- *Corresponding Author:
- Lakshmi Prasanthi Nori
Department of Pharmacy, Vignan College of Pharmacy, Vadlamudi, India
E-mail: prasanthi_pharm@yahoo.com
| Date of Received | 03 March 2020 |
| Date of Revision | 04 February 2021 |
| Date of Acceptance | 07 May 2021 |
| Indian J Pharm Sci 2021; 83(3):416-429 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The purpose of this article is to provide a framework for understanding the principles of personalized and precision medicine. Since the advent of the human genome project in 1990, the approach to precision and accurate medicine has been changed. This turns the practice from the “one size fits all” paradigm into one that tailors patient identification, diagnosis and treatment. Personalized medicine has been a disputed vision of the future for two decades. A humanized approach to personalized medicine will offer the possibility of leveraging the physiology of processes and its application of personalized, predictive, participatory, precision and preventive P5 medicine. The predictive variables for the progression of a disease should be investigated within communities to determine preventive measures for at risk people and healthcare can be customized and participative. Accurate diagnosis, surveillance and treatment of diseases includes developments in the identification of biomarkers, the subsequent development of reliable signatures that suit complex disease states, as well as medical approaches that can be continually modified and modulated for dosage and medication selection. This review illustrates significant breakthroughs in the advancement of emerging technology and obstacles of personalized and precise medicine. It will create groundbreaking capabilities in the realization of fully individualized patient care.
Keywords
Personalized medicine, precision medicine, genomics, biomarkers, epigenetics
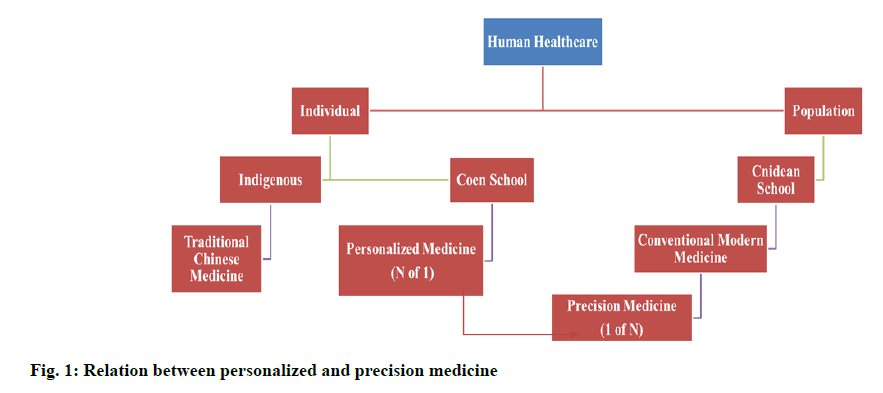
Hippocrates (approximately born in 460 BC), considered the “Father of modern medicine”, believed that disease was a product of environmental conditions, diet and lifestyle habits, the treatment should focus on patient care (prevention) and prognosis (prediction) [1]. He bickered that the human body functioned as a unified organism; it should be treated as a coherent organism. He believed that, in the diagnosis of disease both subjective patient report and objective assessment of the symptoms of the disease should be considered. It helped to found the Coan school of medicine and should be identified more aptly as the “Father of Personalized Medicine”, with a focus on disease prevention, identification, diagnosis and treatment as relating to the individual patient [2]. The ‘protocolisation’ of pharmacy has significantly weakened this tradition over time. The current modus operandi of modern medicine was predicated on the Cnidian school of medicine as shown in fig. 1 and consists of; determination of the symptoms of an individual and a related diagnosis, comparison with a statistically similar and relevant dataset or database of the population of patients, therapeutic treatment and subsequent response to that particular intervention. A particular diagnosis of illness as it applies to compartmentalized tissue and/ or an organ affecting a highly specialized clinician is also the subject. The current health care system tends to be reactive, providing post appearance treatment for the disease with limited prevention and prediction attempts. All of this show reliance on individual’s comparative analysis against a defined population which neglected and disregarded human individuality, complexity and variability. Most recently, such a medical system has been back lashed, as physicians and consumers have started to seek most personalized care. The list of projects in personalized medicine (PM) covers personalized, precise, preventive, predictive, pharmacotherapeutic and participatory treatment for patients. Modern conventional medicine, based on the population model mentioned above, aims to treat post onset disease. In contrast, PM attempts in part to identify pre-set problems of the disease and somewhat reminiscent of the Coan philosophy of medicine sponsored by Hippocrates before expression of specific clinical pathologies [3-5].
