- *Corresponding Author:
- Yinhui Lu
Health Management Center,
Jiangxi Provincial People’s Hospital,
The First Affiliated Hospital of Nanchang Medical College,
Nanchang,
Jiangxi 330006
E-mail: 1307960615@qq.com
| This article was originally published in a special issue, “Modern Applications in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(3) Spl Issue “105-124” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Non-coding ribonucleic acids were thought to be non-functional transcription products decades ago. But recently they have been found to play an essential role in programmed regulation in a variety of diseases, including cancer. They include micro ribonucleic acids and their upstream long non-coding ribonucleic acids and circular ribonucleic acids. Long non-coding ribonucleic acids or circular ribonucleic acids can form a regulatory network through sponging micro ribonucleic acids. The network regulates biological functions of various tumors such as proliferation, invasion and drug resistance. In addition, other non-coding ribonucleic acids such as piwi-interacting ribonucleic acids, transfer ribonucleic acids, ribosomal ribonucleic acids, small nuclear ribonucleic acids and small nucleolar ribonucleic acids can also act as regulators of the biological characteristics of tumor cells. Ferroptosis is a programmed cell death, which is distinct from the classical caspase splicing and has received increasing attention in recent years with regard to the tumorigenesis and treatment of cancer. Interestingly, non-coding ribonucleic acids have been found to regulate ferroptosis in tumors and to maintain the biological function of tumor cells. Herein, we summarize the related research and emphasize that the regulation of ferroptosis in tumor cells by non-coding ribonucleic acids may provide a basis for future targeted cancer therapy.
Keywords
Micro ribonucleic acids, long non-coding ribonucleic acid, circular ribonucleic acid, ferroptosis, cancer
In 1965, Jacob and Monod proposed a central principle of gene coding: Nuclear Deoxyribonucleic Acid (DNA) is transcribed into messenger RNA (mRNA), which is translated into proteins in the cytoplasm to enable gene expression in the body and regulate biological functions[1].
Non-Coding Ribonucleic Acid (ncRNA)
With the improvement of gene sequencing technology, many unique RNA products encoded by the genome have been identified that they cannot be translated into proteins[2]. These were called non-coding RNAs (ncRNAs) where ncRNAs have previously been considered nonfunctional “junk”[3]. But this view was reversed by the Encyclopedia of DNA Elements (ENCODE) project, which revealed that the genetic code that regulated biological function was not quite simple[4]. ncRNAs include micro RNA (miRNA or miR), long non-coding RNA (lncRNA), circular RNA (circRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), small nucleolar RNA (snoRNA) and piwi-interacting RNA (piRNA)[5]. The most widely studied ncRNA is miRNA. MiRNAs are endogenous single-stranded RNAs approximately 22 nucleotides (nt) in length encoded by eukaryotic nuclear DNA[6]. MiRNAs are estimated to regulate 60 % or more genes and are involved in regulating many biological processes, including cell proliferation, apoptosis and differentiation[7]. The production of miRNAs is a strictly transcriptional process. The nuclear-coding gene, which is mostly located in intron regions are transcribed into primary transcripts (pri-miRNA) by binding to RNA Polymerase II (Pol II)[8]. Drosha and Dicer, as two types of Ribonuclease III (RNase III), play an important role in the pri-miRNA maturation. Drosha and double-stranded RNA (dsRNA)-Binding Proteins (dsRBPs), for example DiGeorge Syndrome Critical Region 8 (DGCR8), bind and splice pri-miRNA transcripts to produce a ~70 nt precursor hairpin (pre-miRNA). Exportin-5 is a Ras-related nuclear protein-Guanosine Triphosphate (Ran-GTP) complex. Pre-miRNA is transforted from the nucleus to the cytoplasm by it. With the help of Trans-Activation Response (TAR) RNA-Binding Protein (TRBP), Dicer splices the premiRNA into a miRNA duplex in the cytoplasm[9,10]. The Argonaute (AGO) protein family is the core component of miRNA-Induced Silencing Complex (miRISC), which regulates the miRNA double-strand to a singlestrand[ 11]. Interestingly, miRNAs recognize the miRNA Recognition Elements (mREs) of protein-coding mRNAs in order to degrade mRNAs[12]. MREs are typically located in the 3’-Untranslated Regions (UTR) of the Coding Sequences (CDS), but may also be found in the 5’-UTR and within CDS. Finally, mature single strand miRNAs can bind to mREs via miRISC to inhibit mRNAs translation and lead to mRNAs degradation[13] (fig. 1a). This form of genetic regulation occurs not only in normal cell populations, but also in diseases such as neurodegeneration, metabolic diseases and especially in cancer[14]. LncRNAs are RNA transcripts of more than 200 nucleotides in length which have secondary and tertiary structures. These allow them to perform functions similar to proteins[15]. They may be located in the nucleus or cytoplasm, they may be polyglandular and are usually transcribed from any strand within a protein-coding gene site[4]. The discovery of Homeobox (Hox) Antisense Intergenic RNA (HOTAIR) is a classic accomplishment of lncRNAs research[16]. The interaction between HOTAIR and Polycomb Repressive Complex 2 (PRC2) negatively regulates transcription across the Homeobox D (HoxD) site in the cis-gene[17]. Since its discovery, a growing number of researchers have become increasingly interested in exploring the function and regulation of lncRNAs. However, it may be incorrect to define lncRNAs as simply unable to encode proteins. Despite the fact that ncRNAs cannot theoretically be translated into proteins, some long strands of ncRNAs have been found to contain an Open Reading Frame (ORF) that can be translated into small peptides[18]. A lncRNA generally falls into the following five categories: Sense, overlapping at least one exon in a transcript on the same strand; antisense, overlapping at least one exon in another transcript in a complementary strand; bidirectional, reversely transcribing on a complementary chain with adjacent transcripts and sharing the same promoter; intronic, locating in the intron region of a protein-coding gene and not overlapping with exons of the transcript and intergenic, independently transcribing between two protein-coding genes[19]. The function of lncRNAs has been explored since the discovery of ncRNAs as a biological regulator, but only a small portion of these has been discovered (fig. 1b). LncRNAs can interact with one or more proteins to perform biological functions[20]. They can bind to their own transcription sites to regulate the expression of cis genes or bind to a transcription factor to prevent DNA transcription[21]. It is worth noting that lncRNAs contain predicted miRNA binding sites, which allows lncRNAs to act as negative regulators of miRNAs, reducing the number of miRNAs in the cell and thus regulating the expression of protein transcripts. This type of RNA is known as competitive endogenous RNA (ceRNA)[22] and the interaction is currently known as lncRNA-miRNAmRNA. A ceRNA network (CeRNAnet) is gradually formed based on the lncRNA/miRNA/mRNA axis[23]. LncRNAs play an important role in many diseases including cardiovascular disease, bone disease and cancer[24-26].
CircRNA was first discovered in RNA viruses in 1976 and for a long time they were believed to be the result of RNA splicing errors[27]. Subsequently, with the development of high-throughput RNA sequencing (RNA-seq), hundreds of circRNAs were identified in a variety of eukaryotes. The mechanism of circRNAs production and gene regulation has gradually been revealed[28]. Pre-mRNA is spliced by RNA Pol II to form a covalently closed continuous loop (circRNA)[29]. Compared to linear RNAs, circRNA is usually obtained by nonstandard splicing (back-splicing) and has no 3’ or 5’ polarity[30]. There are three types of circRNA: Exon circRNA (ecircRNA) composed of exons; circular intronic RNA (ciRNA) composed of introns and exonintron circRNA (elciRNA) composed of exons and introns[31]. In recent years, the search for the function of circRNA has never stopped. CircRNA is found to bind to RNA Pol II and regulates transcription efficiency at the promoter region of the parent gene. In addition to the transcriptional regulation of parental genes, it has been revealed that there is a competitive relationship between parental genes splicing and circRNAs splicing[32] (fig. 1c). Interestingly, several binding sites of miRNAs have been identified on circRNAs. Recently, circRNA has also been reported to modulate ferroptosis in tumor cells.
PiRNAs are a class of small ncRNAs (sncRNAs) recently discovered in germ cells. PiRNAs consist of 24-31 nt, with a 5′-terminal uridine or a 10th position adenosine bias, lacking a clear secondary structural motif[33]. Unlike miRNAs, piRNAs are not spliced by the RNase III enzyme Dicer and are integrated into the Piwi subfamily of AGO proteins[34]. PiRNAs bind to Piwi proteins to form the piRNA-Piwi complex, which affects transposon silencing, spermatogenesis, genome rearrangement, germ cell maintenance, epigenetic and protein regulation[35]. Recently, increasing evidence has shown that piRNAs are involved in the regulation of cell proliferation and apoptosis especially in cancer cells[36]. Changes in the expression of Piwi proteins and piRNAs can increase cell proliferation, decrease apoptosis and promote the growth of lung cancer and glioma cells[37,38]. However, the role of piRNA in ferroptosis of tumor cells has not been well studied. In recent years, a new class of sncRNAs from tRNAs has been discovered. These ncRNAs derived from tRNAs are called tRNA Fragments (tRF)[39]. The tRFs can be divided into four types: 3’-trailer sequences that are removed by ElaC Ribonuclease Z 2 (ELAC2) from pretRNAs during tRNAs maturation, resulting in 1-tRF; 5’-tRF generated at the 5’ end in the D-loop; 3’-tRF generated by the 3’-end in the T-loop; tRNA-derived stress-induced RNAs (tiRNAs) or tRNAs produced by Angiogenin (ANG) via the specific cleavage of the antibacterial loops of mature tRNAs under stress conditions[40,41]. Several tRNAs play an important role in the epigenetic regulation of cancer. The expression of tumor suppressor (ts) 101 and ts-46 (tRF-1s) had been associated with the survival, proliferation and apoptosis of tumor cells[42]. By binding to the mRNA of Ribosomal Protein (RP) (RPS28 and RPS15), the specific tRF-3 (LeuCAG 3’ tsRNA) was inhibited to induce apoptosis in vitro and in xenograft mouse models and promoted protein synthesis[43]. Immunoprecipitation of the Piwi gene (Hiwi2) in Human Breast Cancer Cell Line (MDAMB- 231) cells could enrich piRNAs generated mainly from processed tRNAs, which suggested that the PiwipiRNA pathway and tRF-5 exerted an uncharacterized function[44]. The snoRNA is a conserved nuclear RNA family of 70-200 nt in length. SnoRNAs are generally concentrated in Cajal bodies or nucleoli where they are either involved in the modification of snRNAs or rRNAs and the maturation of ribosomal subunits[45,46]. They are usually processed from debranched and excised introns by exonucleolytic trimming and form small nucleolar Ribonucleoproteins (snoRNPs) to carry out their functions in complexes with specific protein components[45,47]. Some intronic lncRNAs contain snoRNAs at the end of the region. These intron-derived snoRNA-ended RNAs are named sno-lncRNAs. The tissue and species-specific expression patterns have been identified in humans, rhesus monkeys and mice[48]. Its regulatory effect on lncRNAs may be the basis for the regulation of cancer cell apoptosis and even ferroptosis.
Ferroptosis
The origin of ferroptosis:
Cell death is divided into Programmed Cell Death (PCD), which is energy-dependent death with different morphological changes including apoptosis and necrosis, which is regarded as passive toxic and energyindependent process[49]. Although apoptosis, autophagy and necroptosis have previously been identified as three major types of PCD[50], a new type of PCD called ferroptosis has recently been defined.
Ferroptosis, as its name suggests, is a specific form of cell death in which iron-dependent lipid Reactive Oxygen Species (ROS) accumulate in cells. This process is not involved in apoptotic effector proteins such as cysteine-aspartic proteases (caspases) and Bcl- 2-Associated X protein (BAX)[51]. ROS are composed mainly of peroxides (Hydrogen Peroxide (H2O2) and Hydroperoxides (ROOH)), superoxide (O2•–) and free radicals (Hydroxyl (HO•) and Alkoxy RO•)). Under normal circumstances, the body can transfer electrons through mitochondrial electron transport chain to produce ROS. It can also be activated during the production of uric acid in hypoxanthine and during the oxidation of fatty acids[52]. The oxidation-reduction imbalance in the body will lead to oxidative stress to produce a large number of free radicals that destroy proteins, DNA and intracellular molecules in cells. Ferrous (Fe) II in the chelatable iron transition pool can catalyze the decomposition of H2O2 to produce ROS such as HO• radicals[53]. The depletion of Polyunsaturated Fatty Acids (PUFAs) and the formation of Lipid Hydroperoxide (L-OOH) provide a new inducer of irondependent mortality. In the presence of Fe (II), L-OOH can form lipid-free radicals that damage the cell surface and intracellular components, such as the alkoxy radical L-O•[54]. Glutamate has been used since the 1990s to treat nerve cells in mice (Immortalized mouse hippocampal cell line (HT-22)). Competing for cystine uptake, this reduces the accumulation of Glutathione (GSH) in cells and ultimately leads to oxidative cell death[55]. Cell death caused by oxidative stress is becoming increasingly more well-known. In 2007, Erastin was found to alter mitochondrial membrane permeability via mitochondrial Voltage-Dependent Anion Channels (VDAC), but did not promote mitochondrial release of cytochrome C or the activation of classical apoptoticcharacteristic caspase-3. It had been suggested that Erastin could promote the death of cancer cells with RAS gene mutation in a non-apoptotic manner[56]. In 2008, two compounds Ras-Selective Lethal small molecule 5 (RSL5) and RSL3 were identified as having similar functions to Erastin, both of which were involved in the activation of RAS signaling pathways. RSL5, like Erastin, also plays an oxidative role through Voltage-Dependent Anion-Selective Channel protein 3 (VDAC3), participating in iron and ROS dependent cell death. They are called Type I RSLs. Type II RSLs such as RSL3 can inhibit regulatory factors leading to cell death[57]. In addition, a new activating factor of cellular oxidative stress was also discovered in 2008. Glutathione Peroxidase 4 (GPX4) gene knock-out in cells can lead to lipid peroxidation and oxidative damage leading to cell death[58]. Finally, in 2012, Dixon et al. formally defined Erastin-induced irondependent cell death as ferroptosis and found that the small molecule Ferrostatin-1 (Fer-1) not only inhibited ferroptosis in cancer cells, but also induced the same effects on glutamate-induced brain cell death in rats[51]. Through high throughput screening of small molecular libraries, it was found that Fer-1 and Liproxstatin-1 (Lip-1) could inhibit the accumulation of L-OOH, likely by preventing the deletion of GPX4 and the inhibition of the xc– system[51,59]. Both compounds are acrylamides which are known to be a good Radical- Trapping Antioxidants (RTA). Fer-1 and Lip-1 are very effective RTAs or inhibition of autoxidation may not be the root cause of its inhibition of ferroptosis. In fact, Fer-1 and Lip-1 may be effective inhibitors of lipoxygenase[60]. In addition, a new concept has been proposed that ferrous itself reduces Fer-1 free radicals and Fer-1 forms complexes with iron to reduce the presence of labile iron[61] (fig. 1d).
