- *Corresponding Author:
- Jihad Jalal Elias
Department of Biology,
College of Education,
Salahaddin University-Erbil,
Erbil,
Iraq
E-mail: jihadjalal990@gmail.com
| This article was originally published in a special issue, “Recent Developments in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(4) Spl Issue “141-147” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Considering the high prevalence of acute myelocytic leukemia in adults, understanding the pathophysiology of this disease and investigating cytogenetic markers play an important role in dealing with the disease for its classification, timely diagnosis, prognosis and predicting the response to the treatment process. The aim of this study was to investigate the molecular and cytogenetic status of patients with acute myelocytic leukemia in the Kurdistan region of Iraq. This study was conducted in 2021 on 40 patients referred to Zheen Erbil international hospital who were sampled using the convenience method. Samples were collected from peripheral blood samples or bone marrow samples of patients, after sampling in the laboratory. The processes of sample centrifugation and deoxyribonucleic acid extraction were performed. Then, different in silico tools were used to check the structure and function of proteins. Fluorescence in situ hybridization method was used to check the presence of p53 deletion. The obtained data was analyzed with statistical package for social sciences version 23 and descriptive statistical tests and regression. The average age of the examined patients was 37.275±22.234 y. 17p deletion was seen in 6 people under study and p53 mutation was seen in 13 out of the examined people and normal in 3 patients. Factors affecting acute myelocytic leukemia based on the linear regression were age, Odds ratio (0.014), p value≤0.001, confidence interval: 0.010-0.019 and factors affecting acute myelocytic leukemia mutations (changes) in gene based on the linear regression were p53 mutation, Odds ratio (0.5), p value≤0.003, confidence interval: 0.33-0.019. Evaluation of p53 gene mutation was significant in acute myelocytic leukemia and age also showed itself as an influential factor in acute myelocytic leukemia disease.
Keywords
Acute myelocytic leukemia, mutations, fluorescence in situ hybridization method, cytogenetic markers
Leukemia is a cancer of the hematopoietic tissues of the body, including the bone marrow and the lymphatic system[1]. Leukemia disrupts and the regular growth process of white blood cells was out of control. In acute leukemia, the body creates a large amount of immature cells and the production of healthy cells stops. Its diagnosis, unlike other cancers that have a mass, is done by examining the colony accumulation and expansion of immature myeloid cells in the bone marrow[2]. The prevalence of Acute Myelocytic Leukemia (AML) is higher among the elderly, as statistics indicate that 14 out of 100 000 adults over the age of 65 in the United States are affected[3]. In this disease, there is a heterogeneous group of neoplastic diseases that differ in the clinical course of the disease, the reaction process to the treatment plan and its genetic and molecular structure[4]. Considering that the mechanisms of cell proliferation in this disease are complex and widespread, it is likely that various mutagenic events occur to cause this disease[5]. Considering that more than 300 chromosomal translocations and genetic mutations have been identified for this AML, cytogenetic and molecular changes and their identification, along with the age factor, provides useful information about the subtypes of this disease and determine the prognosis, plan for related treatment methods and predict their effectiveness[6].
There are different methods to investigate cytogenetic abnormalities. Today, with the progress of the human genome project, there are a variety of specific Deoxyribonucleic Acid (DNA) probes and methods such as Fluorescence In Situ Hybridization (FISH) are used less, except to identify specific abnormalities in specific patients. But this method is accurate, sensitive and fast as a complementary method for conventional chromosome banding studies[7]. In 2002, Frohling et al.have used the FISH method to investigate cytogenetic and chromosomal abnormalities and pointed out its high diagnostic value[8]. Various studies have been conducted in relation to the types of mutations and cytogenetic abnormalities and their diagnostic value in investigating the prognosis of AML[9]. In 2022, Bi et al. in their study, indicated that being a smoker or not, does not affect overall survival in patients with AML with TP53 mutations; of course, they have suggested in conducting more studies[10]. Therefore, considering the importance of examining cytogenetic abnormalities in the context of timely diagnosis of AML disease and follow-up of appropriate treatment, this study was conducted with the aim of examining molecular and cytogenetic and their diagnostic and prognostic value in patients with AML in the Kurdistan region of Iraq.
