- *Corresponding Author:
- Huirong Chen
Shenzhen Key Laboratory of Marine Bioresource and Eco-environmental Science, College of Life Sciences and Oceanography, Shenzhen University, Shenzhen 518071, PR China
E-mail: chenhr@szu.edu.cn
| This article was originally published in a special issue: Special issue on “Animal Models & Experimental Medicine” |
| Indian J Pharm Sci 2020:82(1)spl issue4;19-25 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To provide a potential method for improving microalgal lipid production, an over expression vector that expresses lysophosphatidic acid acyltransferase gene in Phaeodactylum tricornutum was constructed. Total fatty acid production incresed significantly in over-expression transformants and the length of fatty acids ranged from C14 to C22. The response to both lysophosphatidic acid acyltransferase over- and under-expression over the growth phase was also characterized. This study indicated a potential method to increase fatty acids accumulation in microalgae by employing a metabolic engineering approach.
Keywords
Metabolic engineering, Kennedy pathway, fatty acids, particle bombardment, Acrodactylum Tricornutum
The irreversible depletion of fossil fuels and fossil fuel burning-associated global warming have accelerated the search for biofuel production via renewable feedstock over the past few years[1,2]. Due to some obvious advantages which include higher photosynthetic efficiency and biomass production rate, microalgal lipid, especially triacylglycerols (TAGs), have broadly been researched as a hidden biodiesel raw material[3]. In order to cope with environmental stress situations such as nutrient deprivation, changes in salinity, pH variation and increased temperature, photosynthetic microalgae can accumulate high concentrations of storage lipids in the form of droplets. Nutrient deprivation, especially nitrogen depletion is the most commonly used method to trigger lipid storage in oleaginous algae[4]. Microalgae can change the lipid metabolism to the synthesis and storage of neutral or storage lipids to overcome these adverse situations via the fatty acid (FA) assembly pathway. After the synthesis of FAs in the plastid these are exported to the cytosol to enter the triacylglycerol assembly pathway or the Kennedy pathway[5]. The Kennedy pathway is the most understood pathway for TAG assembly from FA and involves stepwise acylation of each hydroxyl group of glycerol which is fairly straight forward. TAGs were produced by the course of the acyl-CoA being incorporated into glycerol-3-phosphate and the reaction process requires the participation of a series of key enzymes, such as glycerol-3-phosphate acyltransferase (GPAT), lysophosphatidic acid acyltransferase (LPAAT), phosphatidic acid phosphatase (PAP) and diacylglycerol acyltransferase (DGAT)[6]. LPAAT is one of the most critical acyltransferases of the FA biosynthesis pathway[7]. In Chlamydomonas reinhardtii, it was shown that LPAAT expression was markedly up-regulated in neutral lipid accumulation phase and knock down mutants of LPAAT gene showed reduced accumulation of neutral lipids[8]. Also, simultaneous heterologous expression of glyceraldehyde-3- phosphate dehydrogenase (G3PDH), GPAT, LPAAT and DGAT, which were cloned from the yeast strains, Saccharomyces cerevisiae and Yarrowia lipolytica in Chlorella minutissima UTEX 2219, improved the lipid content of the algae 3 fold and lipid productivity was increased by 1.5 fold[9]. Thus, LPAAT is a potential target protein to enhance TAG accumulation and productivity in algae. Genetic engineering techniques have been previously used to microalgae to increase lipid accumulation or understand lipid accumulation under stress conditions[10-12]. Genetic transformation is a promising strategy for further improving microalgal lipid production[13].
Among the photosynthetic microalgae, diatoms are eukaryotic phytoplanktons that contribute significantly to oceanic primary productivity and CO2 fixation and has great potential for autotrophic production of biomass[14]. Phaeodactylum tricornutum (P. tricornutum), the model organism of choice for diatoms, is ideal for biodiesel production due to its ease of cultivation, short biomass doubling time, smaller genome size, sequenced genome information, and availability of molecular tools for genetic engineering[15]. Also, P. tricornutum is capable of synthesizing neutral lipids under unfavorable conditions, mainly in the form of TAGs, which accumulate in the late exponential phase[16]. Storage lipids can constitute at least 20-30 % of the dry cell weight of P. tricornutum under standard culture conditions[17]. Literature showed that P. tricornutum was able to be easily genetically transformed[18] and several functional genomics methods such as gene silencing[19] and genome editing[20] have been previously developed. Although tools for genetic modification currently exist for some diatom species[21], these are slightly inefficient compared to the efficient methods used in the model microbials such as E. coli and yeast, which has counteracted both basic diatom research and applied strain development[22]. Moreover, for lipid accumulation in P. tricornutum, the influence of the expression of LPAAT gene on lipid content and FA composition remain poorly understood. Thus, in this study, the effect of over expression and knock down of LPAAT gene in P. tricornutum was studied in relation to TAG synthesis and accumulation and was compared to the wild type.