Cysteine-glutamate antiporter (system xc–):
The antiporter system xc– is a member of the family of heterodimeric amino acid transporters composed of light chain (xCT) and 4F2 cell-surface antigen heavy chain (4F2hc). The system can import Cysteine (Cys) into cells and transport glutamate out of cells at a 1:1 basis[62]. The thiol-containing amino acid Cys is oxidized in the form of cystine (Cys2), which is essential for resistance to cellular oxidative stress[62,63]. Solute Carrier Family 7 Member 11 (SLC7A11) (xCT) is a 12- fold transmembrane transporter that can be linked to Solute Carrier Family 3 Member 2 (SLC3A2) (4F2hc) via a disulfide bridge. Erastin, Sulfasalazine (SAS) and sorafenib can reduce the uptake of cystine, which can lead to lipid oxidation. SAS is an anti-inflammatory drug that eliminates ROS and is often used clinically to treat rheumatoid arthritis inflammatory bowel disease[64]. SAS was also found to inhibit leukocyte movement and the production of Interleukin (IL)-1 and IL-2 and to inhibit Nuclear Factor kappa B (NF-κB)[65,66]. Interestingly, the xc– transporter plays an important role in pancreatic cancer by maintaining or enhancing GSH biosynthesis. SAS also effectively induces GSH depletion and enhances chemotherapy resistance of pancreatic cancer[67]. Sorafenib is the only known first line treatment for patients with advanced liver cancer[68]. Sorafenib, a multi kinase inhibitor, plays an anti-cancer role by inducing apoptosis, inhibiting cell proliferation and angiogenesis[69].
The iron-chelating agent Deferoxamine (DFO) depletes the cells iron stores to protect liver cancer cells from sorafenib’s cellular oxidative stress and cytotoxicity[70]. In terms of mechanism, the study showed that sorafenib could also inhibit ferroptosis induced by SLC7A11[71]. In addition, the occurrence of ferroptosis leads to Endoplasmic Reticulum (ER) stress, which increases ChaC, Cation Transport Regulator Homolog 1 (CHAC1) significantly. It can be regarded as a marker of ferroptosis[71]. The main reason for this is the downregulation of GSH in cells. GSH, a combination of Cys and glutamate, is an important antioxidant molecule in cells. Finally, on this basis, glycine is added to synthesize. The Sulfhydryl group (-SH) of Cys is the most important antioxidant group in GSH[72]. Buthionine Sulfoxamine (BSO) was found to inhibit glutamate-Cys ligase, a rate-limiting enzyme in GSH synthesis, leading to a reduction in GSH levels[73]. In addition, Glutathione Disulfide (GSSG) is involved in the reduction of GSH. GSSG is reduced to GSH by Nicotinamide Adenine Dinucleotide Phosphate (NADPH) and to GSSG reductase in cells to maintain the steady state level of GSH[74]. There are eight species of GPXs in mammals. GPX4 (Phospholipid Hydroperoxide Glutathione Peroxidase (PHGPx)) is a phospholipid catalase that acts to reduce the lipid peroxidation. Therefore, GPX4 plays an important role in inhibiting lipid oxidation in cells[75]. Because GPX4 is a selenoprotein, Selenium (Se) can promote the expression of GPX4 and inhibit ferroptosis induced by endoplasmic reticulum stress[76]. RSL3 does not reduce GSH expression through xc–, but it can inhibit GPX4 enzyme activity by producing lipid ROS and promote ferroptosis[73]. In addition, ML162, ML210 and anti-cancer agent altretamine can also inhibit the activity of GPX4[77,78].
Lipid peroxidation and Transferrin (TF):
Acetyl-Coenzyme A (Acyl-CoA) Synthetase Longchain family member 4 (ACSL4) has been found to be an important promoter of cellular lipid oxidation. The knock-out of both ACSL4 and GPX4 genes result in a marked inhibition of ferroptosis. Doll et al. further demonstrated that anti-diabetic medication thiazolidinediones could inhibit ferroptosis in mouse tissues by targeting ACSL4[79]. In addition, Phosphatidylethanolamines (PEs) containing Arachidonoyl Acid (AA) or Adrenoyl Acid (AdA) is the preferred substrates for oxidation involved in ferroptosis[80]. In fact, AA/ADA can enter the cell via the Fatty Acid Translocator (FAT), Fatty Acid Transporter Protein (FATP) or via free diffusion. ACSL4 catalyzes AA and AdA bonds to produce AA or AdA acyl Co-A derivatives. They are esterified into PEs (AA-PE and AdA-PE) by Lysophosphatidylcholine Acyltransferase 3 (LPCTA3). Finally, AA-PE and AdA-PE are then oxidized by 15-Lipoxygenase (15-LOX, Arachidonate 15-Lipoxygenase (ALOX15)) to produce OO(O)- AA/AdA-PE. Because some inhibitors such as RSL3 decrease the concentration or activity of GPX4, which cannot detoxify OO (O)-AA/AdA-PE, they can generate Phospholipid Hydroperoxides (PL-OOH) to execute the ferroptosis pathway[81,82]. As mentioned earlier, the formation of lipid free radicals in the presence of iron can promote cell death.
TF plays an important role in human iron metabolism. TF is mainly produced in the liver and binds up to two Fe atoms. TF loaded with Fe atoms binds to the Transferrin Receptor 1 (TFR1) and delivers the atoms to the cell by endocytosis[83]. The major binding site of TF outside the cell is Fe3+, which is transported by TRF1 and is oxidized to Fe2+ by the Six-Transmembrane Epithelial Antigen of the Prostate 3 (STEAP3). The Divalent Metal Transporter 1 (DMT1) then transports the Fe2+ into the cytoplasm[84]. When iron is overloaded, ROS can be produced by the Fenton reaction, to promote the pathological changes observed in hereditary hemochromatosis and other diseases[85]. Ferritin is a 24-subunit protein complex capable of chelating more than 4500 Fe (III) iron atoms. It consists of light and heavy chains (Ferritin Light Chain (FTL), Ferritin Heavy Chain 1 (FTH1))[86]. Ferritin complex cages can oxidize Fe (II) to Fe (III) via FTH1 to store iron, which also results in less ROS generation[87]. When our body needs iron, ferritin can release iron in a way that relies on lysosomal degradation[88]. Furthermore, the Nuclear Coactivator 4 (NCOA4) receptor co-exists with ferritin in autophagosomes and lysosomes. Reducing the expression of NCOA4 blocks the transfer of ferritin to lysosomes, which in turn inhibits the degradation of ferritin. Two subunits of ferritin (FTL, FTH1) of the E3 ubiquitin ligase complex (HECT and RLD Domain Containing E3 Ubiquitin Protein Ligase 2 (HERC2) and Neuralized E3 Ubiquitin Protein Ligase 4 (NEURL4)) can interact to keep iron levels stable[89]. Therefore, changes in NCOA4 expression affect the sensitivity of ferroptosis in cells[90]. In addition, two subunits of ferritin can also play an indirect role. Iron chelation treatment with DFO affects the availability of iron in living organisms to inhibit the sensitivity of ferroptosis[51].
Tumor Protein P53 and Nuclear Factor Erythroid 2-Related Factor 2 (NRF2):
As an important tumor suppressor gene, P53 can inhibit tumor proliferation and growth by mediating oxidative stress and ferroptosis[91]. The P53 gene has been found to inhibit the expression of SLC7A11 and thus promote ferroptosis in tumor cells[92]. Glutamine is another important factor involved in iron-induced mortality. Glutaminase 2 (GSL2) can increase the expression of GSH in cancer cells and inhibit the onset of ferroptosis. P53 has been found to regulate the expression of GSL2, which is closely related to mitochondrial respiration and Adenosine Triphosphate (ATP) production in cancer cells[93]. Therefore, P53 is a starting point for promoting the occurrence of ferroptosis in a variety of cancers and warrants further study.
NRF2 is a transcription factor that plays a crucial role in the oxidative stress response. The harmful effects of ROS and some electrophiles are counteracted by NRF2[94]. Under normal conditions, NRF2 can bind to Kelch-like ECH-associated protein 1 (Keap1) and is subjected to ubiquitination and proteasome degradation. In conditions of oxidative stress, NRF2 enters the nucleus and binds to the Antioxidant Response Element (ARE) on DNA to promote the transcription and expression of anti-oxidative genes[95]. Interestingly, NRF2 can regulate two subunits of ferritin (FTL/FTH1) to regulate the level of iron in cells[96]. Several enzymes are regulated by NRF2 during GSH metabolism, such as Glutathione Synthase (GSS) and a subunit of the Cys glutamate transporter xCT, SLC7A11[97]. In addition, NRF2 plays a fundamental role in maintaining cellular redox homeostasis by regulating antioxidant systems such as GSH and Thioredoxin (TXN) and involves NADPH regeneration as well as heme metabolism[98]. Obviously, NRF2 not only plays an essential role in oxidative stress, but also plays a regulatory role in the mechanism of ferroptosis.
Ferroptosis and diseases:
Many diseases have been found to be associated with ferroptosis. By inhibiting or promoting enzymes or genes involved in ferroptosis, this has an impact on the course of diseases, such as liver disease, renal injury or cardiovascular disease and neurodegeneration[99]. NcRNAs, especially miRNAs, can alter disease progression by modulating ferroptosis. Ding et al. found that miR-182-5p and miR-378A-3p could promote ferroptosis by inhibiting GPX4 and SLC7A11 expression in models of ischemia-reperfusion renal injury[100]. In murine model with intracerebral hemorrhage, miR- 124 was found to modulate Ferroportin 1 (FPN1) and inhibit ferroptosis and apoptosis[101]. Interestingly, the role of ferroptosis in cancer is not quite the same as in other diseases. Ferroptosis can inhibit the proliferation and cell cycle of cancer cells, which suggests that we should focus on ferroptosis-based therapies for cancer that escape other forms of cell death, such as apoptosis or autophagy[102]. The role of ncRNAs in cancer has been extensively studied. Their role in regulating ferroptosis in cancer cells has also recently been discovered and will be reviewed below.
ncRNAs and Ferroptosis in Cancers
MiRNAs and ferroptosis:
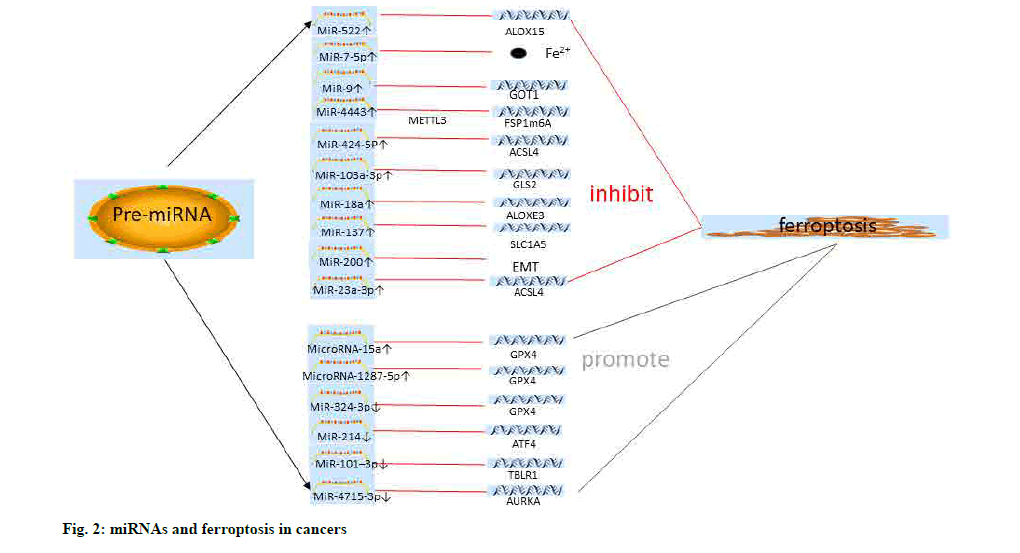
The differential expression of the GPX family of genes in Acute Myeloid Leukemia (AML) might be involved in the regulation of ferroptosis in tumor cells by influencing the expression of miR-202, miR- 381, miR-181 and many other miRNAs, and related transcription factors (Serum Response Factor (SRF) and E2F Transcription Factor 1 (E2F1)) or tumorassociated kinases (Death Associated Protein Kinase 1 (DAPK1) and proto-oncogene c-Src (SRC)). This provides a new direction for exploring the mechanisms of cell proliferation and the cell cycle of AML cells[103]. Tomita et al. found that the expression of miR-7-5p was significantly increased in Radiation-Resistant Cells (CRR) and decreased with the time of irradiation. Interestingly, the study also showed that miR-7-5p could reduce the content of Fe2+ in hepatoma cells and inhibit the occurrence of ferroptosis[104].