Materials and Methods
Study design and settings:
This study was conducted in 2021 on 40 patients with AML referred to Zheen International Hospital, Erbil, Iraq.
The study included 30 AML patients and 10 normal control samples that were grouped according to the clinical characteristics of the patients, including average of age and gender. The sampling method was convenience. The inclusion criteria of patients with AML diagnosed by an oncologist were between the ages of 16 y and 60 y. Patients who did not want to continue to cooperate during the study were excluded from the study. Informed consent was taken from all the participants.
Procedure:
Sampling: Sample used was peripheral blood or bone marrow aspirate on Sodium (Na) heparin. Blood was transferred to a 15 ml conical centrifuge tube; they were centrifuged at 1500 rpm for 10 min; the supernatant was carefully removed; 10 ml of 0.075 M hypotonic Potassium chloride (KCl) were then added (prewarmed at 37°), mixed well and left to stand for 20 min at 37°. At the end of 20 min, the tubes were centrifuged, the supernatant was discarded and then 10 drops of freshly prepared fixative were added in a dropwise manner with good mixing in between. The tubes were then centrifuged at 1500 rpm for 10 min, the supernatant was discarded and the cell pellet was resuspended in 1 ml freshly prepared fixative, added dropwise with shaking. Then, the remaining fixative (9 ml) was slowly added and the suspension was mixed. The tubes were then centrifuged at 1500 rpm for 10 min; the supernatant was discarded and the cell pellet was resuspended again in 10 ml fixative with mixing. The washing steps with the fixative were repeated two more times, until the cell pellet appeared clean and fixative was clear. After that, the last supernatant was discarded, enough fixative was added to obtain a slightly cloudy suspension. Slides were then made or the suspension was kept in the refrigerator at 2°-8° till slide preparation was done.
In brief, the template slide was soaked for 2 min in 100 % methanol and then dried; 4 μl of the cell suspension was added to each of the eight areas of the slide in a sequence of alternating squares. The slides were then dipped in 2× Side Scatter (SSC) for 2 min at room temperature then dehydrated in ascending grades of ethanol (70 %, 90 % and 100 %) for 2 min each, left to air dry then warmed up at 37° on hot plate. A measure of 2 μl of the prewarmed hybridization solution was then added to each square of the eight areas.
The template slide was then carefully inverted on the prewarmed device and placed at 37° in the Hybrite (Vysis serial number 114650) for 10 min followed by denaturation at 75° for 2 min; finally the slide device sandwich was placed in the prewarmed chromophobe hybridization chamber which was left to float at 37°±1° in the water bath overnight (or in the Hybrite). The device was then removed from the slide and the slide was then washed, 4′,6-Diamidino-2-Phenylindole (DAPI)/antifade was applied, cover slip was placed over it and finally examined by the fluorescence microscope (Olympus Microscope BX51/61, Olympus, Japan) and by using Applied Imaging CytoVisionTM system (Microscope model). The blood samples were stored at -20° until further analysis.
Genomic extraction:
Genomic DNA from blood specimens was prepared using a DNA extraction kit (Thermofisher, United States of America (USA)) and following the manufacturer’s instructions with minor modification. Briefly, qualification and quantification of DNA concentration was performed by using NanoDrop spectrophotometer (Biometrica). Samples of genomic DNA with (A260-A320)/(A280-A320) ratio more than 1.7 and outputs more than 30 ng/μl were obtained.
Mutation analysis:
Twist Human Core Monogenic Enzymatic Fragmentation (EF) multiplex complete kit was used for library construction and MGIEasy FS DNA Library Prep Kit was designed for the library to get ready for sequencing on the Mouse Genome Informatics (MGI) system. The library was sequenced on the (MGIDNBSEQG400, China) instrument generating 150 bp paired-end read with 100X mean target coverage. With FastQC, raw fastq files were quality controlled. Then reads were aligned to the reference human genome (hg19) using Burrows-Wheeler Aligner (BWA). Variants were identified with Genome Analysis Toolkit (GATK). Integrative Genomic Viewer (IGV) software was used for variants visualization.
