-83.
[Crossref] [Google Scholar] [PubMed]
- Wang SW, Gao C, Zheng YM, Yi L, Lu JC, Huang XY, et al. Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Mol Cancer 2022;21(1):1-27.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Shi M, Ren Y, Xu H, Weng S, Ning W, et al. Recent advances and applications of CRISPR-Cas9 in cancer immunotherapy. Mol Cancer 2023;22(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Hirano J, Murakami K, Hayashi T. CRISPR-Cas9-based technology for studying enteric virus infection. Front Genome Ed 2022;4:1-8.
[Crossref] [Google Scholar] [PubMed]
- Fang T, Cao X, Ibnat M, Chen G. Stimuli-responsive nanoformulations for CRISPR-Cas9 genome editing. J Nanobiotechnology 2022;20(1):1-28.
[Crossref] [Google Scholar] [PubMed]
- Lakhawat SS, Malik N, Kumar V, Kumar S, Sharma PK. Implications of CRISPR-Cas9 in developing next generation biofuel: A mini-review. Curr Protein Pept Sci 2022;23(9):574-84.
[Crossref] [Google Scholar] [PubMed]
- Alkanli SS, Alkanli N, Ay A, Albeniz I. CRISPR/Cas9 mediated therapeutic approach in huntington’s disease. Mol Neurobiol 2023;60(3):1486-98.
[Crossref] [Google Scholar] [PubMed]
- Dholariya S, Parchwani D, Radadiya M, Singh RD, Sonagra A, Patel D, et al. CRISPR/Cas9: A molecular tool for ovarian cancer management beyond gene editing. Crit Rev Oncog 2022;27(4):1-22.
[Crossref] [Google Scholar] [PubMed]
- Ostrovidov S, Salehi S, Costantini M, Suthiwanich K, Ebrahimi M, Sadeghian RB, et al. 3D bioprinting in skeletal muscle tissue engineering. Small 2019;15(24):1-14.
[Crossref] [Google Scholar] [PubMed]
- Alarcin E, Bal-Öztürk A, Avci H, Ghorbanpoor H, Guzel DF, Akpek A, et al. Current strategies for the regeneration of skeletal muscle tissue. Int J Mol Sci 2021;22(11):1-28.
[Crossref] [Google Scholar] [PubMed]
- Brooks D, Bawa S, Bontrager A, Stetsiv M, Guo Y, Geisbrecht ER. Independent pathways control muscle tissue size and sarcomere remodeling. Dev Biol 2022;490:1-12.
[Crossref] [Google Scholar] [PubMed]
- da Cunha Bataglioli I, de Queiroz JV, Vieira JC, Cavalline NG, Braga CP, Buzalaf MA, et al. Mercury metalloproteomic profile in muscle tissue of Arapaima gigas from the Brazilian Amazon. Environ Monit Assess 2022;194(10):705.
[Crossref] [Google Scholar] [PubMed]
- Kato MK, Muro S, Kato T, Miyasaka N, Akita K. Spatial distribution of smooth muscle tissue in the female pelvic floor and surrounding the urethra and vagina. Anat Sci Int 2020;95(4):516-22.
[Crossref] [Google Scholar] [PubMed]
- Aryeetey OJ, Frank M, Lorenz A, Pahr DH. Fracture toughness determination of porcine muscle tissue based on AQLV model derived viscous dissipated energy. J Mech Behav Biomed Mater 2022;135:1-11.
[Crossref] [Google Scholar] [PubMed]
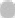 ): No difference; (
): No difference; ( ): Down-regulated genes and (
): Down-regulated genes and ( ): Up-regulated genes
): Up-regulated genes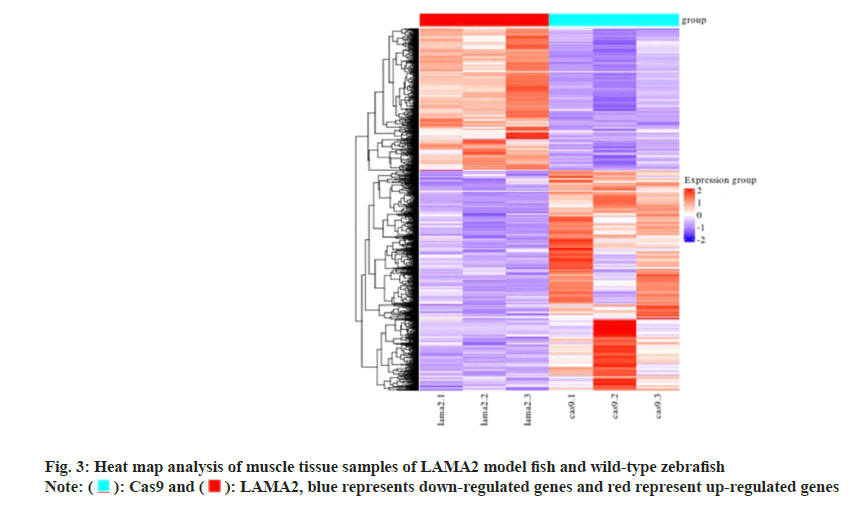
 ): Cas9 and (
): Cas9 and ( ): LAMA2, blue represents down-regulated genes and red represent up-regulated genes
): LAMA2, blue represents down-regulated genes and red represent up-regulated genes