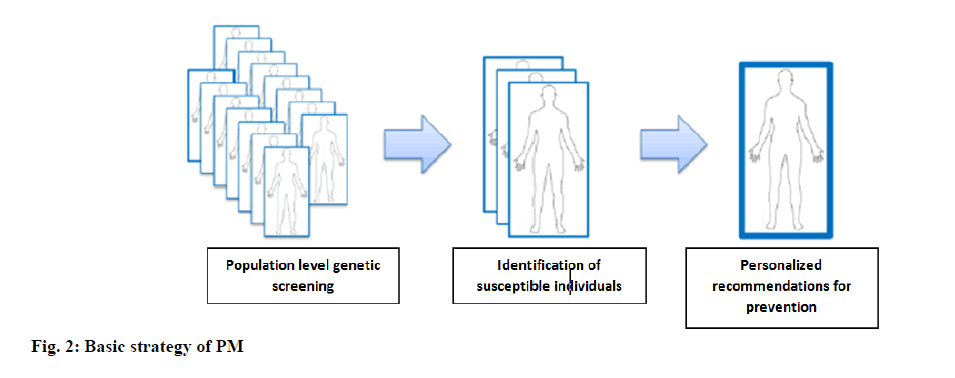
Medicine is moving away from the paradigm of “one size fits all”. Physicians know that people with the same disease will respond differently to everything, from medication to biomaterials and now it is the time to better understand this reaction and treat the patient as a person. The PM’s vision for the 21st century is to provide the right drug, the right dose to the right patient at the right time. Effective application of PM relies heavily on the availability of rigorous diagnostic tools that enable optimal therapeutic product selection to improve patient outcomes. Manufacturers and Food and Drug Administration (FDA) entities fully regulate the products. PM’s aim, according to the FDA, is to increase benefits and reduce risks for patients by more effectively targeting prevention and treatment [6]. The basic strategy of PM is depicted in fig. 2.
Milestones of PM
Hippocrates said two and a half times ago: it is far more important to know what a person’s illness is than what a person’s disease is. In 1956, Favism was found to be the genetic basis for the systematic toxicity of fava beans due to a mutation in the metabolic enzyme glucose-6-phosphate dehydrogenase (G6PD). In 1985, Renato Dulbecco discovered that sequencing of the human genome was needed to advance cancer research. In 1988 Genentech, Inc. done the sampling of whole human growth hormone (hGH) locus (a world record), showing the viability of human genome sequencing. The human genome project (HGP) was introduced in 1990 and the first draft was released in 2001, while its final version was published in 2003.
Since the early 1990s, individualized therapies personalized to each patient’s genome have been envisaged, but never performed. In 1994, laboratory developed a diagnostic test to assess the efficacy of recombinant human growth hormone (rHGH) replacement therapy, the earliest ever conceived companion molecular diagnostics (CMDx) test. It became the first licensed CMDx discovered in 1998 when the FDA approved Herceptin (Anti-epidermal growth factor receptor (EGFR) monoclonal antibodies (mAb) for EGFR+breast tumors) and Hercep Test (for detecting those tumors). Since then, the FDA has issued a growing list of test packages/customized drugs treatments that identify and prescribe them.