Material and Methods
Organisms and culture conditions:
P. tricornutum FACHB-863, was purchased from the Institute of Hydrobiology, Chinese Academy of Science. P. tricornutum was maintained in f/2 solidified medium containing 1.5 % (w/v) agarose. The salinity of the f/2 culture medium was 35. P. tricornutum was cultured in 250 ml flask with 100 ml medium and was under the condition of 12 h:12 h light and dark cycle, a light density of 60 μm2/s and 20°. Initial pH of the culture medium was controlled at approximately 7.5. The inoculation size was controlled at 5.0×104 cell/ ml. Microalgal samples were collected daily and the cell concentrations were determined using a haemocytometer. Transformants of the wild type strain with LPAAT gene were selected using the herbicide glufosinate at a concentration of 100 μg/ml. E. coli top10 strain was used as a host for recombinant vector construction and plasmid isolation. E. coli strain was generally cultivated in Luria-Bertani (LB) medium at 37° and the shaking speed of 180 rpm. When isolating recombinant strains, ampicillin was used at a concentration of 100 μg/ml.
Plasmid construction and transformation:
The genomic cDNA of P. tricornutum was extracted using tissue DNA Kit (Omega, Shanghai, China) and used as the template of polymerase chain reaction (PCR). Then, the LPAAT gene was cloned with the followed amplification condition, 35 cycles of 20 sec denaturation at 94º, annealing for 30 s at 60° and extension for 30 at 72°. The primer pairs used for amplification are given in Table 1. The cloning vector was pMD18-T was isolated from E. coli Top 10. The obtained plasmidpMD18-LPAAT and pfcpA-MCS/fcpB-Barwere digested with BamH I and Hind III at the position of Nco І. The positive clones after screened by LB medium supplemented with 100 μgmL-1ampicillinwere identified by PCR amplification. Digested products were ligated by T4 DNA Ligase and constructed the over-expression vector Lpaat-pfcpA-MCS/fcpB-Bar and antisense-expression vector antisense-Lpaat-pfcpA-MCS /fcpB-Bar.
Table 1: The Primer Sequences for LPAAT Gene Extraction
| Sense primer | |
|---|---|
| Forward primer | CGGGATCCGAACACTGGTACGCTTATTGC |
| Reverse primer | CAAGCTTAAGATGGCGACGATGATGAG |
| Antisense primer | |
| Forward primer | CAAGCTT CGAACACTGGTACGCTTATTGC |
| Reverse primer | CGGGATCCTTAAGATGGCGACGATGATGAG |
Transformation of microalgal cells: particle bombardment method
The microalgal cells of P. tricornutum were collected after 7 d of cultivation in the mid-exponential phase and were counted with a hemocytometer. The microalgal cell concentration of P. tricornutum was optimized to be below 108 cells/ml on the f/2 agar media, the microalgal cells of P. tricornutum were put on the 47-mm diameter cellulose acetate membrane filter (Sartorius Stedim Biotech, Germany) and were bombarded for one time[23]. Particle bombardment transformation was carried out by using the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The plasmid was linearized by Nco I, and was purified, and was adjusted the concentration to 1 μg/μl by ethanol precipitation. The linearized and concentrated recombinant vectors were coated onto gold particles at 0.6 μm (Bio-Rad, USA)[24]. Five micrograms of purified DNA, 50 μl CaCl2 (2.5 M) and 20 μl spermidine (0.1 M) were completely mixed to 3 mg of 0.6 μm gold particles resuspended in 50 % glycerol. The prepared DNA particles were rinsed with 70 % ethanol, dissolved in 100 μl of 100 % ethanol. After bombardment, the microalgal cells were immediately cultivated in f/2 medium and cultivated at a low light density of 60 μmol/m2s for 8 h. Then coating 100 μl liquid microalga on a solid medium plate and incubated for 1 month.