Erastin could increase ferroptosis of liver hepatocellular carcinoma cell line (HePG2 and Hep3B) cells and inhibit the proliferation of liver cancer cells. PremiR- 214 was revealed to increase the sensitivity to Erastin of liver cancer cells and could promote ferroptosis by targeting Activating Transcription Factor 4 (ATF4). In addition, the role of miR-214 in promoting ferroptosis might also be associated with increased levels of Malondialdehyde (MDA) and ROS[105]. Sorafenib-resistant hepatoma cells showed miR-23a-3p transcriptional regulation of ACSL4 expression in an ETS Proto-Oncogene 1 (ETS1) manner. MiR-23a-3p inhibitors could promote cell ferroptosis and improve drug resistance[106]. This suggests that radiation-induced ferroptosis in liver cancer cells is regulated by miRNAs. It is worth exploring whether other treatments, such as chemotherapy, are also regulated by miRNAs.
Zhang et al. found that miR-9 could target Glutamic- Oxaloacetic Transaminase 1 (GOT1) to inhibit Erastin and RSL3 sensitivity to ferroptosis in melanoma cells. GOT1 was a key enzyme in glutaminolysis, which was inhibited by miR-9. Silencing miR-9 could increase the expression of GOT1 and promote ferroptosis in cancer cells[107]. SLC1A5 is a cell membrane surface protein that mediates glutamine uptake. Recently, it was reported that miR-137 overexpression and Glutamyl- P-Nitroanilide (GPNA) could decrease the expression of SLC1A5 and inhibit the breakdown of glutamine and the ferroptosis of melanoma cells[108]. Glutamine metabolism and uptake play an essential role in the occurrence of ferroptosis in cancer cells.
LOXs play a key role in the lipid metabolism of tumors and the development of many cancers. The decrease of Epidermis-type Lipoxygenase 3 (ALOXE3) expressions might inhibit the occurrence of ferroptosis by decreasing the inhibitory effects of P53 on SLC7A11. Furthermore, miR-18a was found to be an upstream gene of ALOXE3 and could inhibit its role in ferroptosis in glioma cells. In addition, ALOXE3 could promote the migration of glioma cells by stimulating the secretion of 12-Hydroxyeicosatetraenoic Acids (12- HETE) and further activating the Phosphatidylinositol 3-Kinase/Protein Kinase B (PI3K/AKT) pathway[109].
Kirsten Rat Sarcoma Viral Oncogene Homologue (KRAS) mutations are common in the progression, metastasis and drug resistance of colorectal cancer[110]. Park et al. found that Achaete-Scute Family BHLH Transcription Factor 4 (ASCL-4) was significantly up-regulated in KRAS mutant colorectal cancer cells and when bromelain was used to treat colorectal cancer cells, ASCL-4 was significantly down-regulated and ferroptosis was evidenced by MDA and ROS generation. In addition, the differential expression of miRNAs in human colorectal adenocarcinoma cell line (Caco-2 and DLD-1) cells treated with bromelain was analyzed. It was found that miR-19b-3p, miR-130A- 3p, miR-150-5p, miR-144-3p, miR-16-5p, miR-7A- 5p and miR-17-5p were differentially expressed and could target ASCL-4 to regulate ferroptosis[111]. Angius et al. found that the epigenetic regulation of TP53 and SLC7A11 expression played an essential role in the regulation of ferroptosis in colorectal cancer cells. It was worth noting that let-7c, let-7e and miR-150-5p genes could regulate TP53. These miRNAs are worthy of further study in the cell death of colorectal cancer[112].
Aurora Kinase A (AURKA) is a serine/threonine kinase that regulates cell division and proliferation and is upregulated in many cancers. MiR-4715-3p was found to inhibit AURKA expression in upper gastrointestinal cancers and inhibit GPX4 expression to promote ferroptosis in tumor cells. The expression of Poly (ADPribose) Polymerase 1 (PARP) and caspase-3 was also regulated by miR-4715-3p, indicating that the apoptosis of tumor cells was also regulated. There is a link between cell death of various types, apoptosis, autophagy and ferroptosis[113]. Lee et al. found that Epithelial- Mesenchymal Transition (EMT) could play a regulatory role in ferroptosis sensitivity of head and neck tumors. Interestingly, inhibiting miR-200 family expression might promote ferroptosis sensitivity by modulating EMT epigenetics in head and neck tumors[114]. Physcion 8-O-Glucopyranoside (PG) has anti-cancer effects in a variety of cancers[114]. It enhanced GLS2 expression by down-regulating miR-103A-3p, increased the levels of Fe2+ and MDA in cells and significantly promoted ferroptosis[115]. Cancer-Associated Fibroblasts (CAFs) can secrete a variety of biologically active regulatory genes or substances to promote the development and proliferation of tumor cells. Recently, Zhang et al. found that CAF secreted miR-522 could regulate the drug resistance and proliferation of gastric cancer cells. MiR-522 was also found to inhibit ferroptosis of gastric cancer cells by targeting ALOX15[116].
Nanomedicine combined with related mechanisms for targeted delivery and treatment of tumors has attracted increasing attention[117]. Luo et al. used POPO-3 as a fluorescent dye to label miR-101-3p, a miRNA reported to modulate biological characteristics of various tumor cells, to inhibit the progression of A549 lung cancer cells via nanocarriers. MiR-101-3p had also been found to promote tumor cell apoptosis and ferroptosis to inhibit tumor cell proliferation by targeting Transducin (Beta)-like 1X Related protein 1 (TBLR1)[118].
Interestingly, miR-324-3p was revealed to directly target GPX4 and regulate ferroptosis and cisplatin sensitivity in A549 cells[119]. In addition, miR-324-3p/ GPX4 had been shown to modulate ferroptosis not only in lung cancer, but also in breast cancer. Metformin, a drug used to treat hypoglycemia, could promote ferroptosis in breast cancer cells by regulating miR- 324-3p to suppress GPX4 expression[120], whether the same mechanism exists in other cancers remains to be further studied.
Cisplatin is a first-line chemotherapy drug for many tumors, including lung cancer. Cisplatin could induce Fibroblast-Specific Protein 1 (FSP1) expression and promote ferroptosis in lung cancer cells. In addition, the expression of miR-4443 was increased in cisplatinresistant cell lines and inhibited the expression of FSP1 via Methyltransferase-Like 3 (METTL3) regulation of N6-methyladenosine (m6A) and interfered with ferroptosis in A549 cells[121].
Erastin and RSL3 could induce ferroptosis of ovarian cancer cells. Ma et al. determined that miR-424-5P could target ACSL4 to promote ferroptosis. MiR-424- 5p activity as a tumor inhibitor might represent a novel potential therapeutic strategy for ovarian cancer[122].
MiR-15a and miR-1287-5p could promote ferroptosis in prostate cancer and osteosarcoma cells by inhibiting the expression of GPX4[123,124] (fig. 2 and Table 1).
| miRNAs | References | Expression | Target/Mechanisms | Cancers |
|---|---|---|---|---|
| miR-202/miR-181/miR-381 | [103] | / | GPX | AML |
| miR-7-5p | [104] | ↑ | Fe2+ | HCC |
| miR-214 | [105] | ↓ | ATF4 | HCC |
| miR-23a-3p | [106] | ↓ | ACSL4 | HCC |
| miR-9 | [107] | ↑ | GOT1 | Melanoma |
| miR-137 | [108] | ↑ | SLC1A5 | Melanoma |
| miR-18a | [109] | ↑ | ALOXE3 | Glioblastoma |
| miR-19b-3p/miR-130A-3P/miR-150-5p/miR-144-3p/miR -16-5p/miR-7A-5P/miR-17-5p | [111] | ↓ | ASCL-4 | Colorectal cancer |
| Let-7c, let-7e, miR-150-5p | [112] | ↑ | TP53 | colorectal cancer |
| miR-4715-3p | [113] | ↓ | AURKA | Upper gastrointestinal cancers |
| miR-200 | [114] | ↑ | EMT | Head and neck cancer |
| miR-103a-3p | [115] | ↑ | GLS2 | Gastric cancer |
| miR-522 | [116] | ↑ | ALOX15 | Gastric cancer |
| miR-101–3p | [118] | ↓ | TBLR1 | Lung cancer |
| miR-324-3p | [119] | ↓ | GPX4 | Lung cancer |
| miR-324-3p | [120] | ↓ | GPX4 | Breast cancer |
| miR-15a | ||||
| miR-4443 | [121] | ↑ | FSP1 | Lung cancer |
| miR-424-5P | [122] | ↑ | ACSL4 | Ovarian cancer |
| miR-15a | [123] | ↑ | GPX4 | Prostate cancer |
| miR-1287-5p | [124] | ↑ | GPX4 | Osteosarcoma |
Table 1: miRNAs and Ferroptosis in Cancers.
LncRNA and ferroptosis:
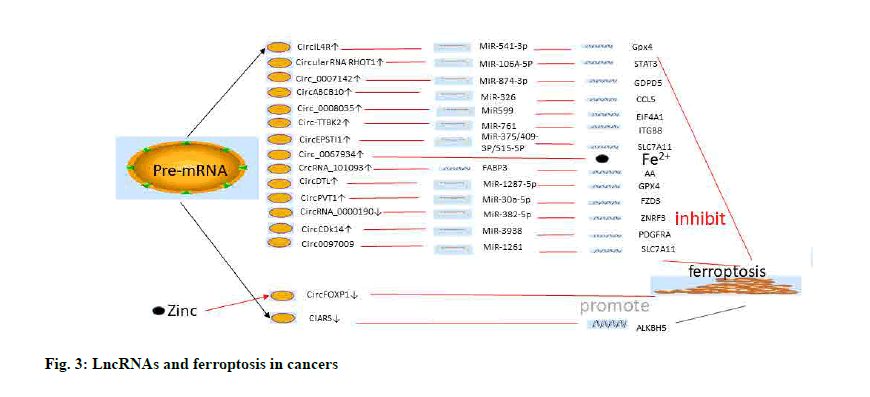
Wang et al. found that the expression of Long Intergenic Non-Protein Coding RNA 618 (LINC00618) was downregulated in leukemia and was significantly inhibited by Vincristine (VCR). In addition, LINC00618 increased the expression of BAX and the cleavage of caspase-3 to enhance apoptosis in leukemic cells. Interestingly, LINC00618 had also been shown to be associated with lipid oxidation of ferroptosis in leukemia cells by detecting ROS levels and intracellular iron deposition.
It could also inhibit the expression of Lymphoid- Specific Helicase (LSH) to inhibit the xc– systemrelated gene SLC7A11 and promote ferroptosis in leukemia cells[125].
In 2016, He et al. reported that LINC00336 was associated with poor prognosis in renal cell carcinoma[126] and also played an important role in ferroptosis of lung cancer cells. LINC00336 could inhibit ferroptosis by binding to ELAV-like RNA binding protein 1 (ELAVL1). In addition, LSH promoted the expression of ELAVL1 and LINC00336 via the P53 signaling pathway. Interestingly, miR6852, a downstream target gene of LINC00336, was found to regulate the ferroptosis-related marker Cystathionine Beta-Synthase (CBS). Thus, there seems to be a regulatory network involving LSH/P53/ELAVL1/ LINC00336 and miR6852/CBS in lung cancer cells[127].
The lncRNA Metallothionein 1D Pseudogene (MT1DP) could interact with miR-365 to enhance cadmium-induced oxidative stress by inhibiting NRF2 expression[128]. Recently, Gai et al. also reported a role for the MT1DP/miR-365a-3p/NRF2 axis in cellular oxidative stress in Non-Small Cell Lung Carcinoma (NSCLC)[129]. Folic acid is widely used as targeted therapy and as optical imaging therapy of tumors. Cancer-targeted optical diagnostics are performed by loading folic acid onto the surfaces of a variety of nanoparticles, including liposomes[130,131]. By detecting MDA and ROS levels, intracellular iron concentrations and GSH, the upregulation of MT1DP could promote NSCLC ferroptosis by inhibiting the expression of NRF2[129].
XAV939, a Tankyrase inhibitor that targets Wnt/beta (β)- catenin signaling was found to act as a small molecule inhibitor by shortening telomeres to promote tumor cell apoptosis[132]. The lncRNA miR503HG might be downregulated by XAV939 to inhibit the proliferation and development of NSCLC by sponging the miR1273c/ SOX4 axis. Furthermore, XAV939 could also inhibit SLC7A11 expression and promote ferroptosis in NSCLC[133]. However, the search for lncRNAs between XAV939 and SLC7A11 able to promote the ferroptosis process has not been confirmed and warrants further exploration.
In lung cancer cells, Wang et al. found that lncRNA Nuclear Paraspeckle Assembly Transcript 1 (lncNEAT1) could inhibit cell ferroptosis by sponging ACSL4. In addition, the levels of ACSL4, SLC7A11 and GPX4 were significantly decreased following silencing of NEAT1 silencing and Erastin co-processing in lung cancer cells, which suggested that this mechanism might facilitate the occurrence of ferroptosis[134].
