- *Corresponding Author:
- S. G. Hwang
Department of Animal Life and Environmental Science, Hankyong National University, Anseong City 456-749, Korea
E-mail: sghwang@hknu.ac.kr
| Date of Received | 24 November 2019 |
| Date of Revision | 27 March 2020 |
| Date of Acceptance | 25 April 2020 |
| Indian J Pharm Sci 2020;82(3):456-464 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In this study, the antimetastatic effect of Chelidonium majus ethanol extract on hepatocellular carcinoma in vitro was investigated. The viability of SK-Hep1 and normal liver cells was determined using a cell counting kit-8 assay. Wound healing assays were performed to investigate SK-Hep1 cell migration and various metastatic characteristics including adhesion, aggregation and invasion were also measured using these cells. Furthermore, the proteolytic activity of extracellular matrix metalloproteinase-9 was measured using gelatin zymography. Expression levels of matrix metalloproteinase-2, matrix metalloproteinase-9, membrane type 1-matrix metalloproteinase and tissue inhibitor of metalloproteinase-1 were measured using reverse transcription-polymerase chain reaction and Western blotting. Chelidonium majus ethanol extract significantly inhibited the proliferation of SK-Hep1 hepatocellular carcinoma cells in a dose-dependent manner. Moreover, metastatic characteristics including adhesion, migration, aggregation and invasion were significantly suppressed by Chelidonium majus ethanol extract treatment. Further, this preparation downregulated the expression of matrix metalloproteinase-2, matrix metalloproteinase-9 and membrane type 1-matrix metalloproteinase, but upregulated tissue inhibitor of metalloproteinase-1 and tissue inhibitor of metalloproteinase-2, in a dose-dependent manner. Additionally, the proteolytic activity of matrix metalloproteinase-9 was greatly diminished with 400 µg/ml of Chelidonium majus ethanol extract. Taken together, it was suggested that Chelidonium majus ethanol extract might exert an antimetastatic effect on hepatocellular carcinoma cells by inhibiting proliferation, adhesion, migration, aggregation and invasion through the downregulation of matrix metalloproteinases and upregulation of tissue inhibitor of metalloproteinases. Thus, this extract could represent a promising therapeutic agent for this disease.
Keywords
Chelidonium majus, SK-Hep1 cells, metastasis, matrix metalloproteinase
Cancer is the second leading cause of global mortality, causing 8.7 million deaths in 2015[1]. In particular, Mongolia has the highest incidence of liver cancer, which is almost eight times higher than the global average[2]. Unsurprisingly, extensive research is being conducted on potential resources to develop optimal drugs for the treatment of malignant tumors. Despite the advances in research and therapeutics, the incidence of cancer has not decreased in recent years and instead, appears to increase annually[1,3]. The spread of cancer cells from the primary site to other body parts, known as metastasis, is mainly responsible for the incurable nature of malignant tumors and 90 % of cancerassociated deaths[4]. Hepatocellular carcinoma (HCC), a primary liver malignancy, is characterized by its ability to spread locally within the liver, invade blood vessels, and subsequently metastasize to distant organs[5-7]. The aggressiveness of HCC has been a dilemma for decades, and the current treatment methods are limited to liver transplantation and surgical resection[5]. Recently, researchers have focused on identifying alternative sources of cancer drugs such as natural products from medicinal plants[8]. Botanical resources are available abundantly and associated with minimal side effects, making them excellent candidates for future anticancer drug discovery.
Chelidonium majus L., an herbal plant belonging to the family Papaveraceae, is an important ingredient in traditional Chinese medicine and Western phytotherapy[9-11]. It is well known that this herbal plant have the phytochemical composition such as alkaloids and phenolic compounds (kaempferol, quercetin, caffeic acid, ferulic acid). Mongolian traditional herbalists have used C. majus to relieve pain, cough, fever, and swelling from microbial infection and to treat stomach and liver diseases[11]. Numerous studies have reported the hepatoprotective[12], antimicrobial[13], immunomodulatory[9] and antiinflammatory[14] effects of crude extracts or purified compounds from C. majus in vitro and in vivo. In addition, this plant has been previously reported to exert potent anticancer effects[15-17].
Cancer drug discovery has primarily been focused on the inhibition of carcinogenesis; however, to date, no antimetastatic drugs are clinically available, and pharmacists are developing an interest in designing drugs with both anticancer and antimetastatic effects[18,19]. Studies have shown Mongolian C. majus to be an excellent candidate for the development of a novel anticancer drug, but its antimetastatic potential has not been investigated. Therefore, in the present study, the effect of C. majus ethanol extract (CME) on the growth, adhesion, migration, aggregation and invasion of the SK-Hep1 human HCC cell line in vitro was investigated.
Materials and Methods
Preparation of CME:
C. majus was collected from Handgait, Ulaanbaatar, Mongolia. The air-dried plants were ground and 50 g of the powdered material was macerated in 500 ml of 80 % ethanol for 48 h. Then, the suspension was filtered, the ethanol was evaporated using a rotary vacuum evaporator and the extract was freeze-dried. A 10-mg/ml stock solution of CME was prepared by dissolving freeze-dried powder in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Rockville, MD, USA).