Real time PCR:
The RNAiso Plus kit for total RNA (Takara Dalian, China) was used to isolate total cellular RNA and the contaminating genomic DNA of which was removed by treating with DNase I. DNA-free RNA was reverse transcribed using SuperScript™ First-Strand (Takara Dalian) in a final volume of 20 μl. The cDNA was dissolved in nuclease-free water and frozen at -20°. Primers were designed to quantify the glufosinate resistance gene (bar) using Primer 5.0. Primers information including sequence and PCR product size are summarized in Table 2. Quantitative PCR was performed in a 50 μl final volume using SYBR Premix Ex TaqTM. The cDNA synthesis was performed with the reverse primer used for genomic PCR with 1 mL RNaseOUT, and the parameter of which was 1 h at 55°, terminated at 75° for 15 min. To reduce the hazard of RNA degradation, all the steps from RNA isolation to cDNA synthesis should be ended in 24 h. PCR amplification was run on PikoReal™ Real-Time PCR System (Thermo Fisher USA) using 96-well optical plates under the following conditions, 6 min at 94°, 30 cycles, 94° for 1 min, 60° for 30 s, 72° for 1 min, and 72° for 10 min[25]. RNA quality was assessed on a 2 % agarose gel.
Table 2: OLIGO Sequences for Fluorescence Quantitative Real-Time PCR Primers
| Target genes | Primer sequence | Size (bp) |
|---|---|---|
| β-actin | F: 5′- ACCCCGTGCTGCTGACTG -3′ | 122 |
| R: 5′- ACGTTGAAGGTCTCGAACA -3′ | ||
| LPAAT | F: 5′- GTCCAGGCCAGTTCATCGTTG -3′ | 212 |
| R: 5′- CTGTTCGGTCAGCACCATTGT -3′ |
FA analysis:
The FAs present in the sample were transformed to fatty acid methyl ester (FAME) to increase the volatility of the sample and clear chromatographic analysis. Therefore, approximately 15 mg lyophilized microalgal biomass was suspended in 1 ml methanol for esterification. During detection, 500 μg/ml C19 as an internal standard for esterification and added to the mixture prior to reaction for 15 min. After completely drying, the pellet was resuspended in 1 ml pf 4 M hydrochloric acid in methanol. In order to saponify the acyl group in the lipid and convert it to FAME, the sample tube was incubated at 100° for 1 h and then the obtained FAME was recovered using n-hexane. The FAME residue contained in the hexane phase recovered by evaporation was reextracted using 100 μl of n-hexane. To measure the concentrations of FAME, the hexane solution was subjected to gas chromatography equipped with a flame-ionization detector (Agilent Technologies 7890 A). The column of VF-23 ms (30.0 m×320 μm×0.25 μm) was employed with the following temperatures, 70° for 4 min, then increased to 195° at a rate of 25°/min and further increased to 205° at a rate of 3°/min, and finally the temperature was increased to 250°.
Laser confocal microscopy:
The microalgal biomass of over-expression and antisense-expression transgenic microalgae at plateau phase was collected for detecting the content of FAs via confocal laser scanning microscope (CLSM). microalgal sample (790 μl) was stained with 10 μl Nile red and 200 μl dimethyl sulfoxide (DMSO) for 6 min.
Results and Discussions
The purpose of this research was to use the genomic technology to create and characterize transgenic marine diatoms for production of useful biofuel FA. The strategy employed was to create two strains of P. tricornutum, one overexpressing and one under expressing the LPAAT via an antisense construct.