Personalized medicine:
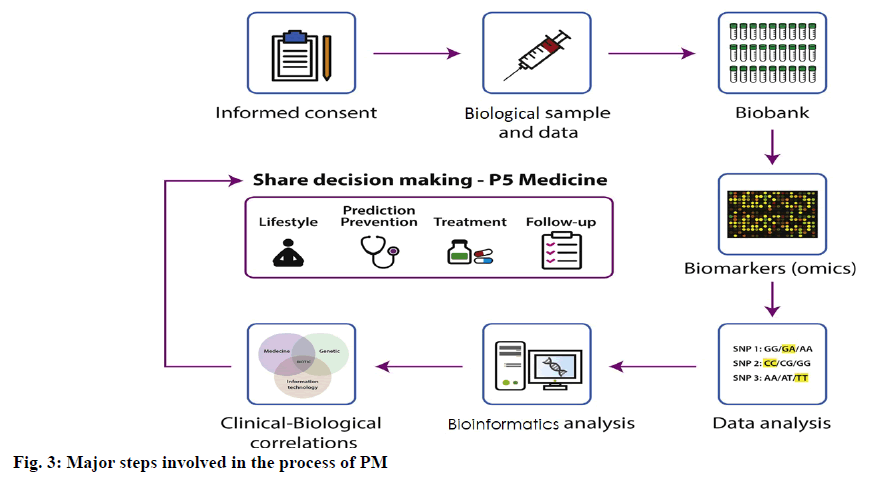
Historically, PM is originated in the early 2000’s based upon the writing of the Francis Collins, “we are witnessing a revolution in the understanding of the human genome and in the subsequent creation of a map of human genetic variation, noted this in an engaging way”. The PM Coalition (in 2004) described PM as “a developing area in which physicians use diagnostic tests to determine the medical treatments which would work best for each patient. Health care providers will create tailored treatment and prevention programs by integrating the evidence from these assessments with the medical history, conditions and beliefs of an individual”. PM comprises a broad and evolving field informed by a patient’s distinctive information and biomarker profile (i.e., clinical, genetic, genomic and epigenetic/environmental). As such, PM is committed to surveying, monitoring and diagnosing the risk of providing and presenting specific treatments to patients from their molecular and specific outline [9]. While PM terminology is changing and there are some qualitative definitions (e.g., customized, individualized, accurate), the core underlying premise is to address and reinvent medicine by using integrative biological markers (biomarkers) to manage non-illness patients as shown in fig. 3. Biomarkers are biological measures of a biological state related to a particular condition. This analysis can help to predict the risk and outcome of a disease as well as classify subtypes within a disease (i.e., sub phenotypes) and patient subgroups with same molecular disease variation. A biomarker should have the trait of being assessed and analyzed independently as a predictor of normal biological, pathogenic or pharmacological processes to a therapeutic intervention. Biomarkers can be used either alone or in conjunction to determine an individual’s state of health or disease. While many studies have identified predictive biomarkers of the reaction to biological therapies, including genomic, immunological (e.g., cytokines, phenotypes of immune cells, immunogenicity) and patient-related characteristics, just a few have been confirmed [10]. The principle characteristics of the biomarkers are highly specific to disease or effect, healthy to assess, easy to measure, low cost, low cost of follow-up research, higher value than traditional biomarkers used in clinical practice, good performance characteristics, corresponds with disease activity levels, demonstrates early and late disease change and consistent across genders and ethnic groups.
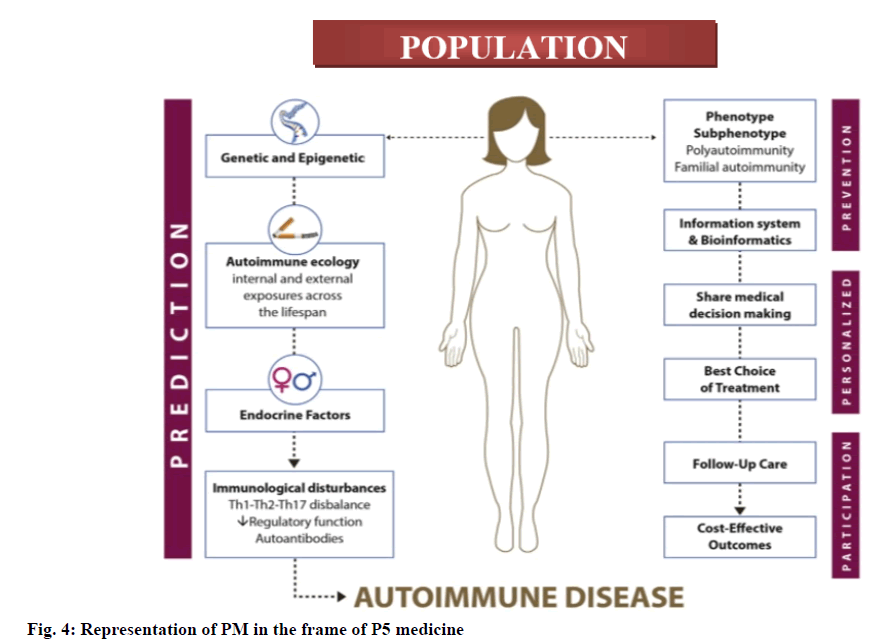
However, the fusion of the digital revolution and device solutions to health and illness is starting to lead a progressive personalized, predictive, participatory, precision and preventive (P5) medicine which is population level predictive, preventive, customized and participatory medicine, its frame work is given in fig. 4. The involvement of population is a matter of concern that is directly related to heritability (i.e., the proportion of phenotypic variation in a population attributable to genetic variation among individuals) that depends on each population, as well as cost-effective public health policies based on evidence-based outcomes that should be clinically valid and useful [11]. The purpose of PM is to recognize which intervention will be most effective on an affected individual’s disease outcome based on ecology/epigenetic ecology and their genetic and molecular landscape. This includes measurement of predisposition to disease, screening and early diagnosis, assessment of prognosis, pharmacogenomic measurements and monitoring of course of disease. All of these approaches may be capable of reducing and in some cases preventing the disease burden of provided population. Recently developed high-throughput omic technologies (i.e., genomics, transcriptomics, proteomics and metabolomics) have contributed to rapid progress in the production of data on healthy and impaired individuals, but the silver lining for interaction studies coming from these methods is far from general acceptance and given the current lack of clinical use [12]. Therefore, the evaluation of genetic testing for a particular illness should be endorsed when the presence of a high risk of disease occurs, disease relevant morbidity and mortality rates and the likelihood of meaningful intervention. The regulatory authorities controlling genomics in various countries are given in Table 1.