Cell culture:
Human SK-Hep1 HCC cells were obtained from the Korean Cell Line Bank and Chang liver cells were obtained from Konkuk University in Korea. Both cell lines were cultured in DMEM containing 10 % fetal bovine serum (FBS; Gibco), and 1 % penicillinstreptomycin (Gibco) in a humidified 5 % CO2 atmosphere at 37°.
Cell viability analysis:
Cells were seeded at 1×105 cells/well in a 96-well plate containing DMEM with 10 % FBS and incubated for 24 h. Cells were incubated with different concentrations of CME (0, 50, 100, 200, and 400 μg/ml) for 24 h. Viability was measured using a cell counting kit (CCK)-8 assay (Dojindo, Tokyo, Japan) according to the manufacturer’s instructions. Optical density was measured at 450 nm using an Infinite F50 microplate reader (Tecan, Mannedorf, Switzerland). Viability of cells treated with CME was expressed as a percentage of untreated cell viability.
Cell adhesion assay:
In brief, a 6-well plate was coated with 0.1 % gelatin overnight at 37° and air-dried for 30 min. SK-Hep1 cells (105 cells/ml) were suspended in DMEM containing CME (0, 100, 200, and 400 μg/ml), plated into the wells and incubated at 37° with 5 % CO2 for 6 h. Then, the medium was removed and cells were washed twice with phosphate-buffered saline (Gibco). The attached cells were counted under a microscope (Olympus CK40, Tokyo, Japan).
Cell aggregation assay:
SK-Hep1 cells (105 cells/ml) were suspended in DMEM containing CME (0, 100, 200, and 400 μg/ml). Then, the cell suspension (20 μl) was incubated on the undersurface of a 60-mm culture dish lid (Sigma-Aldrich, St. Louis, MO, USA). Cell aggregates were monitored under a microscope after 0, 2, 4, 6, 12, and 24 h. Single and multiple cell aggregates were counted as a single particle.
Cell migration assay:
The mobility of cancer cells was studied using the wound healing assay. SK-Hep1 cells (105 cells/ml) were plated in 6-well plates; when cells were 100 % confluent, a wound was created using a plastic pipette tip in the middle of the wells. Then, cells were treated with increasing concentrations of CME (0, 100, 200, 400 μg/ml). Wound closure was subsequently observed under a microscope and images were acquired at 0, 24, and 48 h.
Cell invasion assay:
Transwell chambers with 8-μm pore-size filters (Sigma- Aldrich) in a 6-well plate were used to examine the invasion of SK-Hep1 cells. Matrigel (BD Biosciences, Bedford, MA, USA) was diluted in cold DMEM (1:20) and added to the filters, which were air-dried under a laminar hood overnight. SK-Hep1 cells (105 cells/ml) were suspended in DMEM containing CME (0, 100, 200, and 400 μg/ml) and added to the upper compartment of invasion chambers. DMEM containing 10 % FBS was added to the lower compartment as a chemoattractant. After 24 h, cells on the lower surface were fixed with 70 % ethanol and stained with 0.2 % crystal violet for 10 min. Invading cells were observed using a microscope. Then, crystal violet was extracted with 10 % acetic acid for 5 min and the absorbance (nm) was measured at 595 using an ELISA plate reader (Tecan). The invasion of treated cells was expressed as the percentage of control cells.
Gelatin zymography:
Secreted matrix metalloproteinase (MMP)-9 activity was studied using gel zymography as described in the Gelatin Zymography protocol by Abcam, UK. SKHep1 cells (105 cells/ml) were exposed to CME (0, 100, 200, and 400 μg/ml) for 48 h and activity in treated cells was expressed as a percentage of that in control cells.
Reverse transcription-polymerase chain reaction (RT-PCR):
Total RNA was isolated from SK-Hep1 cells (105 cells/ ml) exposed to the indicated concentrations of CME (0, 100, 200, and 400 μg/ml) using Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Then, extracted RNA (1 μg) was reversed transcribed into cDNA using M-MuLV reverse transcriptase (Fermentas, Vilnius, Lithuania) and metastasis-related gene expression was quantified using the Maxime PCR PreMix (i-MAX II, iNtRON Biotechnology, Seongnam, Korea). PCR conditions consisted of an initial denaturation step at 95° for 5 min, followed by 30 amplification cycles consisting of denaturation for 40 s at 95°, annealing for 40 s (temperature 56-62°), and extension for 1 min at 72°. Primer sequences are shown in Table 1 and their relative expression was normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels, as a control.