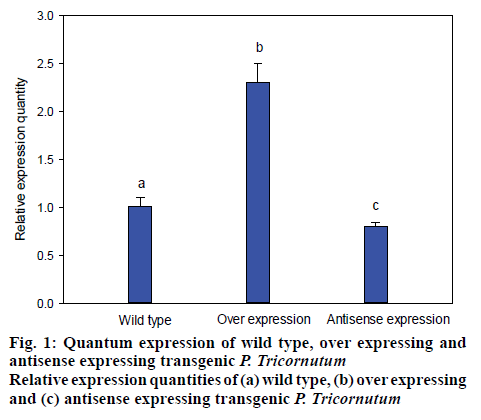
The transformed P. tricornutum FACHB-863 were screened by growing in the presence of the herbicide glufosinate, for which there is a resistance gene on the plasmid. Monoclonal transformants grew in about a month. The selected transformants were cultivated on solid medium containing herbicides, the microalgal cells that resisted the herbicide successfully were cultivated with f/2 medium. Then, microbial genomic DNA was subsequently extracted and dyes with fluorescent dyes for evaluating and quantifying the over-expression and antisense-expression transformants by detecting the strength of fluorescent signal (figure 1). LPAAT gene in the transformant was approximately 240 % compared to wild type microalgae and the antisense-expression transformant had 35 % lower expression compared the wild type microalgae with the relative expression quantity of 65 % (figure 1).
The monoclonal transformants was screened and inoculated on the solid medium for purification and the grown transformants were switched to liquid medium for cultivation. After cultivation for 20 d, the microalgae concentrations reached the maximum cell concentrations of approximately 32×105 for the 3 strains tested, LPAAT over-expression, LPAAT antisenseexpression and wild type P. tricornutum FACHB-863 (figure 2). The maximum cell mass obtained was similar for all the 3 strain, while the transformants has a prolonged lag phase of 2 d, with the wild type reached maximum cell density in about 11 d, the transformants reached maximum cell density in about 13 d.
The FA contents of the transgenic microalgal cells were measured using the esterification method that can saponify the acyl groups and convert into FAME. The FAMEs were detected by GC-MS and results are shown as Table 3. During the lag phase, the total accumulated lipid of LPAAT-overexpressing P. tricornutum FACHB-863 was 248.7 mg/g dry microalgal biomass. This was 85.9 % higher than the result of wild type microalgae (133.8 mg/g). For the antisense-expression microalgae, the total lipid was significantly decreased to 34.7 mg/g with a decrease of 74.1 %. At the early plateau phase, the total lipid content of wild type, overexpression, and antisense-expression microalgae were 214.8, 267.3 (24.4 % increased), and 140.8 mg g-1 (34.5 % decreased), respectively. At the late plateau phase, the total lipid increased was 40.5 % over expression (365.5 mg/g) and decreased 91.5 % (22.2 mg/g dry weight) under antisense expression.
Table 3: Fatty Acids Content of over Expression, Antisense Expression Compared with Wild type P. tricornutum. At different Cultivation Phases
| Fatty acids | Lag phase | Early plateau phase | Late plateau phase | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Wild type | Overexpression | Antisense expression | Wild type | Overexpression | Antisense expression | Wild type | Overexpression | Antisense expression | |
| C14 | 7.71±0.002 | 16.15±0.112 | 2.35±0.004 | 11.49±0.005 | 17.65±0.012 | 7.56±0.145 | 17.61±0.001 | 25.59±0.012 | 15.31±0.020 |
| C15 | 0.57±0.001 | 0.87±0.002 | 0.17±0.002 | 0.78±0.012 | 0.97±0.023 | 0.56±0.023 | 1.02±0.004 | 1.48±0.005 | 0.91±0.030 |
| C16 | 15.3±0.012 | 33.74±0.254 | 4.73±0.125 | 23.01±0.0167 | 34.64±0.210 | 17.35±0.104 | 34.63±0.033 | 50.97±0.105 | 26.74±0.040 |