In silico analysis:
Different in silico tools were used to predict the effect of mutation on the structural features or protein function. Polymorphism Phenotyping (PolyPhen-2) and Sorting Intolerant From Tolerant (SIFT) were used to assess the functional effects of variants. MutationTaster was used for the evaluation of mutation effect on protein function and structure. Align-Grantham Variation and Grantham Difference (GVGD) was used to compute a biochemical distance score[11].
Interphase FISH technique:
The Cytocell Multiprobe AML panel (UKPI027/CE v005; Cytocell Technologies Ltd., Cambridge, England) was designed to detect p53 and FISH probes were used on a single slide in a single hybridization experiment which was used for the p53 deletion.
Data analysis:
Data was analyzed with Statistical Package for the Social Sciences (SPSS) version 23 statistical software and descriptive statistical tests, regression for determining factors affecting (prognosis) AML and mutations (changes) in gene.
Results and Discussion
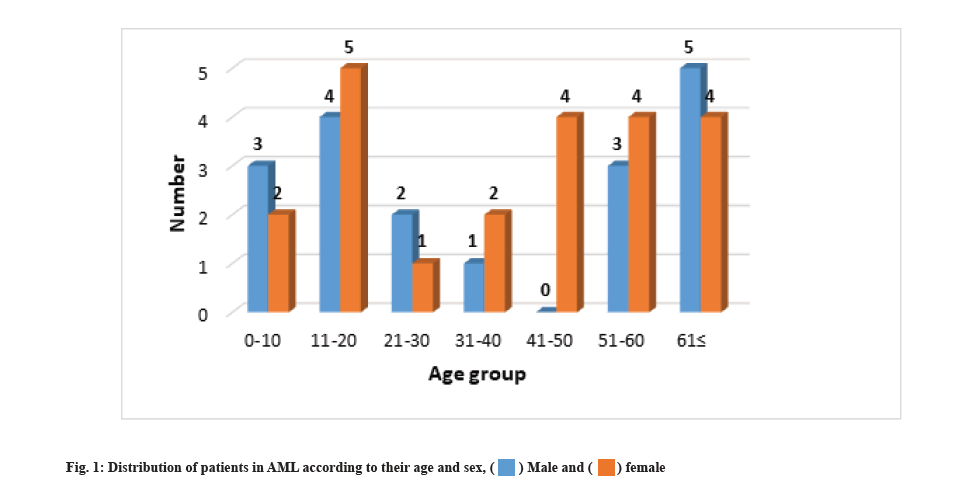
Clinical and demographic characteristics were explained here. This study aimed to investigate the effect of deletion of chromosome 17p using FISH and p53 gene mutation in the diagnosis of AML in Kurdistan region of Iraq. Fig. 1 shows the gender of the participants in the study according to age groups. The results showed that (45 %) 18 people are male and (55 %) 22 are female. Most of the patients are in the age groups of 11 y-20 y and 50 y and above. The average age of the examined patients was 37.275±22.234 y.
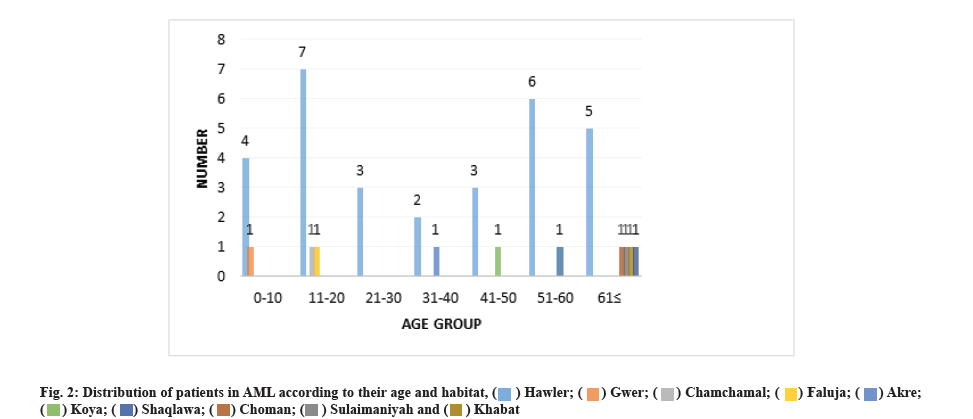
Examining the residence of patients according to age groups showed that most of the patients (75 %) 30 live in Hawler city. 7 of the patients in the age group of 11- 20 y, 6 of the patients in the age group of 51-60 y, and 5 of the patients in the age group of 61 y and above also live in Hawler city. And the patients are also seen in other cities of Kurdistan (fig. 2).