| Name | Forward primer (5ʹ→3ʹ) | Reverse primer (5ʹ→3ʹ) |
|---|---|---|
| GAPDH | CGAGATCCCTCCAAAATCAA | AGGTCCACCACTGACACGTT |
| MMP-2 | GGCCCTGTCACTCCTGAGAT | GGCATCCAGGTTATCGGGGA |
| MMP-9 | CGGAGCACGGAGACGGGTAT | TGAAGGGGAAGACGCACAGC |
| MT1-MMP | TGGGTAGCGATGTCTTC | AGTAAGCAGTCTGGGT |
| TIMP1 | GATCCAGCGCCCAGAGAGACACC | TTCCACTCCGGCATT |
Table 1: List of Primers to Detect Metastasis-Related Genes
Western blot analysis:
SK-Hep1 cells (105 cells/ml) were allowed to adhere overnight in 6-well plates and cultured with 0, 100, 200, and 400 μg/ml CME. After 24 h, protein was extracted from cells using a protein extraction solution (iNtRON Biotechnology) and the protein concentration was standardized using a modified Bradford assay. Then, 25 μg of each protein sample was separated using 10 % SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 5 % skim milk and probed with the following primary antibodies (1:1000 dilution), antiGAPDH, antiMMP-2, antiMMP-9, antimembrane type 1 (MT1)-MMP, and antitissue inhibitor of metalloproteinase-1 (TIMP1, Cell Signaling Technology, Danvers, MA, USA). After incubation with horseradish peroxidase-conjugated secondary antibodies (1:2000 dilution), the membranes were treated with enhanced chemiluminescence Westsave Gold reagent (AbFrontier, Seoul, Korea) for 10 min and exposed to radiographic film (Agfa HealthCare, Greenville, SC, USA). Relative expression was normalized to that of GAPDH, which served as the control.
Statistical analysis:
All experiments were performed in triplicate and repeated at least three times. Quantitative data were expressed as means±standard deviations. Differences between means were calculated using an analysis of variance (ANOVA) followed by Duncan’s multiple range test. P<0.05 was considered statistically significant and these were calculated using SAS/STAT® software.
Results and Discussion
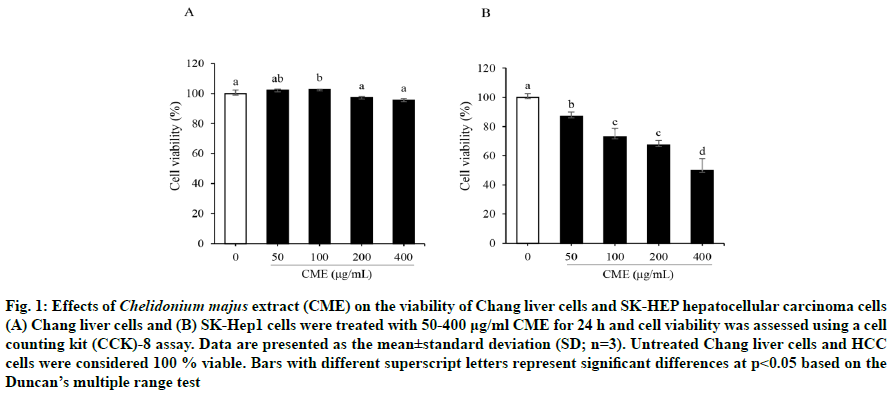
Chang liver cell viability was not altered by CME upto 400 μg/ml (fig. 1A); however, CME significantly inhibited SK-Hep1 HCC cell proliferation in a dosedependent manner (fig. 1B). Moreover, this preparation exerted a stronger antiproliferative effect against SKHep1 cells (half-maximal inhibitory concentration (IC50) = 369.35 μg/ml) than against Chang normal liver cells (IC50 = 4407.69 μg/ml) after a 24-h treatment. Therefore, the following experiments were performed using 100, 200, and 400 μg/ml CME.
Fig. 1: Effects of Chelidonium majus extract (CME) on the viability of Chang liver cells and SK-HEP hepatocellular carcinoma cells (A) Chang liver cells and (B) SK-Hep1 cells were treated with 50-400 μg/ml CME for 24 h and cell viability was assessed using a cell counting kit (CCK)-8 assay. Data are presented as the mean±standard deviation (SD; n=3). Untreated Chang liver cells and HCC cells were considered 100 % viable. Bars with different superscript letters represent significant differences at p<0.05 based on the Duncan’s multiple range test
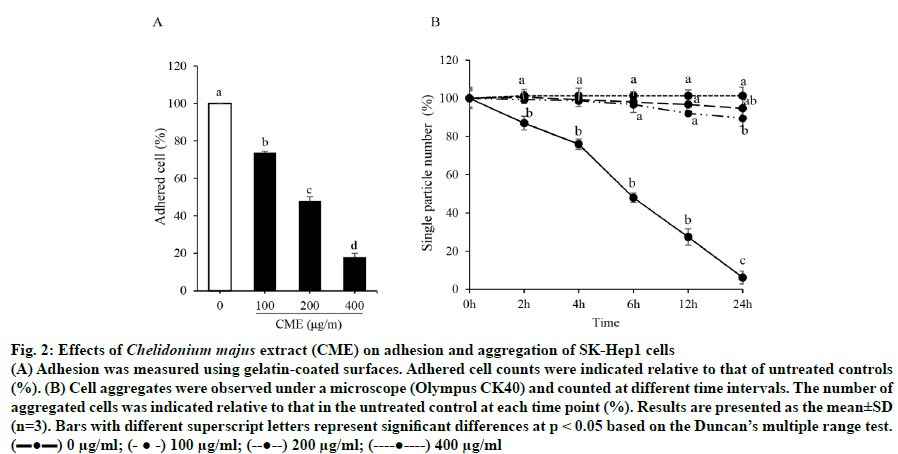
As shown in fig. 2A, cell adhesion to gelatincoated surfaces was significantly decreased by 52.01±5.91 % and 82.04±2.09 % following a 6-h treatment with 200 and 400 μg/ml CME, respectively, compared to that in untreated cells. Additionally, 100 μg/ml of CME inhibited the colony formation of SKHep1 cells by 89.61±4.33 % after a 24-h treatment, based on a cell aggregation assay (fig. 2B).