| C16:1 | 25.6±0.104 | 51.1±0.212 | 6.73±0.032 | 35.93±0.105 | 52.85±0.165 | 26.8±0.235 | 47.55±0.066 | 70.55±0.016 | 35.25±0.010 |
| C16:2 | 7.75±0.021 | 14.9±0.004 | 2.27±0.008 | 13.49±0.001 | 18.24±0.004 | 8.78±0.041 | 16.24±0.033 | 21.01±0.007 | 15.56±0.123 |
| C16:3 | 8.64±0.057 | 17.18±0.007 | 2.36±0.014 | 16.67±0.007 | 5.47±0.005 | 10.38±0.105 | 19.74±0.004 | 23.42±0.001 | 18.17±0.014 |
| C16:4 | 8.65±0.014 | 2.57±0.004 | 0.57±0.001 | 16.64±0.013 | 0.33±0.004 | 2.56±0.017 | 2.61±0.003 | 3.71±0.012 | 2.99±0.010 |
| C18 | 0.77±0.004 | 1.56±0.027 | 0.27±0.001 | 1.39±0.001 | 1.67±0.110 | 1.28±0.008 | 2.32±0.041 | 2.60±0.013 | 1.69±0.005 |
| C18:1c | 6.13±0.002 | 2.56±0.007 | 0.45±0.002 | 2.87±0.004 | 4.27±0.200 | 1.94±0.109 | 3.17±0.194 | 7.12±0.004 | 3.64±0.001 |
| C18:1t | 1.57±0.001 | 2.15±0.001 | 0.2±0.003 | 1.86±0.009 | 2.16±0.103 | 1.56±0.010 | 2.08±0.081 | 2.45±0.005 | 2.09±0.014 |
| C18:2t | 2.27±0.102 | 4.26±0.103 | 0.7±0.003 | 4.14±0.014 | 5.59±0.021 | 2.65±0.013 | 5.64±0.074 | 8.04±0.002 | 5.57±0.011 |
| C18:3n3 | 0.66±0.007 | 0.96±0.006 | 0.1±001 | 0.83±0.005 | 1.27±0.012 | 0.76±0.002 | 1.29±0.007 | 1.44±0.003 | 1.16±0.001 |
| C18:3n6 | 0.54±0.025 | 0.96±0.003 | 0.24±0.003 | 0.92±0.004 | 1.13±0.111 | 0.67±0.004 | 1.25±0.029 | 3.19±0.101 | 1.44±0.081 |
| C20 | 0.40±0.006 | 0.47±0.001 | 0.17±0.002 | 0.60±0.001 | 0.69±0.019 | 0.58±0.023 | 4.04±0.026 | 0.75±0.009 | 0.55±0.029 |
| C20:2 | 0.45±0.011 | 0.53±0.013 | 0.17±0.005 | 0.64±0.081 | 0.76±0.005 | .057±0.017 | 0.65±0.032 | 1.60±0.003 | 0.76±0.001 |
| C20:3 | 0.34±0.002 | 0.36±0.024 | 0.00±0.000 | 0.55±0.049 | 0.58±0.013 | 0.48±0.018 | 0.37±0.011 | 0.61±0.006 | 0.47±0.002 |
| C20:4(5,8,11,14) | 0.41±0.004 | 0.52±0.021 | 0.14±0.012 | 0.81±0.067 | 0.78±0.022 | 0.5±0.003 | 0.88±0.002 | 1.71±0.010 | 0.91±0.030 |
| C20:4(8,11,14,17) | 0.54±0.025 | 0.96±0.006 | 0.24±0.004 | 0.95±0.32 | 1.27±0.101 | 0.77±0.001 | 1.29±0.004 | 2.13±0.102 | 1.32±0.021 |
| C20:5 | 39.92±0.521 | 79.4±0.214 | 11.3±0.315 | 68.48±0.297 | 98.82±0.421 | 46.62±0.204 | 86.04±0.178 | 111.74±0.304 | 70.54±0.026 |
| C22 | 0.84±0.021 | 0.90±0.002 | 0.23±0.012 | 1.27±0.002 | 1.48±0.040 | 0.96±0.007 | 1.16±0.005 | 1.76±0.011 | 1.25±0.011 |
| C22:1 | 1.24±0.002 | 4.84±0.002 | 0.27±0.038 | 1.01±0.051 | 1.83±0.002 | 1.16±0.003 | 1.84±0.00 | 2.25±0.012 | 1.66±0.010 |
| C22:6 | 3.56±0.209 | 5.26±0.105 | 0.95±0.001 | 4.87±0.067 | 7.64±0.031 | 3.80±.012 | 6.14±0.016 | 9.28±0.004 | 6.97±0.021 |
| C24 | 3.74±0.001 | 6.72±0.004 | 1.21±0.002 | 5.09±0.089 | 7.30±0.029 | 3.8±0.001 | 7.21±0.414 | 11.87±0.025 | 7.52±0.022 |
| C24:1 | 0.74±0.003 | 0.81±0.001 | 0.12±0.012 | 1.08±0.031 | 1.31±0.028 | 0.89±0.120 | 0.93 | 1.39±0.010 | 1.02±0.014 |
| Total | 133.84 | 248.7 | 34.7 | 214.76 | 267.32 | 140.77 | 261.06 | 365.54 | 222.25 |
The different FAs that increased or decreased in the LPAAT-overexpression were measured by GC-MS and antisense expression microalgae are shown in figure 3. The overexpression and under-expression of LPAAT gene in P tricornutum has a significant influence on the FA component of the transgenic strains. From the overall view point, it can be seen that the FA synthesis and accumulation with a chain length between C14-C20, normally considered as long chain FAs is affected by the overexpression or under expression of