The role of Erastin in the induction of ferroptosis in Hepatocellular Carcinoma (HCC) cells is well established. Qi et al. found that Erastin promoted the up-regulation of lncRNA GA-Binding Protein Transcription Factor Subunit Beta 1-Antisense RNA 1 (GABPB1-AS1) and inhibited the expression of GABPB1. Down-regulation of GABPB1 translation also resulted in the down-regulation of the gene encoding Peroxiredoxin-5 (PRDX5) peroxidase, which in turn reduced the cell antioxidant capacity and increased the susceptibility to ferroptosis in HCC cells[135].
LncRNA P53RRA was down-regulated in cancer cells and had been associated with the inhibition of cancer progression. P53RRA interacted with Ras GTPaseactivating protein-Binding Protein 1 (G3BP1) to induce the translocation of P53 from the G3BP1 complex and the retention of P53 in the nucleus, which led to lung cancer cells apoptosis and ferroptosis[136].
Recently, several studies had begun to identify lncRNAs associated with ferroptosis in different cancer types. For colon cancer, Cai et al. used RNA-seq results to query the Ferroptosis-Disease associations database (FerrDb) and The Cancer Genome Atlas (TGCA) database. Cox model analysis was used to construct a seven ferroptosisrelated lncRNAs-signature (LINC01503, ARRDC1 Antisense RNA 1 (ARRDC1-AS1), OIP5 Antisense RNA 1 (OIP5-AS1), AC004687.1, AC010973.2, AP001189.3 and NCK1 Divergent Transcript (NCK1- DT)). The results also revealed the potential molecular mechanisms involving ferroptosis-related lncRNAs, including the Mitogen-Activated Protein Kinase (MAPK), mammalian Target of Rapamycin (mTOR) and GSH pathways[137]. Furthermore, Tang et al. analyzed the TGCA database and found that 25 significantly different expressed lncRNAs, such as LINC01963, were associated with ferroptosis in head and neck tumors. The related mechanisms such as Notch pathway and phagocytosis mediated by FC gamma R were analyzed by Gene Set Enrichment Analysis (GSEA)[138]. High expression of lncRNA Brain-Derived Neurotrophic Factor Antisense (BDNF-AS) was revealed in Gastric Cancer (GC) and Peritoneal Metastasis (PM). LncRNA BDNF-AS could recruit WD repeat-containing protein 5 (WDR5) to modulate F-box and WD repeat domain containing 7 (FBXW7) expression to protect GC cells from ferroptosis[139]. LncRNA SLC16A1-AS1 was overexpressed in renal cell carcinoma and could inhibit ferroptosis via miR-143-3P/SLC7A11 axis[140]. LncRNA RP11-89 could promote iron export through miR-129-5P/Prominin2 (PROM2) axis and inhibit ferroptosis in bladder cancer cells[141].
Recently some compounds have been found to modulate the biological characteristics of cancer cells and the occurrence of ferroptosis by regulating lncRNA. Wenyujin’s effective ingredient, curcumenol, has been found to have anti-cancer potential. Curcumenol was revealed to induce cells ferroptosis and inhibit cells proliferation in lung cancer cells via lncRNA Imprinted Maternally Expressed Transcript (H19)/miR-19B- 3P/FTH1 signal transduction[142]. In addition, the researchers found that metformin inhibits autophagy and growth in breast cancer by inducing ferroptosis through lncRNA H19[143] (fig. 3 and Table 2).
| LncRNAs | References | Expression | Target/Mechanisms | Cancers |
|---|---|---|---|---|
| LINC00618 | [125] | ↓ | SLC7A11 | AML |
| LINC00336 | [127] | ↑ | miR6852/CBS | Lung cancer |
| MT1DP | [129] | ↓ | miR-365a-3p/NRF2 | Lung cancer |
| LncRNA miR503HG | [133] | ↑ | miR1273c/SOX4 | Lung cancer |
| LncNEAT1 | [134] | ↑ | ACSL4, SLC7A11 and GPX4 | Lung cancer |
| LncRNA GABPB1-AS1 | [135] | ↓ | GABPB1/PRDX5 | HCC |
| LncRNA P53RRA | [136] | ↓ | G3BP1/P53 | Lung cancer |
| LINC01503, ARRDC1-AS1, OIP5-AS1, AC004687.1, AC010973.2, AP001189.3 and NCK1-DT | [137] | ↑ | MAPK, mTOR and GSH pathways | Colon cancer |
| LINC01963 | [138] | / | Notch pathway | Head and neck cancer |
| LncRNA BDNF-AS | [139] | ↑ | WDR5/FBXW7 | Gastric cancer |
| LncRNA SLC16A1-AS1 | [140] | ↑ | miR143-3P/SLC7A11 | Renal cell carcinoma |
| LncRNA RP11-89 | [141] | ↑ | miR-129-5P/PROM2 | Bladder cancer |
| Metformin/Curcumenol/lncRNAH19 | [142,143] | ↓ | miR-19b-3p/FTH1 | Lung cancer/breast cancer |
Table 2: LncRNAs and Ferroptosis in Cancers.
CircRNA and ferroptosis:
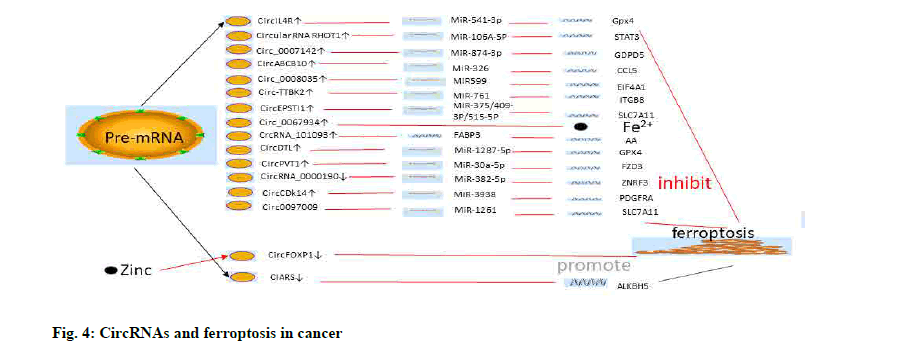
Sorafenib, a commonly used chemotherapy drug for advanced HCC, had been found to promote ferroptosis in liver cancer cells. Further, the inhibition of ferroptosis was also associated with the promotion of sorafenib resistance[144]. Analysis of three sorafenibtreated HCC cell lines by RNA-seq and Sanger sequencing revealed that cIARS (Human Serum Albumin (HSA_circ_0008367)) was highly expressed in hepatoma cells. cIARS could bind with as Alpha (α)-Ketoglutarate-Dependent Dioxygenase (ALKB) Homolog 5 (ALKBH5) to inhibit the expression of P62 and increase levels of the lipidation of autophagyrelated gene (LC3). cIARS could increase the level of MDA and Fe2+ and decrease the expression of GSH by promoting sorafenib and Erastin-mediated ferroptosis. Interestingly, cIARS also modulated ferroptosis in HCC cells by inhibiting ALKBH5 expression[145]. Circ0097009 are significantly up-regulated in HCC tissues and cell lines. The knock-down of circ0097009 could inhibit the proliferation and invasion of HCC cells and promote ferroptosis through the miR-1261/ SLC7A11 axis[146].
CircIL4R was also differentially expressed in HCC tissues and cells. Low expression of circIL4R not only inhibited the proliferation of HCC, but also promoted the ferroptosis of HCC cells. In addition, it was found that circIL4R could promote the expression of GPX4, a ferroptosis suppressor gene, by sponging miR-541-3p[147], which modulated the biological characteristics of prostate cancer[148]. The role of circIL4R in regulating ferroptosis in cancer cells is rarely reported, but it is worth of further study. In addition, quantitative Reverse Transcription Polymerase Chain Reaction (qRT-PCR) and luciferase assays showed that circIL4R was overexpressed in HCC and could also sponge miRNA- 541-3p. GPX4 had been identified as a downstream target gene for miR-541-3p. It is well known that GPX4 is a key regulator of ferroptosis. CircIL4R was also found to promote GPX4 expression and inhibit the role of ferroptosis in HCC[147].
SLC7A11 is an essential subunit of the xc– system as mentioned above. Knocking down the expression of SLC7A11 could promote the occurrence of ferroptosis. Wu et al. found that circRNA Epithelial Stromal Interaction 1 (circEPSTI1) could promote SLC7A11- mediated the production of GSH and inhibit ferroptosis in cervical cancer cells using the GSH/GSSG assay. MiR-375/409-3p/515-5p were downstream genes of circEPSTI1 and might regulate ferroptosis of cervical cancer cells[149]. CircRNA Tau Tubulin Kinase 2 (circ- TTBK2) was revealed to promote the proliferation and invasion of glioma cells via the miR-761/Integrin Subunit Beta 8 (ITGB8) axis. At the same time, the involvement of circ-TTBK2 in ferroptosis inhibition of glioma was determined by analyzing the content of Fe2+ and the generation of ROS in cells[150].
circRNA Cyclin Dependent Kinase 14 (CircCDK14) could promote glioma progression by regulating the miR-3938/Platelet-Derived Growth Factor Receptor Alpha (PDGFRA) axis and protect against ferroptosis[151]. qRT-PCR revealed that circ_0008035 expression was significantly higher in gastric cancer cells and tissues and its expression was inversely proportional to Overall Survival (OS). Circ_0008035 could promote proliferation and inhibit apoptosis of gastric cancer cells by regulating the miR-599/ Eukaryotic Translation Initiation Factor 4A1 (EIF4A1) axis. Further, silencing circ_0008035 expression using small interfering (si)-circ_0008035 could promote Erastin or RSL3-mediated cell death of gastric cancer. Knocking down circ_0008035 also increased the production of MDA and lipid ROS by promoting the occurrence of ferroptosis[152]. In addition, circRNA_0000190 has been shown to further regulate Zinc and Ring Finger 3 (ZNRF3) in gastric cancer cells by sponging miR-382-5p to promote ferroptosis[153]. CircRNA ATP Binding Cassette Subfamily B Member 10 (CircABCB10) had been reported to regulate the proliferation, invasion and metastasis of tumor cells in several tumors, including lung and breast cancer[154,155]. Further, circABCB10 also modulated the apoptosis and ferroptosis of rectal cancer cells via the miR-326/ Chemokine (C-C motif) Ligand 5 (CCL5) axis[156].
Circ_0007142 was revealed to be highly expressed in colorectal cancer. Circ_0007142 could promote Glycerophosphodiester Phosphodiesterase Domain Containing 5 (GDPD5) expression by sponging miR- 874-3p to inhibit colorectal cancer cell apoptosis and ferroptosis[157], thus, providing further support that there is a direct correlation between the types of cell death.
In addition, circRNA Ras Homolog Family Member T1 (RHOT1) was reported to promote breast cancer proliferation, invasion and migration. Further, circRNA RHOT1 also inhibited ferroptosis in breast cancer cells by regulating the miR-106A-5P/Signal Transducer and Activator of Transcription 3 (STAT3) axis[158]. It has been reported that there is a close relationship between STAT3 expression and genes involved in ferroptosis, such as NRF2 and GPX4[159]. Thus, there are good prospects for identifying a regulatory mechanism of ferroptosis by targeting the STAT3 gene.
Circ_0067934 has been found to act as a regulator of thyroid cancer. Circ_0067934 silence could raise the level of iron and ROS in cells[160]. Recently, exosome circRNA_101093 could interact with Fatty Acid- Binding Protein 3 (FABP3) and increase the reaction of arachidonic acid with taurine, which was critical for desensitization of lung adenocarcinoma cells to ferroptosis[161]. In addition, circRNA (Denticleless E3 Ubiquitin Protein Ligase Homolog) circDTL could act as a oncogene in lung cancer cells. The researchers found that knockdown of circDTL promoted apoptosis and ferroptosis in NSCLC cells through the miR-1287- 5P/GPX4 axis[162]. Zinc acts as a regulator of a key gene for ferroptosis. Zinc could inhibit the proliferation of lung cancer cells and promote ferroptosis in lung cancer cells with high expression of circRNA Forkhead Box P1 (circFOXP)[163].
Recently, researchers have found that circPVT1 Oncogene (PVT1) could upregulate and increase the expression of multidrug resistance-related proteins (including Multidrug Resistance Protein 1 (MRP1) and P-Glycoprotein (P-GP)) in 5-Fluorouracil (5- FU) resistant Esophageal Squamous Cell Carcinoma (ESCC) cells. The knockdown of circPVT1 was revealed to enhance chemical sensitivity to 5-FU by miR-30a-5p/Frizzled Class Receptor 3 (FZD3) axis and the occurrence of ferroptosis[164] (fig. 4 and Table 3).
| CircRNAs | References | Expression | Target/Mechanisms | Cancers |
|---|---|---|---|---|
| cIARS | [145] | ↓ | ALKBH5 | HCC |
| CircIL4R | [147] | ↑ | miR-541-3p/GPX4 | HCC |
| Circ0097009 | [146] | ↑ | miR-1261/SLC7A11 | HCC |
| CircEPSTI1 | [149] | ↑ | miR-375/409-3P/515-5P/SLC7A11 | Cervical cancer |
| Circ-TTBK2 | [150] | ↑ | miR-761/ITGB8 | Glioma |
| CircCDk14 | [151] | ↑ | miR-3938/PDGFRA | Glioma |
| Circ_0008035 | [152] | ↑ | miR599/EIF4A1 | Gastric cancer |
| CircRNA_0000190 | [153] | ↓ | miR-382-5p/ZNRF3 | Gastric cancer |
| CircABCB10 | [156] | ↑ | miR-326/CCL5 | Colorectal cancer |
| Circ_0007142 | [157] | ↑ | miR-874-3p/GDPD5 | Colorectal cancer |
| CircRNA RHOT1 | [158] | ↑ | miR-106A-5P/STAT3 | Breast cancer |
| Circ_0067934 | [160] | ↑ | miR-545-3p/SLC7A11 | Thyroid cancer |
| Eosome CircRNA_101093 | [161] | ↑ | FABP3 | Lung adenocarcinoma |
| CircDTL | [162] | ↑ | miR-1287-5p/GPX4 | Lung cancer |
| Zinc/CircFOXP1 | [163] | ↓ | / | Lung cancer |
| CircPVT1 | [164] | ↑ | miR-30a-5p/FZD3 | ESCC |
Table 3: CircRNAs and Ferroptosis in Cancers.