Fig. 2: Effects of Chelidonium majus extract (CME) on adhesion and aggregation of SK-Hep1 cells
(A) Adhesion was measured using gelatin-coated surfaces. Adhered cell counts were indicated relative to that of untreated controls (%). (B) Cell aggregates were observed under a microscope (Olympus CK40) and counted at different time intervals. The number of
aggregated cells was indicated relative to that in the untreated control at each time point (%). Results are presented as the mean±SD
(n=3). Bars with different superscript letters represent significant differences at p < 0.05 based on the Duncan’s multiple range test.
(▬●▬) 0 μg/ml; (- ● -) 100 μg/ml; (--●--) 200 μg/ml; (----●----) 400 μg/ml
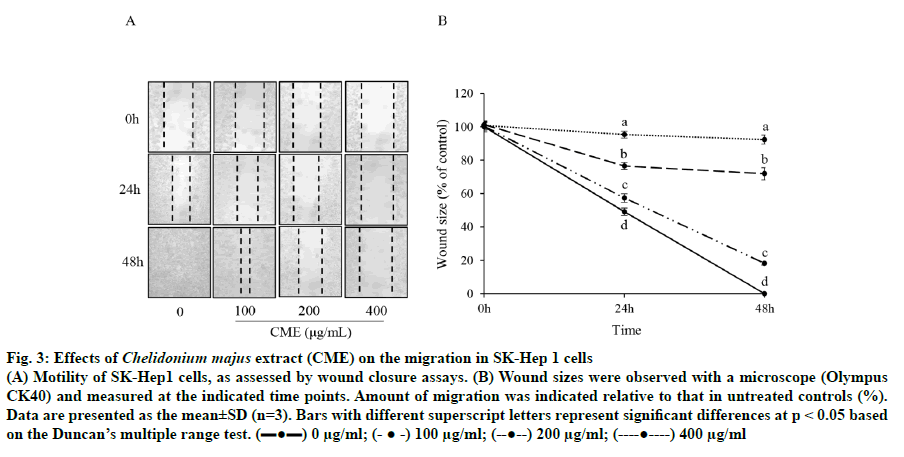
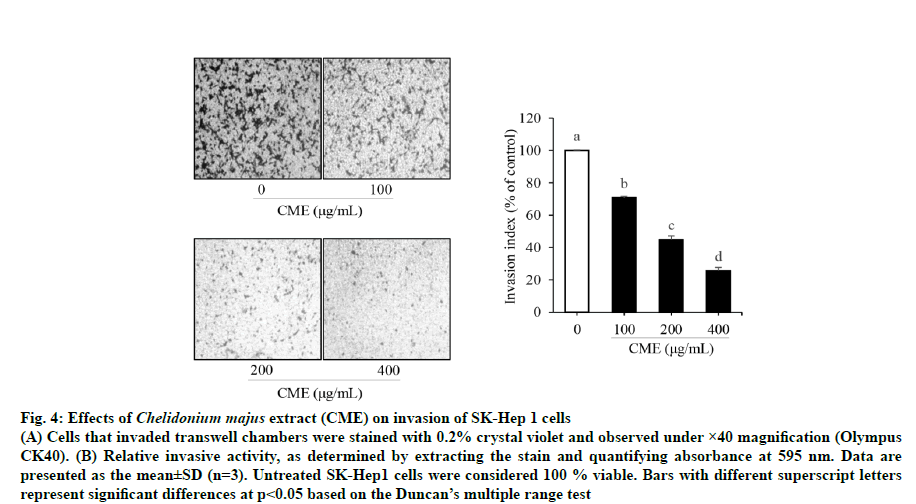
Cell motility was assessed to determine whether CME could inhibit HCC migration using a scratch wound healing assay. As shown in fig. 3, the wound size was reduced by 28.09±3.64 % in cells treated with 200 μg/ml CME compared to that in untreated cells after 24 h; moreover, untreated wounds healed completely after 48 h. Next, a Matrigel-coated chamber assay was performed to confirm the inhibitory effect of CME on HCC cell migration. CME significantly suppressed HCC cell migration in a dose-dependent manner (fig. 4). Further, invasion was reduced by 29.23±0.89, 55.13±2.25, and 74.43±2.10 % after CME treatment with 100, 200, and 400 μg/ml, respectively (fig. 4).