LPAAT gene and the degree of unsaturation increased in the exponential phase. In summary, with the overexpression of LPAAT gene, the accumulation of unsaturated long chain FA increased. There was a 4 fold increase in C22:1 content for LPAAT-overexpressing P. tricornutum, which is significantly higher and other increases were mainly for the chain length of C14 to C20 under exponential phase. Exponential phase in not the lipid accumulation phase and usually late exponential phase is where lipid accumulation starts. Upon reaching the plateau phase, mainly fatty acids of chain length C20 were increased. This result may be relevant with the report that glycolipids of eukaryotic origin contain long chain FAs mainly[26]. For antisense-expression microalgae, in exponential phase, FAs with chain length between C14 to C20 were significantly decreases, with C16 and C18 FAs mainly affected. The effect of reduced expression of LPAAT gene was less pronounced in the stationary phase. In addition, when the LPAAT gene was over expressed, the unsaturated fatty acid content was mainly increased, while the chain length of the unsaturated fatty acids increased from C16 to C20 when the culture phase in changes from exponential to stationary phase.
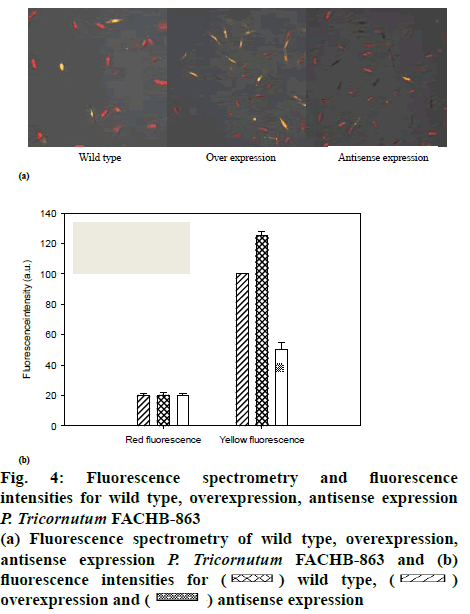
The effect of LPAAT overexpression on lipid production at the cellular level by fluorescence spectrometry (figure 4a), yellow fluorescence and red fluorescence described the cells with or without lipids, respectively. The fluorescence intensities are shown as figure 4b. There was no significant difference in gross cell morphology between wild type and over or antisense expression microalgae. However, the lipid content of LPAATexpressing cells was clearly higher in microalgal cells under plateau phase compared with wild type cells. Consistent with the hypothesis that LPAAT overexpression increases lipid production directly, LPAAT gene inhibition, led to a clear reduction in lipid production.
Figure 4: Fluorescence spectrometry and fluorescence intensities for wild type, overexpression, antisense expression P. tricornutum FACHB-863
(a) Fluorescence spectrometry of wild type, overexpression, antisense expression P. tricornutum FACHB-863 and (b) fluorescence intensities for  wild type,
wild type,  overexpression and
overexpression and  antisense expression
antisense expression
In conclusion, this study confirmed the importance of LPAAT gene in intracellular fatty acid accumulation in the diatom P. tricornutum. Overexpression of LPAAT gene in P. tricornutum resulted in overall increase in fatty acid production and the chain length increased from C14 to C22. Engineering the fatty acid biosynthesis pathway is a potential metabolic approach to enhance fatty acid accumulation in marine microalgae.
Acknowledgements
The authors gratefully acknowledge the support of the development special funds of Shenzhen strategic emerging industries and future industries (NO. 201708021308) and Shenzhen science and technology application demonstration project (NO.KJYY20180201180253571) to Shuangfei Li, Shenzhen Grant Plan for Science and Technology (NO. JCYJ20170818092708470, JCYJ20170302144706218) to Huirong Chen and Shuangfei Li, and the Natural Science Foundation of Guangdong Province (NO. 2018A030310446) to Yue Wang.