Other ncRNAs and ferroptosis:
To date, there are no reports evaluating the role of piRNA in ferroptosis. In breast cancer, piRNA-36712 was found to be significantly less expressed than in normal breast cells. RNAs produced by SEPW1 Pseudogene (SEPW1P) interacted with piRNA-36712 to inhibit the expression of P53[165]. PiRNA is known to associate with Piwi proteins to form effector complexes and regulate gene expression through epigenetics. Piwi-like protein 2 (Piwil2) was reported to form a Piwil2/STAT3/c-Src triple protein complex by directly associating with STAT3 protein via its Piwi Argonaut and Zwille (PAZ) domain. In addition, STAT3 is phosphorylated by c-Src and translocates to the nucleus, where it represses P53 gene transcription by binding to its promoter region of target genes in tumor cells[166]. Jiang et al. also reported that Piwil2 could inhibit tumor cell apoptosis by repressing P53 phosphorylation via P38[167]. These findings indicate that that Piwil2 may or may not inhibit ferroptosis in tumor cells. As a piRNA-like small RNA, piR-L-138 might interact with p60-MDM2 Proto- Oncogene (MDM2) to inhibit apoptosis and promote cisplatin resistance in P53-mutated lung cancer[168]. Recently, it had been reported that overexpression of piR-31470 could inhibit the levels of Glutathione S-Transferase Pi 1 (GSTP1) through DNA methylation modification and increase the vulnerability of RWPE1 cells (human prostate epithelial cells) to DNA damage and oxidative stress[169]. Overall, these finding suggest that piR-31470 might promote ferroptosis in cancer cells.
Loss of Cysteinyl-tRNA Synthetase (CARS) could inhibit ferroptosis by promoting transport in the xc- system and induction of the transsulfuration pathway[170]. In addition, Shimada et al. found that inhibition of tRNA synthetase might compensate for the loss of Cys in tumor cells, promote endogenous Cys biosynthesis and subsequently inhibit ferroptosis[171], tRNAs may regulate the formation of the cytochrome C-mediated apoptosome and further alter the apoptotic sensitivity of tumor cells[172].
The prospect of N6-methyladenosine (m6A) regulation:
With the development of high-throughput sequencing, over 170 types of RNA modifications have been discovered, of which methylation is the most common in eukaryotes. RNA modifications make RNA more complex and diverse, and are involved in the regulation of a variety of biological functions[173]. m6A was first reported to modulate mRNA expression in Novikoff hepatoma cells[174]. Since then, over 7000 mRNA types have been identified that are regulated by m6A in mammals, and a new type of epigenetics emerged. m6A is considered the most common methylation modification[175]. In addition, m6A modification has been identified in ncRNAs including miRNA, lncRNA, circRNA, ribosomal RNA (rRNA) and tRNA[176]. m6A modification occurs mainly in adenosine-rich RRACH sequences and is achieved by RNA methyltransferase (writers) such as METTL3 and METTL14; RNA demethylase (erasers) such as ALKBH5 and m6A binding protein (readers) such as YTH N6-Methyladenosine RNA Binding Protein 1 (YTHDF1) and YTHDF2. Likewise, m6A plays a key role in regulating the stability or splicing of cancer genes[177]. Two subunits SLC7A11 and SLC3A2 in the xc– system, which are important regulatory mechanisms of ferroptosis, are regulated by the m6A reader YT521-B Homology Containing 2 (YTHDC2) in lung adenocarcinoma. YTHDC2 could inhibit the expression of SLC3A2 by enhancing the m6A modification of the Homeo Box A13 (HOXA13) mRNA 3’-UTR and promote the ferroptosis of lung cancer cells[178]. Recently, METTL3 was identified as the target gene of miR-4443. Interestingly, miR- 4443 could regulate FSP1 m6A modification via METLL3, which inhibited ferroptosis and promoted NSCLC cisplatin resistance[121]. Both writers and readers can modify the function and death of cancer cells, including ferroptosis via the regulation of target genes. Bioinformatics analysis identified the upstream ncRNAs of YTHDC2 and METLL3 could regulate the ferroptosis of cancer cells via mRNA m6A. FSP1, SLC3A2 and SLC7A11 are essential regulatory genes for ferroptosis. Whether their m6A modification is regulated by other ncRNAs is worth further study. The role of m6A modification of ncRNAs in ferroptosis has not been reported. However, it has recent reports indicate that m6A regulates ncRNAs expression in apoptosis and autophagy. Sorafenib has been shown to be a promoter of ferroptosis. The stability of circRNA-SORE in sorafenib-resistant hepatoma cells was regulated by m6A and might inhibit cells apoptosis via the β-catenin signaling pathway[179]. The m6A modification of Forkhead box O3 (FOXO3) was revealed to maintain the stability of FOXO3 mRNA via METLL3 activity and promote the sensitivity of sorafenib in hepatoma cells by inhibiting autophagy[180]. Further studies had validated the role of circRNA-SORE m6A in regulating ferroptosis in sorafenib-resistant hepatoma cells and searched for upstream ncRNAs of FOXO3. The interaction between cIARS (HSA) and ALKBH5 might modulate ferroptosis in hepatoma cells through autophagy and apoptosis, although the authors only used ALKBH5 as an autophagy inhibitor.
ALKBH5 can be used as an eraser of m6A activity to explore the role of mRNAs methylation or of ncRNAs demethylation in ferroptosis.
The prospect of immunotherapy:
In the past few decades, cancer immunotherapy has made great progress from scientific exploration to clinical practice. Immune checkpoint therapy and the use of blocking antibodies have produced clinically significant efficacy[181]. Inhibition or activation of T cell function is directly controlled by certain immune checkpoint molecules, these include Programmed Cell Death protein 1 (PD-1) and its corresponding ligand PD-L1, Cytotoxic T Lymphocyte Associated Antigen 4 (CTLA-4), T Cell Membrane Protein 3 (TIM3), B and T Lymphocyte Attenuator (BTLA) and Lymphocyte Activating Gene 3 (LAG3)[182]. In the tumor’s microenvironment, these molecules are often expressed abnormally, which can induce excessive cellular immune tolerance and immune escape[183]. In the study of lncRNA-mediated ferroptosis in head and neck cancer, there were also differences in the expression of PD-1, CTLA4 and LAG3 between the high and low-risk groups[138]. It is suggested that lncRNA may modulate the occurrence of ferroptosis and change the immune escape of tumor cells. In addition, tumor cells are also regulated by intrapacket or vesicular ncRNAs in the process of tumor immune escape[184]. Circ-Carboxypeptidase A4 (CPA4) was found to inhibit apoptosis of NSCLC cells via let-7 miRNA/PD-L1 axis and promote tumor immune escape through inactivated CD8 T cells[185]. However, immunotherapy for cancer has been found to restore or enhance the effect of CD8 T cells to enhance the specific lipid peroxidation of ferroptosis in the tumor microenvironment. The release of Interferon gamma (IFNγ) from CD8 T cells could decrease the expression of SLC7A11 and SLC3A2[186]. NcRNAs may inhibit immune escape and become an important target of immunotherapy while regulating tumor cell death (ferroptosis).
Discussion
As a type of cell death, ferroptosis is closely associated with other forms of cell death. The correlation between ferroptosis, apoptosis or autophagy can be identified by detecting the related proteins or by observing changes in mitochondria. Ferroptosis is also a genetically regulated form of cell death.
The role of ncRNAs in the regulation of gene expression represents a novel direction to be explored in ferroptosis. There is no doubt that the most studied ncRNAs have focused on miRNA and its upstream lncRNA and circRNA. These form a network called the CeRNAnet, which regulates gene expression, especially in various cancers. But most ceRNA network regulation mechanisms investigated are still limited to apoptosis, autophagy or other metastatic pathways and drug resistance in cancer cells. We should continue to explore the occurrence of ferroptosis in different cancer types for the following reasons: Due to their potential role in apoptosis and autophagy; to identify marker genes associated with ferroptosis; and to search for upstream genes such as lncRNAs and circRNAs based on known miRNAs. For other ncRNAs such as piRNAs and tRNAs, there is still much to be explored in terms of tumor cell ferroptosis. PiRNA has been found to play a regulatory role in many mechanisms of ferroptosis. Further research could be carried out to investigate piRNA interacting proteins such as Piwil1 or Piwil2. We can also study the role of ferroptosis in tumor cells apoptosis by piRNAs. The regulatory networks involving ncRNAs such as tRNAs, rRNAs and piRNAs are still relatively unknown, and further research is needed to identify the differential expression of these ncRNAs based on gene sequencing and the functional role of ferroptosis. m6A, a type of gene editing, can promote the stability and maturation of mRNAs or ncRNAs. This suggests that there is a critical role for gene regulation of PCD including ferroptosis in cancer. Readers, writers and erasers of m6A may be the potential regulators of ferroptosis.
Current ncRNAs and ferroptosis researches are still lacking in terms of improving cancer chemoresistance. Further studies should address genes or pathways associated with chemoresistance such as ATPBinding Cassette (ABC) transporters, DNA damage repair, exosomes, KRAS signaling pathways. These mechanisms could be combined with ncRNAs regulation of ferroptosis to modulate the resistance towards chemotherapeutic drugs such as cisplatin and sorafenib. The use of bioinformatics will easily approach to screening of ncRNAs and at the same time, in future studies, ferroptosis activators and inhibitors such as Fe-1 or Erastin should be investigated for their role on ncRNAs. In addition, we wish to see further exploration of gene therapy (ncRNAs) and immunotherapy in tumor PCD to improve chemotherapy resistance.
Author’s contributions:
Yun Gong and Yue Lv contributed equally to this work and share first authorship.
Funding:
The article was funded by General Program of Jiangxi Natural Science Foundation (20202BABL206100) and the Science and Technology Projects of Jiangxi Provincial Department of Education (GJJ190117).
Conflict of interests:
The authors have no competing interests.
References
- Crick F. Central dogma of molecular biology. Nature 1970;227(5258):561-3.
- Johnsson P, Lipovich L, Grandér D, Morris KV. Evolutionary conservation of long non-coding RNAs; sequence, structure, function. Biochim Biophys Acta Gen Subj 2014;1840(3):1063-71.
[Crossref] [Google Scholar] [PubMed]
- Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer 2018;18(1):5-18.
[Crossref] [Google Scholar] [PubMed]
- ENCODE Project Consortium. Identification and analysis of functional elements in 1 % of the human genome by the ENCODE pilot project. Nature 2007;447(7146):799-816.
[Crossref] [Google Scholar] [PubMed]
- Saw PE, Xu X, Chen J, Song EW. Non-coding RNAs: The new central dogma of cancer biology. Sci China Life Sci 2021;64(1):22-50.
[Crossref] [Google Scholar] [PubMed]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993;75(5):843-54.
[Crossref] [Google Scholar] [PubMed]
- He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5(7):522-31.
[Crossref] [Google Scholar] [PubMed]
- Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell 2009;136(4):642-55.
[Crossref] [Google Scholar] [PubMed]
- Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 2010;11(9):597-610.
[Crossref] [Google Scholar] [PubMed]
- Rupaimoole R, Slack FJ. MicroRNA therapeutics: Towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov 2017;16(3):203-22.
[Crossref] [Google Scholar] [PubMed]
- Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol 2009;21(3):452-60.
[Crossref] [Google Scholar] [PubMed]
- Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell 2009;136(2):215-33.
[Crossref] [Google Scholar] [PubMed]
- Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat Struct Mol Biol 2010;17(10):1169-74.
[Crossref] [Google Scholar] [PubMed]
- Acunzo M, Romano G, Wernicke D, Croce CM. MicroRNA and cancer-a brief overview. Adv Biol Regul 2015;57:1-9.
[Crossref] [Google Scholar] [PubMed]
- Novikova IV, Hennelly SP, Sanbonmatsu KY. Tackling structures of long noncoding RNAs. Int J Mol Sci 2013;14(12):23672-84.
[Crossref] [Google Scholar] [PubMed]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007;129(7):1311-23.
[Crossref] [Google Scholar] [PubMed]
- Jathar S, Kumar V, Srivastava J, Tripathi V. Technological developments in lncRNA biology. Adv Exp Med Biol 2017;1008:283-323.
[Crossref] [Google Scholar] [PubMed]
- Tautz D. Polycistronic peptide coding genes in eukaryotes-how widespread are they? Brief Funct Genomic Proteomic 2009;8(1):68-74.
[Crossref] [Google Scholar] [PubMed]
- Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009;136(4):629-41.
[Crossref] [Google Scholar] [PubMed]
- Zhu J, Fu H, Wu Y, Zheng X. Function of lncRNAs and approaches to lncRNA-protein interactions. Sci China Life Sci 2013;56(10):876-85.
[Crossref] [Google Scholar] [PubMed]
- Ferre F, Colantoni A, Helmer-Citterich M. Revealing protein–lncRNA interaction. Brief Bioinform 2016;17(1):106-16.
[Crossref] [Google Scholar] [PubMed]
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011;146(3):353-8.