Fig. 3: Effects of Chelidonium majus extract (CME) on the migration in SK-Hep 1 cells
(A) Motility of SK-Hep1 cells, as assessed by wound closure assays. (B) Wound sizes were observed with a microscope (Olympus CK40) and measured at the indicated time points. Amount of migration was indicated relative to that in untreated controls (%).
Data are presented as the mean±SD (n=3). Bars with different superscript letters represent significant differences at p < 0.05 based
on the Duncan’s multiple range test. (▬●▬) 0 μg/ml; (- ● -) 100 μg/ml; (--●--) 200 μg/ml; (----●----) 400 μg/ml
Fig. 4: Effects of Chelidonium majus extract (CME) on invasion of SK-Hep 1 cells
(A) Cells that invaded transwell chambers were stained with 0.2% crystal violet and observed under ×40 magnification (Olympus
CK40). (B) Relative invasive activity, as determined by extracting the stain and quantifying absorbance at 595 nm. Data are
presented as the mean±SD (n=3). Untreated SK-Hep1 cells were considered 100 % viable. Bars with different superscript letters
represent significant differences at p<0.05 based on the Duncan’s multiple range test
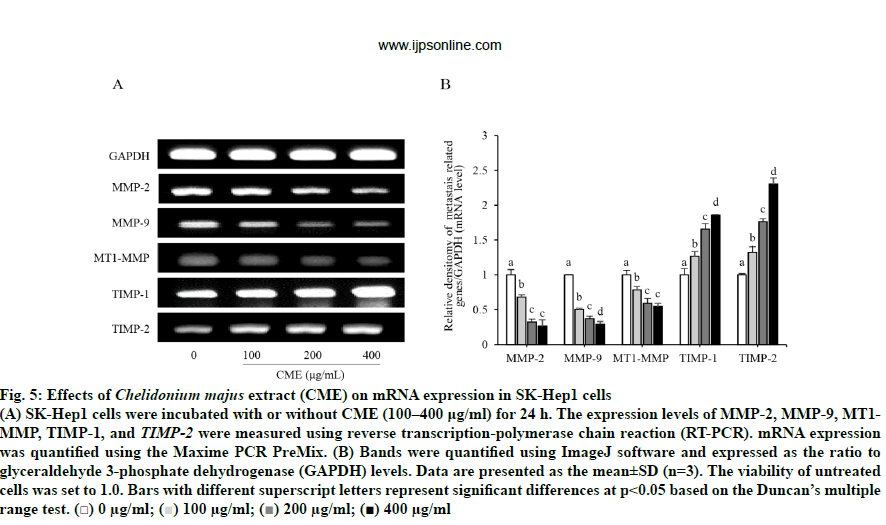
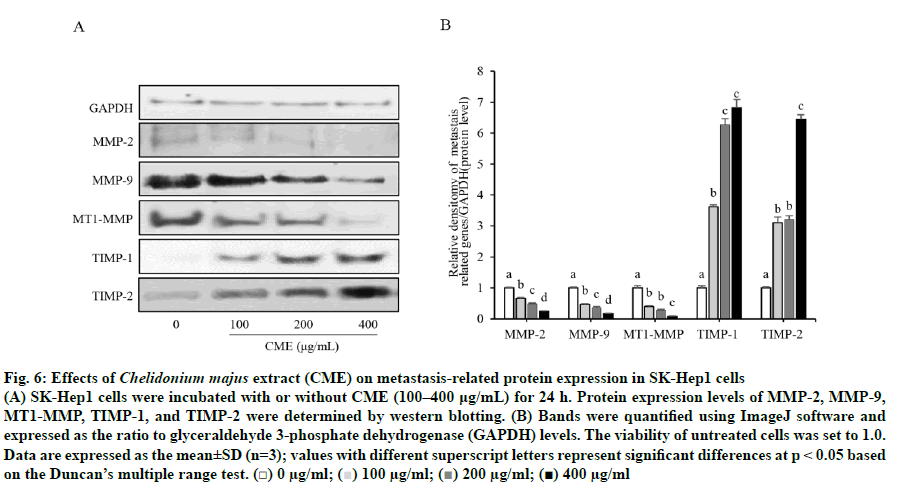
As shown in fig. 5, CME treatment suppressed the expression of MMP-2 and MMP-9 in a dose-dependent manner. MMP-2 mRNA expression was significantly decreased to 32, 67, and 73 % of levels in untreated cells, in response to 100, 200, and 400 μg/ml CME, respectively. Similarly, MMP-9 mRNA expression was significantly and gradually reduced with increasing concentrations of CME (100-400 μg/ml). Moreover, the mRNA expression of MT1-MMP was significantly reduced in a dose-dependent manner to 21.73±5.05, 40.83±6.79, and 45.57±4.78 % of control levels after treatment with 100, 200, and 400 μg/ml CME, respectively. In contrast, the mRNA expression of TIMPs (TIMP-1 and TIMP-2) was significantly increased by CME treatment, compared to that in untreated cells, in a dose-dependent manner. Furthermore, protein levels of MMPs, MTI-MMP, and TIMPs followed the same trend in treated HCC cells (fig. 6). Specifically, CME treatment significantly increased the expression of TIMP-1 and TIMP-2 compared to levels in untreated cells. However, TIMP-1 expression in cells treated with 200 μg/ml CME was not significantly changed compared to that in cells treated with 400 μg/ml CME. Additionally, TIMP-2 expression in cells treated with 100 μg/ml CME was not significantly different from that in cells treated with 200 μg/ml CME.