References
- Jia B, Song,Y Wu M, Lin B, Xiao K, Hu Z, et al. Characterization of long-chain acyl-CoA synthetases which stimulate secretion of fatty acids in green algae Chlamydomonas reinhardtii. Biotechnol Biofuels 2016;9:184.
- Shemesh Z, Leu S, Khozin-Goldberg I, Didi-Cohen S, Zarka A, Boussiba S. Inducible expression of Haematococcus oil globule protein in the diatom Phaeodactylum tricornutum: Association with lipid droplets and enhancement of TAG accumulation under nitrogen starvation. Algal Res 2016;18:321-31.
- Misra N, Panda PK, Parida B, Mishra BK. dEMBF: A Comprehensive Database of Enzymes of Microalgal Biofuel Feedstock. PLoS One 2016;11(1):e0146158.
- Sharma KK, Schuhmann H, Schenk PM. High Lipid Induction in Microalgae for Biodiesel Production. Energies 2012;5:1532-53.
- Liu BS, Benning C. Lipid metabolism in microalgae distinguishes itself. Current Opinion in Biotechnology 2013;24(2):300-9.
- Gong Y, Zhang J, Guo X, Wan X, Liang Z, Hu CJ, et al. Identification and characterization of PtDGAT2B, an acyltransferase of the DGAT2 acyl-coenzyme A: diacylglycerol acyltransferase family in the diatom Phaeodactylum tricornutum. FEBS Lett 2013;587(5):481-7.
- Celikten I, Koca A, Arslan MA. Comparison of performance and emissions of diesel fuel, rapeseed and soybean oil methyl esters injected at different pressures. Renew Energy 2010;35:814-20.
- Lv H, Qu G, Qi X, Lu L, Tian C, Ma Y. Transcriptome analysis of Chlamydomonas reinhardtii during the process of lipid accumulation. Genomics 2013;101(4): 229-37.
- Hsieh HJ, Su CH, Chien LJ. Accumulation of lipid production in Chlorella minutissima by triacylglycerol biosynthesis-related genes cloned from Saccharomyces cerevisiae and Yarrowia lipolytica. J Microbiol 2012;(50):526-34.
- Li Y, Han D, Hu G, Dauvillee D, Sommerfeld M, Ball S, et al. Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab Eng 2010;12(4):387-91.
- Moellering ER, Benning C. RNA interference silencing of a major lipid droplet protein affects lipid droplet size in Chlamydomonas reinhardtii. Eukaryot Cell 2010;9: 97-106.
- Radakovits R, Jinkerson RE, Fuerstenberg SI, Tae H, Settlage RE, Boore JL, et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropis gaditana. Nat Commun 2012;3:686.
- Muto M, Fukuda Y, Nemot M, Yoshino T, Matsunaga T, Tanaka T. Establishment of a genetic transformation system for the marine pennate diatom Fistulifera sp. strain JPCC DA0580-a high triglyceride producer. Mar Biotechnol (NY) 2013;15(1):48-55.
- Hildebrand M, Davis AK, Smith SR, Traller JC, Abbriano R. The place of diatoms in the biofuels industry. Biofuels 2012;3:221-40.
- Peng KT, Zheng CN, Xue J, Chen XY, Yang WD, Liu JS, et al. Delta 5 fatty acid desaturase upregulates the synthesis of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum. J Agric Food Chem 2014;62:8773-6.
- Mata TM, Martins AA, Caetano NS. Microalgae for biodiesel production and other applications: A review. Renewable & Sustainable Energy Reviews 2010;14:217-32.
- Chisti Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol 2008;26:126-31.
- Zaslavskaia LA, Lippmeier JC, Kroth PG, Grossman AR, Apt KE. Transformation of the diatom Phaeodactylum tricornutum (Bacillariophyceae) with a variety of selectable marker and reporter genes. Journal of Phycology 2000;36:379-86.
- De Riso V, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res 2009;37:96.
- Weyman PD, Beeri K, Lefebvre SC, Rivera J, McCarthy JK, Heuberger AL, et al. Inactivation of Phaeodactylum tricornutum urease gene using transcription activator-like effector nuclease-based targeted mutagenesis. Plant Biotechnol J 2015;13:460-70.
- Miyagawa A, Okami T, Kira N, Yamaguchi H, Ohnishi K, Adachi M. Research note: High efficiency transformation of the diatom Phaeodactylum tricornutum with a promoter from the diatom Cylindrotheca fusiformis. Phycological Research 2009;57:142-6.