[Crossref] [Google Scholar] [PubMed]
- Chan JJ, Tay Y. Noncoding RNA: RNA regulatory networks in cancer. Int J Mol Sci 2018;19(5):1310.
[Crossref] [Google Scholar] [PubMed]
- Huang Y. The novel regulatory role of lncRNA-mi RNA-mRNA axis in cardiovascular diseases. J Cell Mol Med 2018;22(12):5768-75.
[Crossref] [Google Scholar] [PubMed]
- Li D, Yang C, Yin C, Zhao F, Chen Z, Tian Y, et al. LncRNA, important player in bone development and disease. Endocr Metab Immune Disord Drug Targets 2020;20(1):50-66.
[Crossref] [Google Scholar] [PubMed]
- Li J, Li Z, Zheng W, Li X, Wang Z, Cui Y, et al. Lnc RNA-ATB: An indispensable cancer-related long noncoding RNA. Cell Prolif 2017;50(6):e12381.
[Crossref] [Google Scholar] [PubMed]
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 1976;73(11):3852-6.
[Crossref] [Google Scholar] [PubMed]
- Kristensen LS, Andersen MS, Stagsted LV, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 2019;20(11):675-91.
[Crossref] [Google Scholar] [PubMed]
- Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 2003;72(1):291-336.
[Crossref] [Google Scholar] [PubMed]
- Hentze MW, Preiss T. Circular RNAs: Splicing's enigma variations. EMBO J 2013;32(7):923-5.
[Crossref] [Google Scholar] [PubMed]
- Meng X, Li X, Zhang P, Wang J, Zhou Y, Chen M. Circular RNA: An emerging key player in RNA world. Brief Bioinform 2017;18(4):547-57.
[Crossref] [Google Scholar] [PubMed]
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA-new member of noncoding RNA with novel functions. Exp Biol Med 2017;242(11):1136-41.
[Crossref] [Google Scholar] [PubMed]
- Liu P, Dong Y, Gu J, Puthiyakunnon S, Wu Y, Chen XG. Developmental piRNA profiles of the invasive vector mosquito Aedes albopictus. Parasit Vectors 2016;9(1):1-5.
- Hombach S, Kretz M. Non-coding RNAs: Classification, biology and functioning. Adv Exp Med Biol 2016;937:3-17.
[Crossref] [Google Scholar] [PubMed]
- Han YN, Li Y, Xia SQ, Zhang YY, Zheng JH, Li W. PIWI proteins and PIWI-interacting RNA: Emerging roles in cancer. Cell Physiol Biochem 2017;44(1):1-20.
[Crossref] [Google Scholar] [PubMed]
- Zeng Q, Wan H, Zhao S, Xu H, Tang T, Oware KA, et al. Role of PIWI-interacting RNAs on cell survival: Proliferation, apoptosis and cycle. IUBMB Life 2020;72(9):1870-8.
[Crossref] [Google Scholar] [PubMed]
- Fathizadeh H, Asemi Z. Epigenetic roles of PIWI proteins and piRNAs in lung cancer. Cell Biosci 2019;9(1):1-8.
- Tamtaji OR, Behnam M, Pourattar MA, Hamblin MR, Mahjoubin-Tehran M, Mirzaei H, et al. PIWI-interacting RNAs and PIWI proteins in glioma: Molecular pathogenesis and role as biomarkers. Cell Commun Signal 2020;18(1):168.
[Crossref] [Google Scholar] [PubMed]
- Guzzi N, Bellodi C. Novel insights into the emerging roles of tRNA-derived fragments in mammalian development. RNA Biol 2020;17(8):1214-22.
[Crossref] [Google Scholar] [PubMed]
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA Biol 2013;10(12):1798-806.
[Crossref] [Google Scholar] [PubMed]
- Yu M, Lu B, Zhang J, Ding J, Liu P, Lu Y. tRNA-derived RNA fragments in cancer: Current status and future perspectives. J Hematol Oncol 2020;13(1):1-4.
- Balatti V, Nigita G, Veneziano D, Drusco A, Stein GS, Messier TL, et al. tsRNA signatures in cancer. Proc Natl Acad Sci USA 2017;114(30):8071-6.
[Crossref] [Google Scholar] [PubMed]
- Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, et al. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 2017;552(7683):57-62.
[Crossref] [Google Scholar] [PubMed]
- Keam SP, Young PE, McCorkindale AL, Dang TH, Clancy JL, Humphreys DT, et al. The human Piwi protein Hiwi2 associates with tRNA-derived piRNAs in somatic cells. Nucleic Acids Res 2014;42(14):8984-95.
[Crossref] [Google Scholar] [PubMed]
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J 2001;20(14):3617-22.
[Crossref] [Google Scholar] [PubMed]
- Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol 2007;8(7):574-85.
[Crossref] [Google Scholar] [PubMed]
- Kiss T, Filipowicz W. Exonucleolytic processing of small nucleolar RNAs from pre-mRNA introns. Genes Dev 1995;9(11):1411-24.
[Crossref] [Google Scholar] [PubMed]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, et al. Long noncoding RNAs with snoRNA ends. Mol Cell 2012;48(2):219-30.
[Crossref] [Google Scholar] [PubMed]
- Elmore S. Apoptosis: A review of programmed cell death. Toxicol Pathol 2007;35(4):495-516.
[Crossref] [Google Scholar] [PubMed]
- Ouyang L, Shi Z, Zhao S, Wang FT, Zhou TT, Liu B, et al. Programmed cell death pathways in cancer: A review of apoptosis, autophagy and programmed necrosis. Cell Prolif 2012;45(6):487-98.
[Crossref] [Google Scholar] [PubMed]
- Dixon SJ, Lemberg KM, Lamprecht MR, Skouta R, Zaitsev EM, Gleason CE, et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012;149(5):1060-72.
[Crossref] [Google Scholar] [PubMed]
- Frey PA, Reed GH. The ubiquity of iron. ACS Chem Biol 2012;7(9):1477-81.
[Crossref] [Google Scholar] [PubMed]
- Petrat F, Rauen U, de Groot H. Determination of the chelatable iron pool of isolated rat hepatocytes by digital fluorescence microscopy using the fluorescent probe, phen green SK. Hepatology 1999;29(4):1171-9.
[Crossref] [Google Scholar] [PubMed]
- Cheng Z, Li Y. What is responsible for the initiating chemistry of iron-mediated lipid peroxidation: An update. Chem Rev 2007;107(3):748-66.
[Crossref] [Google Scholar] [PubMed]
- Tan S, Wood M, Maher P. Oxidative stress induces a form of programmed cell death with characteristics of both apoptosis and necrosis in neuronal cells. J Neurochem 1998;71(1):95-105.
[Crossref] [Google Scholar] [PubMed]
- Yagoda N, von Rechenberg M, Zaganjor E, Bauer AJ, Yang WS, Fridman DJ, et al. RAS-RAF-MEK-dependent oxidative cell death involving voltage-dependent anion channels. Nature 2007;447(7146):865-9.
[Crossref] [Google Scholar] [PubMed]
- Yang WS, Stockwell BR. Synthetic lethal screening identifies compounds activating iron-dependent, nonapoptotic cell death in oncogenic-RAS-harboring cancer cells. Chem Biol 2008;15(3):234-45.
[Crossref] [Google Scholar] [PubMed]
- Seiler A, Schneider M, Förster H, Roth S, Wirth EK, Culmsee C, et al. Glutathione peroxidase 4 senses and translates oxidative stress into 12/15-lipoxygenase dependent-and AIF-mediated cell death. Cell Metab 2008;8(3):237-48.
[Crossref] [Google Scholar] [PubMed]
- Friedmann Angeli JP, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 2014;16(12):1180-91.
[Crossref] [Google Scholar] [PubMed]
- Zilka O, Shah R, Li B, Friedmann Angeli JP, Griesser M, Conrad M, et al. On the mechanism of cytoprotection by ferrostatin-1 and liproxstatin-1 and the role of lipid peroxidation in ferroptotic cell death. ACS Cent Sci 2017;3(3):232-43.
[Crossref] [Google Scholar] [PubMed]
- Miotto G, Rossetto M, Di Paolo ML, Orian L, Venerando R, Roveri A, et al. Insight into the mechanism of ferroptosis inhibition by ferrostatin-1. Redox Biol 2020;28:101328.
[Crossref] [Google Scholar] [PubMed]
- Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, et al. The cystine/glutamate antiporter system xc-in health and disease: From molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal 2013;18(5):522-55.
[Crossref] [Google Scholar] [PubMed]
- Cao JY, Dixon SJ. Mechanisms of ferroptosis. Cell Mol Life Sci 2016;73(11):2195-209.
[Crossref] [Google Scholar] [PubMed]
- Zenlea T, Peppercorn MA. Immunosuppressive therapies for inflammatory bowel disease. World J Gastroenterol 2014;20(12):3146-52.
[Crossref] [Google Scholar] [PubMed]
- Nielsen OH, Verspaget HW, Elmgreen J. Inhibition of intestinal macrophage chemotaxis to leukotriene B4 by sulphasalazine, olsalazine, and 5-aminosalicylic acid. Aliment Pharmacol Ther 1988;2(3):203-11.
[Crossref] [Google Scholar] [PubMed]
- Wahl C, Liptay S, Adler G, Schmid RM. Sulfasalazine: A potent and specific inhibitor of nuclear factor kappa B. J Clin Invest 1998;101(5):1163-74.
[Crossref] [Google Scholar] [PubMed]
- Lo M, Ling V, Low C, Wang YZ, Gout PW. Potential use of the anti-inflammatory drug, sulfasalazine, for targeted therapy of pancreatic cancer. Curr Oncol 2010;17(3):9-16.
[Crossref] [Google Scholar] [PubMed]
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359(4):378-90.
[Crossref] [Google Scholar] [PubMed]
- Liu L, Cao Y, Chen C, Zhang X, McNabola A, Wilkie D, et al. Sorafenib blocks the RAF/MEK/ERK pathway, inhibits tumor angiogenesis and induces tumor cell apoptosis in hepatocellular carcinoma model PLC/PRF/5. Cancer Res 2006;66(24):11851-8.
[Crossref] [Google Scholar] [PubMed]
- Louandre C, Ezzoukhry Z, Godin C, Barbare JC, Mazière JC, Chauffert B, et al. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int J Cancer 2013;133(7):1732-42.
[Crossref] [Google Scholar] [PubMed]
- Dixon SJ, Patel DN, Welsch M, Skouta R, Lee ED, Hayano M, et al. Pharmacological inhibition of cysteine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis. Elife 2014;3:e02523.
[Crossref] [Google Scholar] [PubMed]
- Meister A. Biosynthesis and functions of glutathione, an essential biofactor. J Nutr Sci Vitaminol 1992;38:1-6.
[Crossref] [Google Scholar] [PubMed]
- Yang WS, Sri Ramaratnam R, Welsch ME, Shimada K, Skouta R, Viswanathan VS, et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014;156(1-2):317-31.
[Crossref] [Google Scholar] [PubMed]
- Griffith OW. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic Biol Med 1999;27(9-10):922-35.
[Crossref] [Google Scholar] [PubMed]
- Forcina GC, Dixon SJ. GPX4 at the crossroads of lipid homeostasis and ferroptosis. Proteomics 2019;19(18):1800311.
[Crossref] [Google Scholar] [PubMed]
- Alim I, Caulfield JT, Chen Y, Swarup V, Geschwind DH, Ivanova E, et al. Selenium drives a transcriptional adaptive program to block ferroptosis and treat stroke. Cell 2019;177(5):1262-79.
[Crossref] [Google Scholar] [PubMed]
- Weïwer M, Bittker JA, Lewis TA, Shimada K, Yang WS, MacPherson L, et al. Development of small-molecule probes that selectively kill cells induced to express mutant RAS. Bioorg Med Chem Lett 2012;22(4):1822-6.
[Crossref] [Google Scholar] [PubMed]
- Woo JH, Shimoni Y, Yang WS, Subramaniam P, Iyer A, Nicoletti P, et al. Elucidating compound mechanism of action by network perturbation analysis. Cell 2015;162(2):441-51.
[Crossref] [Google Scholar] [PubMed]
- Doll S, Proneth B, Tyurina YY, Panzilius E, Kobayashi S, Ingold I, et al. ACSL4 dictates ferroptosis sensitivity by shaping cellular lipid composition. Nat Chem Biol 2017;13(1):91-8.
[Crossref] [Google Scholar] [PubMed]
- Kagan VE, Mao G, Qu F, Angeli JP, Doll S, St Croix C, et al. Oxidized arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem Biol 2017;13(1):81-90.
- Latunde-Dada GO. Ferroptosis: Role of lipid peroxidation, iron and ferritinophagy. Biochim Biophys Acta Gen Subj 2017;1861(8):1893-900.
[Crossref] [Google Scholar] [PubMed]
- Doll S, Conrad M. Iron and ferroptosis: A still ill-defined liaison. IUBMB Life 2017;69(6):423-34.
[Crossref] [Google Scholar] [PubMed]
- Gammella E, Buratti P, Cairo G, Recalcati S. The transferrin receptor: The cellular iron gate. Metallomics 2017;9(10):1367-75.
[Crossref] [Google Scholar] [PubMed]
- Kawabata H. Transferrin and transferrin receptors update. Free Radic Biol Med 2019;133:46-54.
[Crossref] [Google Scholar] [PubMed]
- Sadrzadeh SM, Graf E, Panter SS, Hallaway PE, Eaton JW. Hemoglobin: A biologic fenton reagent. J Biol Chem 1984;259(23):14354-6.