| S. No | Country | Authority | Role |
|---|---|---|---|
| 1 | United states of America | ||
| Generic information non-discrimination act (GINA), 2008 | It prohibits some types of genetic discrimination. | ||
| 2 | European Union | ||
| Estonia | Human gene research law, 2000 | It regulates genetic research and DNA samples, handling and procedure. | |
| Personal data protection Act, 2007 | It covers the area of protection and handling of personal data including genetic data. | ||
| Ireland | Disability act, 2005 | It governs genetic testing in persons and provides that testing shall not be carried out on a person. | |
| Data protection Act, 2018 | It provides the protection of personal data, which includes genetic data. | ||
| Sweden | Act on bio banks in Healthcare 2002:297 | Relevant to genetic research. | |
| Act on genetic integrity 2006:351 | Relevant to genetic research. | ||
| Latvia | Human genome research law, 2003 | It regulates the establishment and operation of a single genome database for genetic research and ensures the voluntary nature and confidentiality of gene donation. | |
| Iceland | Law of insurance contracts, 2004 | It prohibits companies from using results of genetic testing and any associated risks of developing certain diseases. | |
| Privacy act, 2000 | It governs the handling, storage and access of genetic information in a confidential way. | ||
| Lithuania | Law on ethics in biomedical research, 2002 | It specifies that biomedical studies are only carried out for prevention, diagnosis or treatment but not to alter the progeny genome. | |
| Netherlands | Law on medical research on humans , 1998 | It regulates the medical research on humans. | |
| Norway | Act on human medicine use of biotechnology, 2003 | It provides, how and who is allowed to access the genetic information. | |
| Finland | Medical and research act | It specifies that an ethics committee must approve the research proposals. | |
| Luxembourg | Health code in memorial A-343 | It establishes an ethics committee concerning gene therapy. | |
| France | Public health code | It concerning genetics identified in the review. | |
| Portugal | Personal genetic information and health information | It creates a comprehensive regulatory frame work for genetic research, storage of genetic information. | |
| Slovenia | Slovenia medical law, 2014 | It regulates medicines and clinical trials. | |
| Spain | Spanish health code | To regulate the procedures of genetic analysis. | |
| Italy | The code for the protection of personal data, 2003 | Genetics as personal data and persons consents are necessary when the genetic data are being used. | |
| Croatia | Protection of patient’s rights, 2004 | No interventions are allowed with the view to improve the patient’s germ line. | |
| Greece | Law 2619/1998 | The convention for the protection human rights and dignity of the human being about the application of biology and medicine. | |
| Cyprus | Bioethics law, 2001 | Analyze and evaluates the projects relating to genetics. | |
| Personal data processing act, 2001 | It covers the data protection of sensitive personal data. | ||
| Romania | Civil code, 2009 | It prohibits any medical intervention intended to modify the genetic information. | |
| Austria | Gene technology act Data protection act |
Regulates the use of gene technologies and licensing. | |
| Bulgaria | Health law, 2005 | It covers the use of genetics in health care and medical research. | |
| Hungary | XXI Law on the protection of human genetic data on the rules of human genetic testing and research and operation of bio banks, 2008 | It aims to protect people’s genetic information. It also provides how to handle the genetic data. | |
| Czech republic | Law N. 202/2017 | It relates to specific medical services. It regulates genetic testing in patients. | |
| Law N. 66/2013 | It governs the provision of medical services. | ||
| Law N. 101/2000 | It specifies the obligations on the storing or processing confidential personnel information. | ||
| Germany | Genetic testing act, 2009 | It regulates genetic testing in humans. | |
| Switzerland | Federal act on human genetic testing, 2004 | It stipulates the conditions under which human genetic testing to be performed in the medical, employment, insurance and liability contexts. | |
| 3 | Asia | ||
| Singapore | Human biomedical research act, 2015 Human cloning and other prohibited practices act, 2005 |