Fig. 5: Effects of Chelidonium majus extract (CME) on mRNA expression in SK-Hep1 cells
(A) SK-Hep1 cells were incubated with or without CME (100–400 μg/ml) for 24 h. The expression levels of MMP-2, MMP-9, MT1-MMP, TIMP-1, and TIMP-2 were measured using reverse transcription-polymerase chain reaction (RT-PCR). mRNA expression
was quantified using the Maxime PCR PreMix. (B) Bands were quantified using ImageJ software and expressed as the ratio to
glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels. Data are presented as the mean±SD (n=3). The viability of untreated
cells was set to 1.0. Bars with different superscript letters represent significant differences at p<0.05 based on the Duncan’s multiple
range test. (□) 0 μg/ml;  100 μg/ml;
100 μg/ml;  200 μg/ml; (■) 400 μg/ml
200 μg/ml; (■) 400 μg/ml
Fig. 6: Effects of Chelidonium majus extract (CME) on metastasis-related protein expression in SK-Hep1 cells
(A) SK-Hep1 cells were incubated with or without CME (100–400 μg/mL) for 24 h. Protein expression levels of MMP-2, MMP-9, MT1-MMP, TIMP-1, and TIMP-2 were determined by western blotting. (B) Bands were quantified using ImageJ software and
expressed as the ratio to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) levels. The viability of untreated cells was set to 1.0.
Data are expressed as the mean±SD (n=3); values with different superscript letters represent significant differences at p < 0.05 based on the Duncan’s multiple range test. (□) 0 μg/ml;  100 μg/ml;
100 μg/ml;  200 μg/ml; (■) 400 μg/ml
200 μg/ml; (■) 400 μg/ml
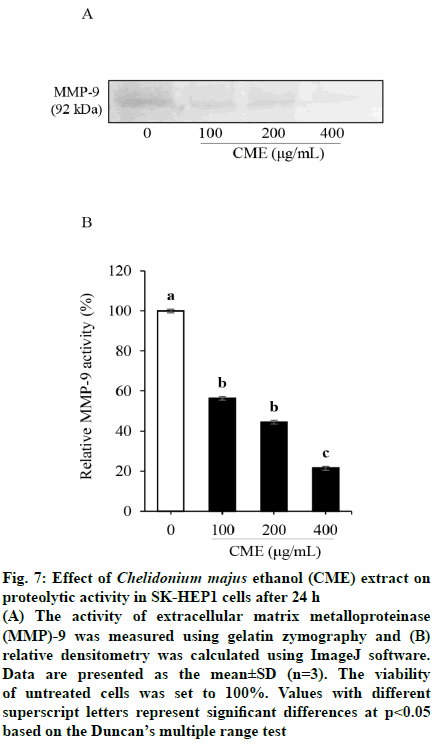
MMP activation was measured by gelatin zymography to elucidate the inhibitory effect of CME on the production of gelatinases in HCC cells (fig. 7). The patterns of MMP-9 secretion, based on zymography assays, was similar to those of mRNA and protein expression. MMP-9 activity was significantly suppressed after CME treatment when compared to that in untreated cells. As shown in fig. 7, MMP-9 activity was decreased to 40.22±5.44, 48.56±3.36, and 81.38±4.09 % of control levels in response to 100, 200, and 400 μg/ml of CME, respectively. However, MMP-9 activity was not significantly different between 100 and 200 μg/ml CME treatment groups.
Fig. 7: Effect of Chelidonium majus ethanol (CME) extract on
proteolytic activity in SK-HEP1 cells after 24 h
(A) The activity of extracellular matrix metalloproteinase (MMP)-9 was measured using gelatin zymography and (B)
relative densitometry was calculated using ImageJ software.
Data are presented as the mean±SD (n=3). The viability
of untreated cells was set to 100%. Values with different
superscript letters represent significant differences at p<0.05
based on the Duncan’s multiple range test
HCC, the most common hepatic malignancy, is an aggressive tumor associated with frequent intrahepatic and extrahepatic invasion[20]. Consequently, its prognosis remains poor in the late stage[21]. Extrahepatic metastasis is a multi-step process consisting of migration from the primary liver site, invasion of the blood vessels or lymph nodes, the formation of aggregates to facilitate travel, and adherence to distant secondary sites such as the lung, bones, and adrenal gland[22-25]. Cancer cells exhibit altered expression of extracellular matrix (ECM)-degradative proteins, which mainly include MMPs[25] that are highly expressed in HCC cells and are associated with the growth, expansion, and invasion of malignant tumors[26]. MMPs comprise a family of proteolytic enzymes that create space for cell migration by rearranging the structure of the ECM and manipulating intracellular signaling and junctions[27]. Among family members, MMP-2 and MMP-9 are gelatinases that play a key role in angiogenesis by upregulating vascular endothelial growth factors and breaking down basement membrane components to reduce cell-to-cell interactions and facilitate cell motility[28]. In this study, an examination of MMP-2 and MMP-9 expression revealed that CME down-regulated their transcription in a concentration-dependent manner. Huang et al. reported that β-mangostin, a dietary xanthone, exerts antimetastatic activity through the inhibition of MMP-2 and MMP-9 mRNA and protein expression in human HCC cells[29]. These results are consistent with our findings showing that CME could reduce colony formation, adhesion, invasion, and migration via the suppression of MMP-2 and MMP-9 expression.