[Google Scholar] [PubMed]
- Arosio P, Ingrassia R, Cavadini P. Ferritins: A family of molecules for iron storage, antioxidation and more. Biochim Biophys Acta Gen Subj 2009;1790(7):589-99.
[Crossref] [Google Scholar] [PubMed]
- Philpott CC. The flux of iron through ferritin in erythrocyte development. Curr Opin Hematol 2018;25(3):183-8.
[Crossref] [Google scholar] [PubMed]
- Radisky DC, Kaplan J. Iron in cytosolic ferritin can be recycled through lysosomal degradation in human fibroblasts. Biochem J 1998;336(1):201-5.
[Crossref] [Google scholar] [PubMed]
- Mancias JD, Wang X, Gygi SP, Harper JW, Kimmelman AC. Quantitative proteomics identifies NCOA4 as the cargo receptor mediating ferritinophagy. Nature 2014;509(7498):105-9.
[Crossref] [Google scholar] [PubMed]
- Hou W, Xie Y, Song X, Sun X, Lotze MT, Zeh III HJ, et al. Autophagy promotes ferroptosis by degradation of ferritin. Autophagy 2016;12(8):1425-8.
[Crossref] [Google scholar] [PubMed]
- Jiang L, Hickman JH, Wang SJ, Gu W. Dynamic roles of p53-mediated metabolic activities in ROS-induced stress responses. Cell Cycle 2015;14(18):2881-5.
[Crossref] [Google scholar] [PubMed]
- Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, et al. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015;520(7545):57-62.
[Crossref] [Google scholar] [PubMed]
- Hu W, Zhang C, Wu R, Sun Y, Levine A, Feng Z. Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc Natl Acad Sci USA 2010;107(16):7455-60.
[Crossref] [Google scholar] [PubMed]
- Tonelli C, Chio II, Tuveson DA. Transcriptional regulation by Nrf2. Antioxid Redox Signal 2018;29(17):1727-45.
[Crossref] [Google scholar] [PubMed]
- Zhang DD. Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab Rev 2006;38(4):769-89.
[Crossref] [Google scholar] [PubMed]
- Harada N, Kanayama M, Maruyama A, Yoshida A, Tazumi K, Hosoya T, et al. Nrf2 regulates ferroportin 1-mediated iron efflux and counteracts lipopolysaccharide-induced ferroportin 1 mRNA suppression in macrophages. Arch Biochem Biophys 2011;508(1):101-9.
[Crossref] [Google scholar] [PubMed]
- Dodson M, Castro-Portuguez R, Zhang DD. NRF2 plays a critical role in mitigating lipid peroxidation and ferroptosis. Redox Biol 2019;23:101107.
[Crossref] [Google scholar] [PubMed]
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov 2013;12(12):931-47.
[Crossref] [Google scholar] [PubMed]
- Han C, Liu Y, Dai R, Ismail N, Su W, Li B. Ferroptosis and its potential role in human diseases. Front Pharmacol 2020;11:239.
[Crossref] [Google scholar] [PubMed]
- Ding C, Ding X, Zheng J, Wang B, Li Y, Xiang H, et al. miR-182-5p and miR-378a-3p regulate ferroptosis in I/R-induced renal injury. Cell Death Dis 2020;11(10):1-4.
- Bao WD, Zhou XT, Zhou LT, Wang F, Yin X, Lu Y, et al. Targeting miR?124/Ferroportin signaling ameliorated neuronal cell death through inhibiting apoptosis and ferroptosis in aged intracerebral hemorrhage murine model. Aging Cell 2020;19(11):e13235.
[Crossref] [Google scholar] [PubMed]
- Bebber CM, Müller F, Prieto Clemente L, Weber J, von Karstedt S. Ferroptosis in cancer cell biology. Cancers 2020;12(1):164.
[Crossref] [Google scholar] [PubMed]
- Wei J, Xie Q, Liu X, Wan C, Wu W, Fang K, et al. Identification the prognostic value of glutathione peroxidases expression levels in acute myeloid leukemia. Ann Transl Med 2020;8(11):678.
[Crossref] [Google scholar] [PubMed]
- Tomita K, Fukumoto M, Itoh K, Kuwahara Y, Igarashi K, Nagasawa T, et al. miR-7-5p is a key factor that controls radioresistance via intracellular Fe2+ content in clinically relevant radioresistant cells. Biochem Biophys Res Commun 2019;518(4):712-8.
[Crossref] [Google scholar] [PubMed]
- Bai T, Liang R, Zhu R, Wang W, Zhou L, Sun Y. MicroRNA?214?3p enhances erastin?induced ferroptosis by targeting ATF4 in hepatoma cells. J Cell Physiol 2020;235(7):5637-48.
[Crossref] [Google scholar] [PubMed]
- Lu Y, Chan YT, Tan HY, Zhang C, Guo W, Xu Y, et al. Epigenetic regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates sorafenib resistance in human hepatocellular carcinoma. J Exp Clin Cancer Res 2022;41(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Zhang K, Wu L, Zhang P, Luo M, Du J, Gao T, et al. miR?9 regulates ferroptosis by targeting glutamic?oxaloacetic transaminase GOT1 in melanoma. Mol Carcinog 2018;57(11):1566-76.
[Crossref] [Google scholar] [PubMed]
- Luo M, Wu L, Zhang K, Wang H, Zhang T, Gutierrez L, et al. miR-137 regulates ferroptosis by targeting glutamine transporter SLC1A5 in melanoma. Cell Death Differ 2018;25(8):1457-72.
[Crossref] [Google scholar] [PubMed]
- Yang X, Liu J, Wang C, Cheng KK, Xu H, Li Q, et al. miR-18a promotes glioblastoma development by down-regulating ALOXE3-mediated ferroptotic and anti-migration activities. Oncogenesis 2021;10(2):1-3.
[Crossref] [Google scholar] [PubMed]
- Porru M, Pompili L, Caruso C, Biroccio A, Leonetti C. Targeting KRAS in metastatic colorectal cancer: Current strategies and emerging opportunities. J Exp Clin Cancer Res 2018;37(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Park S, Oh J, Kim M, Jin EJ. Bromelain effectively suppresses Kras-mutant colorectal cancer by stimulating ferroptosis. Anim Cells Syst 2018;22(5):334-40.
[Crossref] [Google scholar] [PubMed]
- Angius A, Uva P, Pira G, Muroni MR, Sotgiu G, Saderi L, et al. Integrated analysis of miRNA and mRNA endorses a twenty miRNAs signature for colorectal carcinoma. Int J Mol Sci 2019;20(16):4067.
[Crossref] [Google scholar] [PubMed]
- Gomaa A, Peng D, Chen Z, Soutto M, Abouelezz K, Corvalan A, et al. Epigenetic regulation of AURKA by miR-4715-3p in upper gastrointestinal cancers. Sci Rep 2019;9(1):1-11.
[Crossref] [Google scholar] [PubMed]
- Lee J, You JH, Kim MS, Roh JL. Epigenetic reprogramming of epithelial-mesenchymal transition promotes ferroptosis of head and neck cancer. Redox Biol 2020;37:101697.
[Crossref] [Google scholar] [PubMed]
- Niu Y, Zhang J, Tong Y, Li J, Liu B. Physcion 8-O-β-glucopyranoside induced ferroptosis via regulating miR-103a-3p/GLS2 axis in gastric cancer. Life Sci 2019 237:116893.
[Crossref] [Google scholar] [PubMed]
- Zhang H, Deng T, Liu R, Ning T, Yang H, Liu D, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer 2020;19(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Chow EK, Ho D. Cancer nanomedicine: From drug delivery to imaging. Sci Transl Med 2013;5(216):216.
[Crossref] [Google scholar] [PubMed]
- Luo Y, Niu G, Yi H, Li Q, Wu Z, Wang J, et al. Nanomedicine promotes ferroptosis to inhibit tumour proliferation in vivo. Redox Biol 2021;42:101908.
[Crossref] [Google scholar] [PubMed]
- Deng SH, Wu DM, Li L, Liu T, Zhang T, Li J, et al. miR-324-3p reverses cisplatin resistance by inducing GPX4-mediated ferroptosis in lung adenocarcinoma cell line A549. Biochem Biophys Res Commun 2021;549:54-60.
[Crossref] [Google scholar] [PubMed]
- Hou Y, Cai S, Yu S, Lin H. Metformin induces ferroptosis by targeting miR-324-3p/GPX4 axis in breast cancer. Acta Biochim Biophys Sin 2021;53(3):333-41.
[Crossref] [Google scholar] [PubMed]
- Song Z, Jia G, Ma P, Cang S. Exosomal miR-4443 promotes cisplatin resistance in non-small cell lung carcinoma by regulating FSP1 m6A modification-mediated ferroptosis. Life Sci 2021;276:119399.
[Crossref] [Google scholar] [PubMed]
- Ma LL, Liang L, Zhou D, Wang SW. Tumor suppressor miR-424-5p abrogates ferroptosis in ovarian cancer through targeting ACSL4. Neoplasma 2021;68(1):165-73.
[Crossref] [Google scholar] [PubMed]
- Xu P, Wang Y, Deng Z, Tan Z, Pei X. MicroRNA?15a promotes prostate cancer cell ferroptosis by inhibiting GPX4 expression. Oncol Lett 2022;23(2):1-8.
[Crossref] [Google scholar] [PubMed]
- Xu Z, Chen L, Wang C, Zhang L, Xu W. MicroRNA-1287-5p promotes ferroptosis of osteosarcoma cells through inhibiting GPX4. Free Radic Res 2021;55(11-12):1119-29.
[Crossref] [Google scholar] [PubMed]
- Wang Z, Chen X, Liu N, Shi Y, Liu Y, Ouyang L, et al. A nuclear long non-coding RNA LINC00618 accelerates ferroptosis in a manner dependent upon apoptosis. Mol Ther 2021;29(1):263-74.
[Crossref] [Google scholar] [PubMed]
- He HT, Xu M, Kuang Y, Han XY, Wang MQ, Yang Q. Biomarker and competing endogenous RNA potential of tumor-specific long noncoding RNA in chromophobe renal cell carcinoma. Onco Targets Ther 2016;9:6399.
[Crossref] [Google scholar] [PubMed]
- Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu N, et al. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ 2019;26(11):2329-43.
[Crossref] [Google scholar] [PubMed]
- Gao M, Li C, Xu M, Liu Y, Cong M, Liu S. LncRNA MT1DP aggravates cadmium?induced oxidative stress by repressing the function of Nrf2 and is dependent on interaction with miR?365. Adv Sci 2018;5(7):1800087.
[Crossref] [Google scholar] [PubMed]
- Gai C, Liu C, Wu X, Yu M, Zheng J, Zhang W, et al. MT1DP loaded by folate-modified liposomes sensitizes erastin-induced ferroptosis via regulating miR-365a-3p/NRF2 axis in non-small cell lung cancer cells. Cell Death Dis 2020;11(9):1-11.
- Farran B, Montenegro RC, Kasa P, Pavitra E, Huh YS, Han YK, et al. Folate-conjugated nanovehicles: Strategies for cancer therapy. Mater Sci Eng C Mater Biol Appl 2020;107:110341.
[Crossref] [Google scholar] [PubMed]
- Alavi M, Hamidi M. Passive and active targeting in cancer therapy by liposomes and lipid nanoparticles. Drug Metab Pers Ther 2019;34(1):1-8.
[Crossref] [Google scholar] [PubMed]
- Tian X, Hou W, Bai S, Fan J, Tong H, Bai Y. XAV939 promotes apoptosis in a neuroblastoma cell line via telomere shortening. Oncol Rep 2014;32(5):1999-2006.
[Crossref] [Google scholar] [PubMed]
- Yu H, Han Z, Xu Z, An C, Xu L, Xin H. RNA sequencing uncovers the key long non?coding RNAs and potential molecular mechanism contributing to XAV939?mediated inhibition of non?small cell lung cancer. Oncol Lett 2019;17(6):4994-5004.
[Crossref] [Google scholar] [PubMed]
- Wu H, Liu A. Long non-coding RNA NEAT1 regulates ferroptosis sensitivity in non-small-cell lung cancer. J Int Med Res 2021;49(3):1-11.
[Crossref] [Google scholar] [PubMed]
- Qi W, Li Z, Xia L, Dai J, Zhang Q, Wu C, et al. LncRNA GABPB1-AS1 and GABPB1 regulate oxidative stress during erastin-induced ferroptosis in HepG2 hepatocellular carcinoma cells. Sci Rep 2019;9(1):1-2. [Crossref]
[Google scholar] [PubMed]
- Mao C, Wang X, Liu Y, Wang M, Yan B, Jiang Y, et al. A G3BP1-interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res 2018;78(13):3484-96.
[Crossref] [Google scholar] [PubMed]
- Cai HJ, Zhuang ZC, Wu Y, Zhang YY, Liu X, Zhuang JF, et al. Development and validation of a ferroptosis-related lncRNAs prognosis signature in colon cancer. Bosn J Basic Med Sci 2021;21(5):569.
[Crossref] [Google Scholar] [PubMed]
- Tang Y, Li C, Zhang YJ, Wu ZH. Ferroptosis-related long non-coding RNA signature predicts the prognosis of head and neck squamous cell carcinoma. Int J Biol Sci 2021;17(3):702-11.
[Crossref] [Google Scholar] [PubMed]
- Huang G, Xiang Z, Wu H, He Q, Dou R, Lin Z, et al. The lncRNA BDNF-AS/WDR5/FBXW7 axis mediates ferroptosis in gastric cancer peritoneal metastasis by regulating VDAC3 ubiquitination. Int J Biol Sci 2022;18(4):1415-33.