The smoking status of the examined patients showed that 34 (90 %) of the patients were non-smokers and only 6 (10 %) were smokers. None of the patients had a history of alcohol consumption. Examining the blood group status of people, it showed that the highest frequency of blood group in patients is O+ve blood group with a frequency (55 %) of 22 people. After that, blood type A+ve was the most common in patients with a frequency of 8 (20 %). Blood group AB+ve had frequency (10 %) of 4 and blood groups AB-ve, A-ve and O-ve had frequency (5 %) of 2 people. The type of drugs used for patients showed that 30 patients (75 %) use sodium heparin and 10 (25 %) use sodium heparin and Ethylenediamine Tetraacetic Acid (EDTA). 17p deletion was seen in 6 people under study and p53 mutation was seen in 13 of the examined people in 3 patients (Table 1).
| Variable | Female patients | Percentage (%) | |
|---|---|---|---|
| Smoker | No | 34 | 90 % |
| Yes | 6 | 10 % | |
| Alcohol consumption | No | 40 | 100 % |
| Yes | 0 | 0 | |
| Blood group | AB+ve | 4 | 10 % |
| AB-ve | 2 | 5 % | |
| A+ve | 8 | 20 % | |
| A-ve | 2 | 5 % | |
| O+ve | 22 | 55 % | |
| O-ve | 2 | 5 % | |
| Type of medicine | Sodium heparin and EDTA | 10 | 25 % |
| Sodium heparin | 30 | 75 % | |
| 17p deletion | Yes | 6 | 85 % |
| No | 34 | 15 % | |
| P53 mutation | Yes | 3 | 23.10 % |
| No | 10 | 76.90 % | |
Table 1: Clinical and Demographic Characteristics of Patients of AML
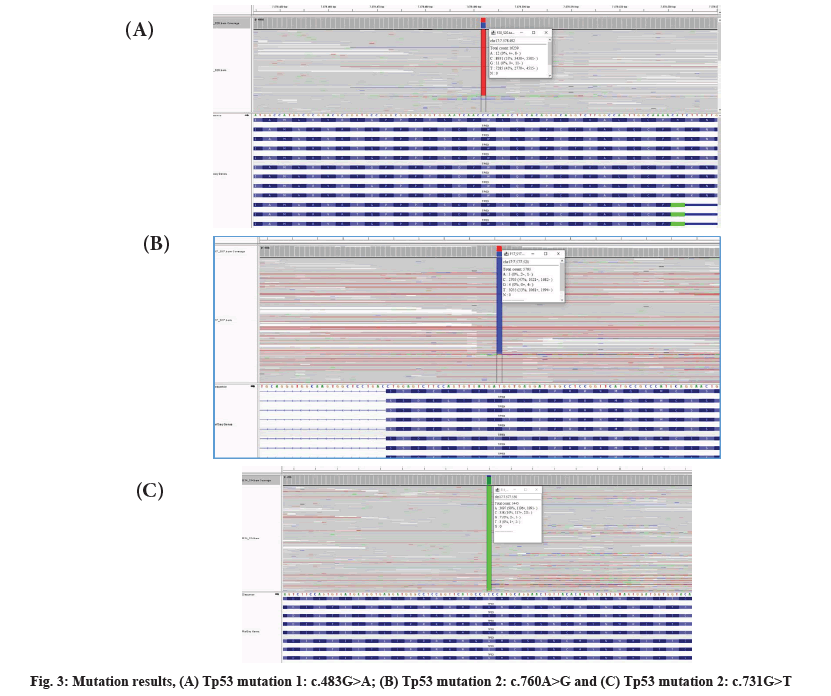
Mutation results were shown as follows. Next generation sequencing was performed on 13 samples (10 AML cases and 3 normal controls). We found mutations in 3 samples (fig. 3).