Moreover, CME treatment suppressed MT1-MMP transcription, whereas it upregulated the expression of TIMP-1 and TIMP-2. MMP-2 and MMP-9 are generally secreted as inactive proenzymes and are either activated by other MMPs or suppressed by their inhibitors[30]. MT1-MMP, a well-known activator of MMP-2, forms a complex with the latent form of MMP- 2 (proMMP-2) and TIMP-2 to activate MMP-2[31]. This trimolecular complex cleaves proMMP-2 to generate the MMP-2/MT1-MMP complex, which subsequently stimulates proMMP-9, as reported previously[32]. Thus, MT1-MMP directly activates MMP-2 but indirectly stimulates MMP-9, which consequently promotes tumour metastasis[33]. In contrast, TIMP-1 is a negative regulator of MMP-9, and the balance between MMP-9 and TIMP-1 interactions has been reported to modulate inhibitory effects on liver cancer cell invasion[34]. This study demonstrated that CME suppresses HCC cell metastatic potential by up-regulating TIMP-1 and TIMP-2 and down-regulating MMP-2 and MMP-9.
MMP-2 and MMP-9 (gelatinase-A and gelatinase-B, respectively) degrade type IV collagen and gelatin, major structural components of the ECM and basement membrane. Particularly, the latent form of MMP-9 (92 kDa) is highly expressed in the human SKHep1 HCC cell line compared to MMP-2 levels[32]. Furthermore, to verify the relationship between MMP expression and the metastasis-inhibitory potential of CME, the proteolytic activity of secreted MMPs was measured by gelatin zymography. CME treatment inhibited the gelatin-degrading activity of MMP-9 in a concentration-dependent manner by 40-81 %.
Despite the steady development of chemotherapeutics with high efficacy and safety, the prognosis of HCC remains poor due to the high rates of recurrence and metastasis. Currently, there is no effective chemotherapeutic regimen that can prevent metastasis. Therefore, the evaluation of traditional medicinal plants and their application to drug discovery has been the center of attention over the past few years[35]. This study was performed to evaluate the antimetastatic effect of C. majus used as a Mongolian traditional medicine. These results suggest that CME suppresses all critical processes related to metastasis such as cell migration, invasion, aggregation, and adhesion by down-regulating the predominant metastasis-related gelatinases and their activators while upregulating their natural inhibitors. Thus, CME, as an MMP inhibitor, might represent a promising antimetastatic agent. Although it was confirmed that the antimetastatic effect of C. majus in vitro, animal studies were not performed in this study. Hence, further studies are needed to address antimetastatic properties of C. majus in a relevant animal model.
Acknowledgment
This study was supported by Konkuk University.
References
- Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability adjusted life-years for 32 cancer groups, 1990 to 2015: A systematic analysis for the Global Burden of Disease Study. JAMA Oncol 2017;3(3):524-48.
- Laursen L. A preventable cancer. Nature 2014;516(7529):S2-S3.
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015;65(2):87-108.
- Guan X. Cancer metastases: challenges and opportunities. Acta Pharmaceutica Sinica B 2015;5(5):402-18.
- Choi KJ, Baik IH, Ye SK, Lee YH. Molecular targeted therapy for hepatocellular carcinoma: present status and future directions. Biol Pharm Bull 2015;38(7):986-91.
- Chan SL, Yeo W. Targeted therapy of hepatocellular carcinoma: Present and future. J Gastroenterol Hepatol 2012;27(5):862-72.
- Stotz M, Gerger A, Haybaeck J, Kiesslich T, Bullock MD, Pichler M. Molecular targeted therapies in hepatocellular carcinoma: past, present and future. Anticancer Res 2015;35(11):5737-44.
- Katiyar C, Gupta A, Kanjilal S, Katiyar S. Drug discovery from plant sources: An integrated approach. Ayu 2012;33(1):10-9.
- Zielińska S, Jezierska-Domaradzka A, Wójciak-Kosior M, Sowa I, Junka A, Matkowski AM. Greater Celandine's Ups and Downs−21 Centuries of Medicinal Uses of Chelidonium majus From the Viewpoint of Today's Pharmacology. Front Pharmacol 2018;9:299.
- Jyoti BS. Chelidonium majus L. - A review of pharmacological activities and clinical effects. Global J Res Med Plants Indigen Med 2013;2(4):238-45.
- Ligaa U. Medicinal plants of Mongolia used in Western and Eastern medicine. JKC printing: Ulaanbaatar; 2006. p. 536-7.