[Crossref] [Google Scholar] [PubMed]
- Li YZ, Zhu HC, Du Y, Zhao HC, Wang L. Silencing lncRNA SLC16A1-AS1 induced ferroptosis in renal cell carcinoma through miR-143-3p/SLC7A11 signaling. Technol Cancer Res Treat 2022;21:1-10.
[Crossref] [Google Scholar] [PubMed]
- Luo W, Wang J, Xu W, Ma C, Wan F, Huang Y, et al. LncRNA RP11-89 facilitates tumorigenesis and ferroptosis resistance through PROM2-activated iron export by sponging miR-129-5p in bladder cancer. Cell Death Dis 2021;12(11):1043.
[Crossref] [Google Scholar] [PubMed]
- Zhang R, Pan T, Xiang Y, Zhang M, Xie H, Liang Z, et al. Curcumenol triggered ferroptosis in lung cancer cells via lncRNA H19/miR-19b-3p/FTH1 axis. Bioact Mater 2022;13:23-36.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Qin C, Zhou Y, Chen Y, Mao M, Yang J. Metformin may induce ferroptosis by inhibiting autophagy via lncRNA H19 in breast cancer. FEBS Open Bio 2022;12(1):146-53.
[Crossref] [Google Scholar] [PubMed]
- Sun X, Niu X, Chen R, He W, Chen D, Kang R, et al. Metallothionein?1G facilitates sorafenib resistance through inhibition of ferroptosis. Hepatology 2016;64(2):488-500.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Wang Q, Wang X, Xu Z, Wei X, Li J. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov 2020;6(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Lyu N, Zeng Y, Kong Y, Chen Q, Deng H, Ou S, et al. Ferroptosis is involved in the progression of hepatocellular carcinoma through the circ0097009/miR-1261/SLC7A11 axis. Ann Transl Med 2021;9(8):675.
[Crossref] [Google Scholar] [PubMed]
- Xu Q, Zhou L, Yang G, Meng F, Wan Y, Wang L, et al. CircIL4R facilitates the tumorigenesis and inhibits ferroptosis in hepatocellular carcinoma by regulating the miR?541?3p/GPX4 axis. Cell Biol Int 2020;44(11):2344-56.
[Crossref] [Google Scholar] [PubMed]
- Long BO, Li NA, Xu XX, Li XX, Xu XJ, Liu JY, et al. Long noncoding RNA LOXL1-AS1 regulates prostate cancer cell proliferation and cell cycle progression through miR-541-3p and CCND1. Biochem Biophys Res Commun 2018;505(2):561-8.
[Crossref] [Google Scholar] [PubMed]
- Wu P, Li C, Ye DM, Yu K, Li Y, Tang H, et al. Circular RNA circEPSTI1 accelerates cervical cancer progression via miR-375/409-3P/515-5p-SLC7A11 axis. Aging 2021;13(3):4663-73.
[Crossref] [Google Scholar] [PubMed]
- Zhang HY, Zhang BW, Zhang ZB, Deng QJ. Circular RNA TTBK2 regulates cell proliferation, invasion and ferroptosis via miR-761/ITGB8 axis in glioma. Eur Rev Med Pharmacol Sci 2020;24(5):2585-600.
[Crossref] [Google Scholar] [PubMed]
- Chen S, Zhang Z, Zhang B, Huang Q, Liu Y, Qiu Y, et al. CircCDK14 promotes tumor progression and resists ferroptosis in glioma by regulating PDGFRA. Int J Biol Sci 2022;18(2):841-57.
[Crossref] [Google Scholar] [PubMed]
- Li C, Tian Y, Liang Y, Li Q. Circ_0008035 contributes to cell proliferation and inhibits apoptosis and ferroptosis in gastric cancer via miR-599/EIF4A1 axis. Cancer Cell Int 2020;20(1):1-5.
[Crossref] [Google Scholar] [PubMed]
- Jiang M, Mo R, Liu C, Wu H. Circ_0000190 sponges miR-382-5p to suppress cell proliferation and motility and promote cell death by targeting ZNRF3 in gastric cancer. J Biochem 2022.
[Crossref] [Google Scholar] [PubMed]
- Tian X, Zhang L, Jiao Y, Chen J, Shan Y, Yang W. CircABCB10 promotes nonsmall cell lung cancer cell proliferation and migration by regulating the miR?1252/FOXR2 axis. J Cell Biochem 2019;120(3):3765-72.
[Crossref] [Google Scholar] [PubMed]
- Zhao Y, Zhong R, Deng C, Zhou Z. Circle RNA circABCB10 modulates PFN2 to promote breast cancer progression, as well as aggravate radioresistance through facilitating glycolytic metabolism via miR-223-3p. Cancer Biother Radiopharm 2021;36(6):477-90.
[Crossref] [Google Scholar] [PubMed]
- Xian ZY, Hu B, Wang T, Cai JL, Zeng JY, Zou Q, et al. CircABCB10 silencing inhibits the cell ferroptosis and apoptosis by regulating the miR-326/CCL5 axis in rectal cancer. Neoplasma 2020;67(5):1063-73.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Chen H, Wei X. Circ_0007142 downregulates miR?874?3p?mediated GDPD5 on colorectal cancer cells. Eur J Clin Invest 2021;51(7):e13541.
[Crossref] [Google Scholar] [PubMed]
- Zhang H, Ge Z, Wang Z, Gao Y, Wang Y, Qu X. Circular RNA RHOT1 promotes progression and inhibits ferroptosis via miR-106a-5p/STAT3 axis in breast cancer. Aging 2021;13(6):8115-26.
[Crossref] [Google Scholar] [PubMed]
- Liu Q, Wang K. The induction of ferroptosis by impairing STAT3/Nrf2/GPx4 signaling enhances the sensitivity of osteosarcoma cells to cisplatin. Cell Biol Int 2019;43(11):1245-56.
[Crossref] [Google Scholar] [PubMed]
- Wang HH, Ma JN, Zhan XR. Circular RNA Circ_0067934 attenuates ferroptosis of thyroid cancer cells by miR-545-3p/SLC7A11 signaling. Front Endocrinol 2021;12:1-10.
[Crossref] [Google Scholar] [PubMed]
- Zhang X, Xu Y, Ma L, Yu K, Niu Y, Xu X, et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun 2022;42(4):287-313.
[Crossref] [Google Scholar] [PubMed]
- Shanshan W, Hongying M, Jingjing F, Yiming Y, Yu R, Rui Y. CircDTL functions as an oncogene and regulates both apoptosis and ferroptosis in non-small cell lung cancer cells. Front Genet 2021:12:1-12.
[Crossref] [Google Scholar] [PubMed]
- Li H, Liu L. Zinc moderates circular RNA CircFOXP1 expression in order to regulate ferroptosis during lung adenocarcinoma. Chem Biol Interact 2022;352:109760.
[Crossref] [Google Scholar] [PubMed]
- Yao W, Wang J, Meng F, Zhu Z, Jia X, Xu L, et al. Circular RNA CircPVT1 inhibits 5-fluorouracil chemosensitivity by regulating ferroptosis through MiR-30a-5p/FZD3 axis in esophageal cancer cells. Front Oncol 2021;11:1-13.
[Crossref] [Google Scholar] [PubMed]
- Tan L, Mai D, Zhang B, Jiang X, Zhang J, Bai R, et al. PIWI-interacting RNA-36712 restrains breast cancer progression and chemoresistance by interaction with SEPW1 pseudogene SEPW1P RNA. Mol Cancer 2019;18(1):1-5.
[Crossref] [Google Scholar] [PubMed]
- Lu Y, Zhang K, Li C, Yao Y, Tao D, Liu Y, et al. Piwil2 suppresses p53 by inducing phosphorylation of signal transducer and activator of transcription 3 in tumor cells. PLoS One 2012;7(1):e30999.
[Crossref] [Google Scholar] [PubMed]
- Jiang S, Zhao L, Lu Y, Wang M, Chen Y, Tao D, et al. Piwil2 inhibits keratin 8 degradation through promoting p38-induced phosphorylation to resist Fas-mediated apoptosis. Mol Cell Biol 2014;34(21):3928-38.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Gable T, Ma MZ, Clark D, Zhao J, Zhang Y, et al. A piRNA-like small RNA induces chemoresistance to cisplatin-based therapy by inhibiting apoptosis in lung squamous cell carcinoma. Mol Ther Nucleic Acids 2017;6:269-78.
[Crossref] [Google Scholar] [PubMed]
- Zhang L, Meng X, Pan C, Qu F, Gan W, Xiang Z, et al. piR-31470 epigenetically suppresses the expression of glutathione S-transferase pi 1 in prostate cancer via DNA methylation. Cell Signal 2020;67:109501.
[Crossref] [Google Scholar] [PubMed]
- Hayano M, Yang WS, Corn CK, Pagano NC, Stockwell BR. Loss of cysteinyl-tRNA synthetase (CARS) induces the transsulfuration pathway and inhibits ferroptosis induced by cystine deprivation. Cell Death Differ 2016;23(2):270-8.
[Crossref] [Google Scholar] [PubMed]
- Shimada K, Stockwell BR. tRNA synthase suppression activates de novo cysteine synthesis to compensate for cystine and glutathione deprivation during ferroptosis. Mol Cell Oncol 2016;3(2):e1091059.
[Crossref] [Google Scholar] [PubMed]
- Mei Y, Yong J, Stonestrom A, Yang X. tRNA and cytochrome C in cell death and beyond. Cell Cycle 2010;9(15):3008-11.
[Crossref] [Google Scholar] [PubMed]
- Huang H, Weng H, Chen J. m6A modification in coding and non-coding RNAs: Roles and therapeutic implications in cancer. Cancer Cell 2020;37(3):270-88.
[Crossref] [Google Scholar] [PubMed]
- Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc Natl Acad Sci USA 1974;71(10):3971-5.
[Crossref] [Google Scholar] [PubMed]
- Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5′ terminus of HeLa cell messenger RNA. Cell 1975;4(4):379-86.
[Crossref] [Google Scholar] [PubMed]
- Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech 2019;1862(3):310-8.
[Crossref] [Google Scholar] [PubMed]
- Ma S, Chen C, Ji X, Liu J, Zhou Q, Wang G, et al. The interplay between m6A RNA methylation and noncoding RNA in cancer. J Hematol Oncol 2019;12(1):1-5.
[Crossref] [Google Scholar] [PubMed]
- Ma L, Zhang X, Yu K, Xu X, Chen T, Shi Y, et al. Targeting SLC3A2 subunit of system XC− is essential for m6A reader YTHDC2 to be an endogenous ferroptosis inducer in lung adenocarcinoma. Free Radic Biol Med 2021;168:25-43.
[Crossref] [Google Scholar] [PubMed]
- Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N6-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer 2020;19(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen X, et al. RNA m6A methylation regulates sorafenib resistance in liver cancer through FOXO 3?mediated autophagy. EMBO J 2020;39(12):e103181.
[Crossref] [Google Scholar] [PubMed]
- Couzin-Frankel J. Breakthrough of the year 2013. Cancer Immunother Sci 2013;342(6165):1432-3.
[Crossref] [Google Scholar] [PubMed]
- Wiener Z, Kohalmi B, Pocza P, Jeager J, Tolgyesi G, Toth S, et al. TIM-3 is expressed in melanoma cells and is upregulated in TGF-beta stimulated mast cells. J Invest Dermatol 2007;127(4):906-14.
[Crossref] [Google Scholar] [PubMed]
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12(4):252-64.
[Crossref] [Google Scholar] [PubMed]
- Huang M, Peng X, Yang L, Yang S, Li X, Tang S, et al. Non-coding RNA derived from extracellular vesicles in cancer immune escape: Biological functions and potential clinical applications. Cancer Lett 2021;501:234-46.
[Crossref] [Google Scholar] [PubMed]
- Hong W, Xue M, Jiang J, Zhang Y, Gao X. Circular RNA circ-CPA4/let-7 miRNA/PD-L1 axis regulates cell growth, stemness, drug resistance and immune evasion in non-small cell lung cancer (NSCLC). J Exp Clin Cancer Res 2020;39(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Wang W, Green M, Choi JE, Gijón M, Kennedy PD, Johnson JK, et al. CD8+ T cells regulate tumour ferroptosis during cancer immunotherapy. Nature 2019;569(7755):270-4.
[Crossref] [Google Scholar] [PubMed]
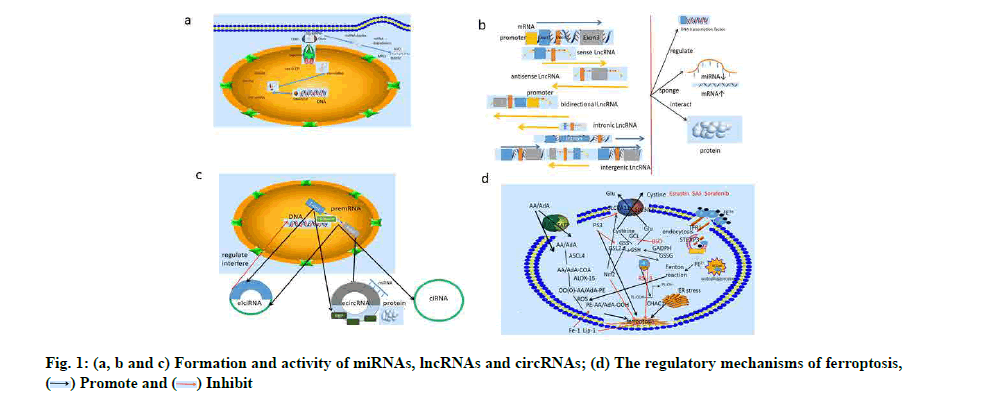
 .
.