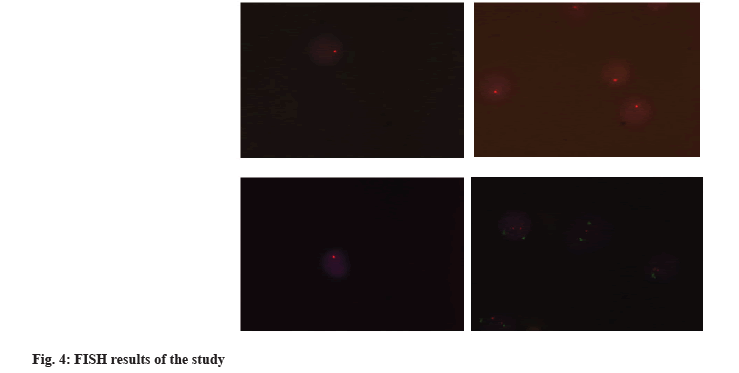
We performed FISH analysis for 17p deletion for 40 samples (30 sample AML cases and 10 normal controls). We detected 17p deletion in 6 samples of AML cases (fig. 4).
Factors affecting AML based on the linear regression were age, Odds Ratio (OR): 0.014, p-value≤0.001, Confidence Interval (CI): 0.010-0.019, which has an increasing role in the AML. These results indicate with an increase in 1 y of life, the risk of AML (1 %) will increase (Table 2).
| Variable | Odds Ratio (OR) | p-value, (CI 95 %)* |
|---|---|---|
| Age | 0.014 | 0.001, CI 95 %: -0.010-0.018 |
| Gender | -0.051 | 0.721, CI 95 %: -0.336-0.235 |
| Habitat | 0.046 | 0.104, CI 95 %: -0.010-0.103 |
| Smoking | 0.294 | 0.132, CI 95 %: -0.092-0.680 |
| Blood group | 0.508 | 0.348, CI 95 %: -0.066-0.182 |
Table 2: Factors Affecting (Prognosis) AML
Factors affecting AML mutations (changes) in gene based on the linear regression were p53 mutation, OR: 0.5, p-value≤0.003, CI: 0.33-0.019 which has an increasing role in the AML. These results indicate with an increase in mutations (changes) in the p53, AML also increases (Table 3).
| Variable | Odds Ratio (OR) | p-value, (CI 95 %)* |
|---|---|---|
| 17p deletion | -0.294 | 0.132, CI 95 %: -0.68-0.092 |
| p53 mutation | 0.5 | 0.03, CI 95 %: 0.33-0.954 |
Table 3: Factors Affecting (Prognosis) AML Mutations (Changes) in Gene
This study investigated the effect of deletion of chromosome 17p using FISH and p53 gene mutation in the diagnosis of AML in Kurdistan region of Iraq. The results of the study showed that p53 mutation can be considered as a suitable evaluation criterion in the evaluation and diagnosis of AML. In addition, the results of the study indicate a significant effect of age in increasing the risk of AML, as the risk of infection will increase with age.
P53 is an important gene that plays an important and key role in the destruction of tumors and it is responsible for the important task of apoptosis, aging and DNA repair. Although it is considered as a rare mutation in AML patients, it should be noted that, it has the worst prognosis for these patients, so that the survival of patients in whom this mutation is diagnosed is less than 1 y[12]. The results of the present study showed that, out of 13 patients in whom chromosomal p53 mutation was investigated, 3 patients had this mutation and the regression results indicated that this mutation can be considered more than in the assessment of AML. In the study by Kim et al.[13] who examined the p53 mutation and its results on AML patients, the results indicated that patients who have this mutation can be better compared to other patients in disease evaluation, prognosis and treatment results. It should be noted that these results are consistent with the results of this study, which also shown the important role of this mutation in the evaluation of the disease.