- Dutt-Roy R, Kayalvizhi E, Manikandan B, Chandrasekhar M. Hepatoprotective effect of Chelidonium majus. L extract against antitubercular drug-induced hepatic damage in Wistar rats. Int J Pharma Bio Sci 2015;6(2):677-81.
- Zuo GY, Meng FY, Hao XY, Zhang YL, Wang GC, Xu GL. Antibacterial alkaloids from Chelidonium majus Linn (Papaveraceae) against clinical isolates of methicillin-resistant Staphylococcus aureus. J Pharm Pharm Sci 2008;11(4):90-4.
- Mikołajczak PL, Kędzia B, Ożarowski M, Kujawski R, Bogacz A, Bartkowiak-Wieczorek J, et al. Evaluation of antiinflammatory and analgesic activities of extracts from herb of Chelidonium majus L. Cent Eur J Immunol 2015;40(4):400-10.
- Jang S, Yoo D. Effects of Chelidonium majus extract on apoptosis induction of MDA-MB-231 human breast cancer cells. J Korean Obstet Gynecol 2015;28(2):15-25.
- Kulp M, Bragina O. Capillary electrophoretic study of the synergistic biological effects of alkaloids from Chelidonium majus L. in normal and cancer cells. Anal Bioanal Chem 2013;405(10):3391-7.
- Park SW, Kim SR, Kim Y, Lee JH, Woo HJ, Yoon YK, et al. Chelidonium majus L. extract induces apoptosis through caspase activity via MAPK-independent NF-κB signaling in human epidermoid carcinoma A431 cells. Oncol Rep 2014;33(1):419-24.
- Stock AM, Troost G, Niggemann B, Zänker KS, Entschladen F. Targets for anti-metastatic drug development. Curr Pharm Des 2013;19(28):5127-34.
- Gandalovičová A, Rosel D, Fernandes M, Veselý P, Heneberg P, Čermák V, et al. Migrastatics - anti-metastatic and anti-invasion drugs: promises and challenges. Trends Cancer 2017;3(6):391-406.
- Balogh J, Victor D III, Asham EH, Burroughs SG, Boktour M, Saharia A, et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma 2016;3:41-53.
- Uchino K, Tateishi R, Shiina S, Kanda M, Masuzaki R, Kondo Y, et al. Hepatocellular carcinoma with extrahepatic metastasis. Cancer 2011;117(19):4475-83.
- van Zijl F, Krupitza G, Mikulits W. Initial steps of metastasis: Cell invasion and endothelial transmigration. Mutat Res 2011;728(1-2):23-34.
- Li S, Li Q. Cancer stem cells and tumor metastasis (Review). Int J Oncol 2014;44(6):1806-12.
- Becker A, Tso D, Harris A, Malfair D, Chang S. Extrahepatic metastases of hepatocellular carcinoma: a spectrum of imaging findings. Can Assoc Radiol J 2014;65(1):60-6.
- Jiang W, Sanders A, Katoh M, Ungefroren H, Gieseler F, Prince M, et al. Tissue invasion and metastasis: molecular, biological and clinical perspectives. Semin Cancer Biol 2015;35 Suppl:S244-S275.
- Roomi MW, Kalinovsky T, Rath M, Niedzwiecki A. Modulation of u-PA, MMPs and their inhibitors by a novel nutrient mixture in human female cancer cell lines. Oncol Rep 2012;28(3):768-76.
- Page-McCaw A, Ewald A, Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat Rev Mol Cell Biol 2007;8(3):221-33.
- Gialeli C, Theocharis A, Karamanos N. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J 2010;278(1):16-27.
- Huang CF, Teng YH, Lu FJ, Hsu WH, Lin CL, Hung CC, et al. β-mangostin suppresses human hepatocellular carcinoma cell invasion through inhibition of MMP-2 and MMP-9 expression and activating the ERK and JNK pathways. Environ Toxicol 2017;32(11):2360-70.
- Cathcart J, Pulkoski-Gross A, Cao J. Targeting matrix metalloproteinases in cancer: Bringing new life to old ideas. Genes Dis 2015;2(1):26-34.
- Sternlicht MD, Werb Z. How matrix metalloproteinases regulate cell behavior. Annu Rev Cell Dev Biol 2001;17:463-516.
- Roomi MW, Monterrey JC, Kalinovsky T, Rath M, Niedzwiecki A. Patterns of MMP-2 and MMP-9 expression in human cancer cell lines. Oncol Rep 2009;21(5):1323-33.
- Li Z, Takino T, Endo Y, Sato H. Activation of MMP-9 by membrane type-1 MMP/MMP-2 axis stimulates tumor metastasis. Cancer Sci 2017;108(3):347-53.
- Avădanei R, Căruntu ID, Amălinei C, Lozneanu L, Balan R, Grigoraş A, et al.: High variability in MMP2/TIMP2 and MMP9/TIMP1 expression in secondary liver tumors. Rom J Morphol Embryol 2013;54(3):479-85.
- Harvey A, Edrada-Ebel R, Quinn R. The re-emergence of natural products for drug discovery in the genomics era. Nat Rev Drug Discov 2015;14(2):111-29