- Nelson DM, Treguer P, Brzezinski MA, Leynaert A, Queguiner. Production and Dissolution of Biogenic Silica in the Ocean - Revised Global Estimates, Comparison with Regional Data and Relationship to Biogenic Sedimentation. GLOBAL BIOGEOCHEM CY 1995;9:359-72.
- Kang NK, Choi GG, Kim EK, Shin SE, Jeon S, Park MS, et al. Heterologous overexpression of sfCherry fluorescent protein in Nannochloropsis salina. Biotechnol Rep 2015;8;10-5.
- Kang NK, Jeon S, Kwon S, Koh HG, Shin SE, Lee B, et al. Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol Biofuels 2015;8:200.
- Radakovits R, Eduafo PM, Posewitz MC. Genetic engineering of fatty acid chain length in Phaeodactylum tricornutum. Metab Eng 2011;13:89-95.
- Yamaoka Y, Achard D, Jang S, Legeret B, Kamisuki S, Ko D, et al. Identification of a Chlamydomonas plastidial 2-lysophosphatidic acid acyltransferase and its use to engineer microalgae with increased oil content. Plant Biotechnol J 2016;14(11): 2158-67.
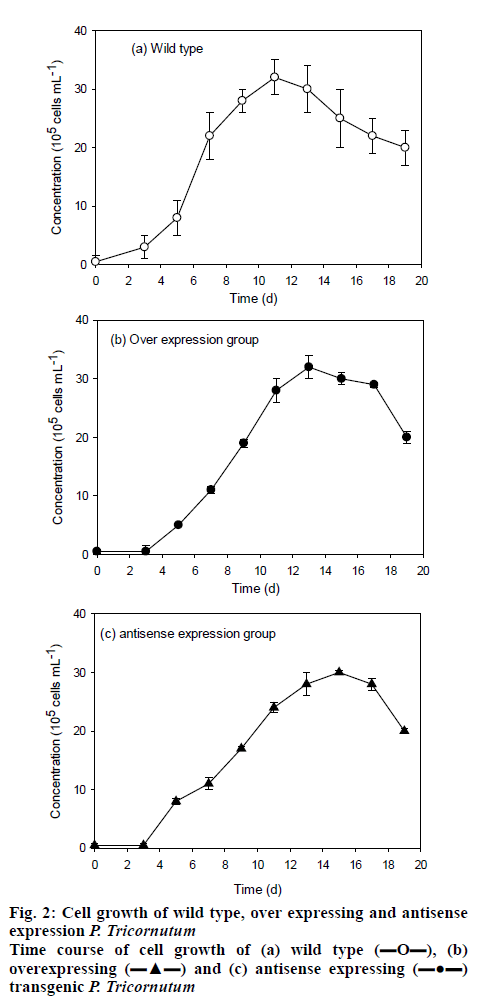
 , (b) overexpressing
, (b) overexpressing  and (c) antisense expressing
and (c) antisense expressing  transgenic P. tricornutum
transgenic P. tricornutum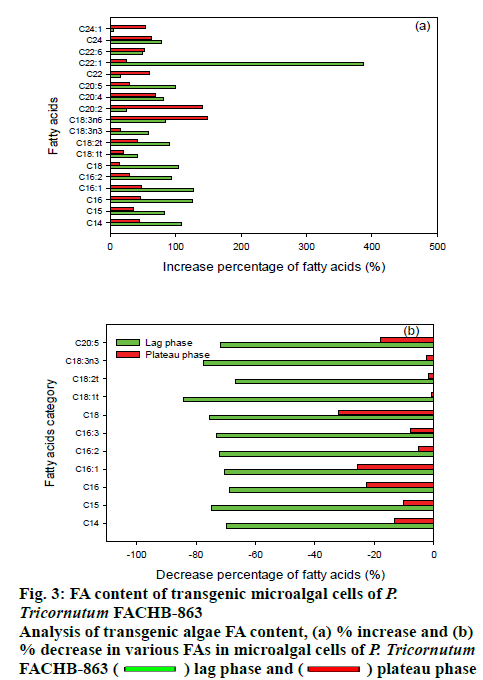
 lag phase and
lag phase and  plateau phase
plateau phase