According to The Cancer Gene Atlas (TCGA) data, about 8 % of AML patients have p53 mutation[14]. The presence of this mutation in AML patients will cause the gene unstoppable cell growth and lead to disruption in the cells work process and eventually lead to sarcoma. This mutation can either be inherited from parents or occur during life. In various studies, the heterogeneity and characteristics of this mutation and its effects have been considered. Apart from the molecular characteristics, it has been shown that this mutation is very important and significant in clinical evaluations, the process of therapeutic decisions, and the final outcome, which is in harmony with the current discussion that revealed the importance of this mutation in the clinical evaluation of AML[15]. Also, in the treatment strategies and management process of AML disease, the removal and mutation of p53 should be given more attention. In most of the studies, this mutation alone and together with other mutations has a negative prognosis in the survival of patients, disease recurrence and many problems for AML patients[16,17].
In AML patients, another common deviation is 17p deletion, which has an unfavorable prognosis for patients. In this study, 6 patients were identified with this mutation, but it did not show a significant trend in the evaluation of AML disease in this study. In other studies, it has been shown that the presence of this mutation and its evaluation in relation to AML disease reduces the survival of patients and interferes in the care and treatment of patients, which is not in line with the results of the present study[18,19].
In the study of demographic variables and the evaluation of these variables, age showed a significant effect and the regression results showed that increasing age brings the risk of AML and this can be effective in the prognosis of the disease, and this result is in agreement with the results of the study of Möricke et al.[20], that increases were associated with poor prognosis of patients, which is consistent. Also, in the study of Kristensen et al.[21], smoking did not show a significant effect on the evaluation of AML. It is not consistent that smoking had a significant effect on the evaluation and prognosis. It has been shown in various studies that the prevalence of AML in men is higher than in women[22]. In this study, most of the participants in the study were women and no significant difference was seen in the evaluation of AML between the two sexes.
There are few limitations in this study. The number of AML patients was less. Also, in this study, due to the lack of time and data, it was possible to measure the survival of patients, which can further show the value of the work.
In this study, 17p deletion played a significant role in the evaluation of AML to achieve a significant result and the evaluation of p53 gene mutation has been significant in AML where the age also showed itself as an influential factor in AML disease. It seems necessary to conduct more studies with a larger sample size and pay more attention to the influencing factors.
Author’s contributions:
All authors passed the criteria for authorship contribution based on recommendations of the international committee of medical journal editors.
Acknowledgements:
We would like to thank all the parents of the patients who patiently helped in this research.
Conflict of interests:
The authors declared no conflict of interest.
References
- Pelcovits A, Niroula R. Acute myeloid leukemia: A review. R I Med J 2020;103(3):38-40.
[Google scholar] [PubMed]
- Döhner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med 2015;373(12):1136-52.
[Crossref] [Google scholar] [PubMed]
- Shallis RM, Wang R, Davidoff A, Ma X, Zeidan AM. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev 2019;36:70-87.
[Crossref] [Google scholar] [PubMed]
- Bullinger L, Döhner K, Döhner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol 2017;35(9):934-46.
[Crossref] [Google scholar] [PubMed]
- Wang M, Yang C, Zhang L, Schaar DG. Molecular mutations and their co-occurrences in cytogenetically normal acute myeloid leukemia. Stem Cells Int 2017;2017:1-11.
[Crossref] [Google scholar] [PubMed]
- Grimwade D, Mrózek K. Diagnostic and prognostic value of cytogenetics in acute myeloid leukemia. Hematol Oncol Clin North Am 2011;25(6):1135-61.
[Crossref] [Google scholar] [PubMed]
- Abbas N, Samra Waheed AJ, Saleem A, Shamsi TS. Acute leukemia of ambiguous lineage with a rare abnormality Del17p by FISH analysis. Int J Health Sci 2020;5(2):79-82.
- Fröhling S, Skelin S, Liebisch C, Scholl C, Schlenk RF, Döhner H, et al. Comparison of cytogenetic and molecular cytogenetic detection of chromosome abnormalities in 240 consecutive adult patients with acute myeloid leukemia. J Clin Oncol 2002;20(10):2480-5.
[Crossref] [Google scholar] [PubMed]
- Velloso ED, Motta CH, Furtado JB, Bacal NS, Silveira PA, Moyses CB, et al. Molecular and cytogenetic abnormalities in acute myeloid leukemia: Review and case studies. Einstein (São Paulo) 2011;9:184-9.