It regulates that every person who got genetic information or human biologic material must take all reasonable steps and safeguards to protect such information or materials against accidental or unlawful loss. | |
| China | China food and drug administration (CFDA) and National Health and Family planning commission (NHFPC) | Medical institution clinical laboratories regulation for amplification based molecular diagnostics. | |
| 4 | Middle east | ||
| United Arab Emirates | Article 2/27 of Federal law 28/2005 | Concerning the personal status. | |
| Lebanon | Law No. 625 of 2004 | Genetic variants, ethnic discrimination, genetic tests, paternity testing etc. | |
| Qatar | Banked DNA | Regulates the issues related to the human genome and gene analysis. | |
Table 1: Regulatory Authorities of Genomics in Various Countries
Precision medicine:
Clayton Christensen first coined the term “precision medicine” in his book “Innovator’s Prescription”, published in 2009. However, this descriptor did not gain widespread acceptance and use until the United States National Research Council (NRC) published a report entitled “Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and New Taxonomy of Disease” in 2011. The Institute of Precision Medicine provided an early definition which stated, “Precision medicine is targeted individualized care tailored to each patient based on their particular genetic profile and medical history. Unlike traditional medicine where one-size-fits-all, precision medicine physicians use genomic sequencing methods to query a patient’s entire genome to identify the specific genetic changes that have triggered and drive their tumor”. Francis Collins stated in 2015 that advances in data science and cost of sequencing the entire genome of an individual make it a perfect time to launch the imitative precision medicine [13]. The approach to precision medicine uses individuals and identifies (sub) population based cohorts which have a specific network of taxonomy for disease (or health). Many factors relate to the varied reactions to pharmacological diagnosis by patients or subsets of patients. Pharmaceutical potency and effectiveness depend on its association with on-target and off-target molecules but how the pharmaceutical is absorbed in the body to reach the pharmacological target depends on its absorption, distribution, metabolism and excretion (ADME). The drug-metabolizing enzymes and transporters (DMETs) are the major part of this system. Variations in drug ADME, common among a defined patient population, influence the effectiveness and/or toxicity of the treatment. In this regard inhibition or activation of the protein functions of DMETs contributes to changes in the pharmacokinetics of a medication, affecting its efficacy and/or toxicity; changes in the transcriptional gene expression of DMETs, mediated by xenobiotic receptors and transcription factors; genetic variations of the ADME genes frequently lead to significant differences in DMET expression and eventually enzyme/protein activity; epigenetics, non-coding RNAs and gut microbiota may modulate ADME gene expression and induce variations in drug metabolism and toxicity [14,15].
The terms customized medicine and precision medicine are frequently used interchangeably, but their meaning may vary. Precision medicine is characterized as refining of the taxonomy of diseases or disorders, by classification of patient subgroups predicted to respond differently to an intervention. For example, lung cancer, which has historically been defined on the basis of histology and clinical stage, can now be defined using genetic testing to identify subgroups of patients who are more likely to respond to therapies targeting a driver mutation. Precision medicine, therefore, does not mean to develop a drug for a single patient, but rather a subset of patients with a disease redefined by a more precise taxonomy. The term is defined less specifically for PM. Many consider it synonymous with precision medicine, but it is also possible to use PM to connote treatment of individual patients, rather than groups. Individual patient PM depends on many contextual factors that are independent of the underlying disease as defined by the precision medicine. These factors may include custom dosing, concomitant therapies, therapy response, drug metabolism and preferences of patients [16].