[Crossref] [Google scholar] [PubMed]
- Bi X, French Z, Palmisiano N, Wen KY, Wilde L. The prognostic impact of cigarette smoking on survival in acute myeloid leukemia with TP53 mutations and/or 17p deletions. Ann Hematol 2022;101(6):1251-9.
[Crossref] [Google scholar] [PubMed]
- Yılmaz H, Toy HI, Marquardt S, Karakülah G, Küçük C, Kontou PI, et al. In silico methods for the identification of diagnostic and favorable prognostic markers in acute myeloid leukemia. Int J Mol Sci 2021;22(17):9601.
[Crossref] [Google scholar] [PubMed]
- Granowicz EM, Jonas BA. Targeting TP53-mutated acute myeloid leukemia: Research and clinical developments. Onco Targets Ther 2022;15:423-36.
[Crossref] [Google scholar] [PubMed]
- Kim K, Maiti A, Loghavi S, Pourebrahim R, Kadia TM, Rausch CR, et al. Outcomes of TP53‐mutant acute myeloid leukemia with decitabine and venetoclax. Cancer 2021;127(20):3772-81.
[Crossref] [Google scholar] [PubMed]
- Molica M, Mazzone C, Niscola P, de Fabritiis P. TP53 mutations in acute myeloid leukemia: Still a daunting challenge? Front Oncol 2021;10:610820.
[Crossref] [Google scholar] [PubMed]
- Grob T, Al Hinai AS, Sanders MA, Kavelaars FG, Rijken M, Gradowska PL, et al. Molecular characterization of mutant TP53 acute myeloid leukemia and high-risk myelodysplastic syndrome. Blood 2022;139(15):2347-54.
[Crossref] [Google scholar] [PubMed]
- Seifert H, Mohr B, Thiede C, Oelschlägel U, Schäkel U, Illmer T, et al. The prognostic impact of 17p (p53) deletion in 2272 adults with acute myeloid leukemia. Leukemia 2009;23(4):656-63.
[Crossref] [Google scholar] [PubMed]
- Sebaa A, Ades L, Baran‐Marzack F, Mozziconacci MJ, Penther D, Dobbelstein S, et al. Incidence of 17p deletions and TP53 mutation in myelodysplastic syndrome and acute myeloid leukemia with 5q deletion. Genes Chromosomes Cancer 2012;51(12):1086-92.
[Crossref] [Google scholar] [PubMed]
- Warnstorf D, Bawadi R, Schienke A, Strasser R, Schmidt G, Illig T, et al. Unbalanced translocation der (5; 17) resulting in a TP53 loss as recurrent aberration in myelodysplastic syndrome and acute myeloid leukemia with complex karyotype. Genes Chromosomes Cancer 2021;60(6):452-7.
[Crossref] [Google scholar] [PubMed]
- Britt A, Mohyuddin GR, McClune B, Singh A, Lin T, Ganguly S, et al. Acute myeloid leukemia or myelodysplastic syndrome with chromosome 17 abnormalities and long-term outcomes with or without hematopoietic stem cell transplantation. Leuk Res 2020;95:106402.
[Crossref] [Google scholar] [PubMed]
- Möricke A, Zimmermann M, Reiter A, Gadner H, Odenwald E, Harbott J, et al. Prognostic impact of age in children and adolescents with acute lymphoblastic leukemia: Data from the trials ALL-BFM 86, 90, and 95. Klin Padiatr 2005;217(06):310-20.
[Crossref] [Google scholar] [PubMed]
- Kristensen D, Nielsen LB, Roug AS, Kristensen TC, El‐Galaly TC, Nørgaard JM, et al. The prognostic effect of smoking status on intensively treated acute myeloid leukaemia-A Danish nationwide cohort study. Br J Haematol 2020;190(2):236-43.
[Crossref] [Google scholar] [PubMed]
- Singh SK, Lupo PJ, Scheurer ME, Saxena A, Kennedy AE, Ibrahimou B, et al. A childhood acute lymphoblastic leukemia genome-wide association study identifies novel sex-specific risk variants. Medicine 2016;95(46):e5300.
[Crossref] [Google scholar] [PubMed]