PM, using the concept of individualized therapy, provides the drug producers and regulators with significant challenges. Firstly, the multidimensional nature of PM exponentially increases the scope of drug development. Imagine running a controlled study with the multiple arms needed to test certain variables. Furthermore, a drug meant for just one user poses realistic as well as economic considerations. Precision medicine splits conventional diseases into several subpopulations, where a small incidence of the target population may be the main challenge. Design of a drug for a single patient requires a study design of N=1 which is not technically feasible. There is no competition for the medication after the first user has been screened for a drug developed, for just one patient. By comparison, individualized dosing of a drug approved for a large population has often been used by cancer, pain and diabetes management. The regulatory approach to these drugs was to give general guidance and leave the rest to medical practice. For example, the insulin product labeling includes the sentence, “the target blood glucose levels as well as the doses and timing of anti-diabetic medicinal products must be decided individually”. The same relates to medicinal products licensed for ovarian stimulation during in vitro fertilization processes, where product labeling notes that “the dosage of gonadotropins should be modified individually”. The FDA has regulated PM mainly through drug substance labeling that recognizes the importance and requires physician discretion in the management of individual patients, rather than demanding permission for each individual patient. Most people would assume that custom medication is made solely because of the term “personalized”. While the definition of Wikipedia centers on the individual patient, the Council of Presidents explicitly refers to subpopulations and the national cancer organization does not specify whether it is for the individual or on a subpopulation. For example the use of human epidermal growth factor receptor 2 (HER2)-trastuzumab, a subgroup of women with early breast cancer has a HER2 positive tumor. This subgroup is therefore qualifying for trastuzumab which is selective for tumors that are HER2 positive. All women in this subgroup are accessing trastuzumab in the same manner, many are seeing this as an indication of subgroup or stratified product. The purist may therefore conclude that most current examples of personalized medications aren’t really personalized [17].
Technologies for Precision Medicine
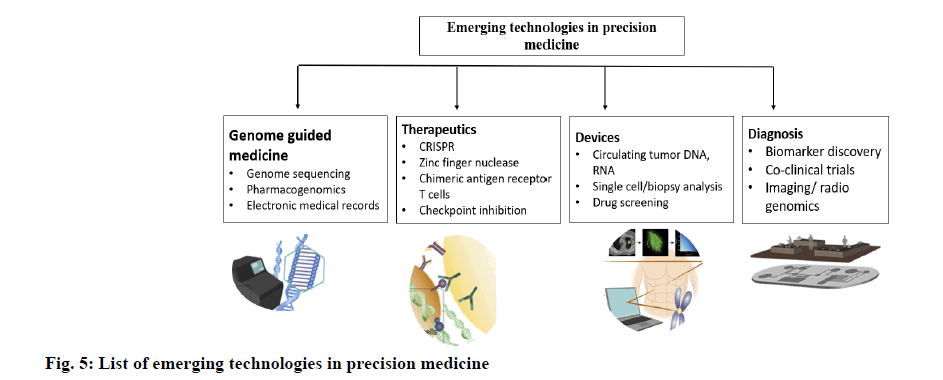
There are actually several clinical trials in place to bring precision medicine to the bedside. On review of patients with lung cancer couples genomic analysis of murine and human specimens and integrates these results with imaging research in a co-clinical trial to classify genomic signatures for optimizing liquid biopsies (NCT02597738). Patients are tested for the genotypes of cytochrome p450 (CYP) 2D6 (CYP2D6) and μ-opioid receptors (OPRM1) to monitor their exposure to opioid therapy (NCT02664350) in an objective test to treat cancer pain. Precision medicine is used to classify genomic and molecular markers to assess patient reaction to 17 α hydroxyprogesterone caproate (17OHPC) as a possible premature birth prevention drug (NCT02173210). Finally, one study integrates four breast cancer predictive genes (HER2; tumor protein 53 (TP53), checkpoint kinase 2 (CHEK2) and retinoblastoma protein (RB1)) with broad parallel genetic screening to create data sets that can be leveraged to individualize treatment regimens (NCT02624973). Nevertheless, precision medicine goes further than genomic medicine. Instead, it is a confluence of biological, physical, technological, computer and health sciences for each person toward data-driven, mechanism based health and health care [18,19]. As such, precision medicine looks at a variety of data across a population and determines the response of patients to a particular disease state and subsequent interventions on the basis of fields such as big data. It is here that innovation plays a powerful role in connecting these different ‘layers’, redefining illness and discovering clear mechanistic underpinnings that can guide therapies of the next generation. Scientists continue to contribute to precision medicine in the following ways: by facilitating the development of biomarkers, by designing testing and sensing systems and by developing new methods of distributing medicines to achieve patient precise therapies as shown in fig. 5.
Technologies for PM
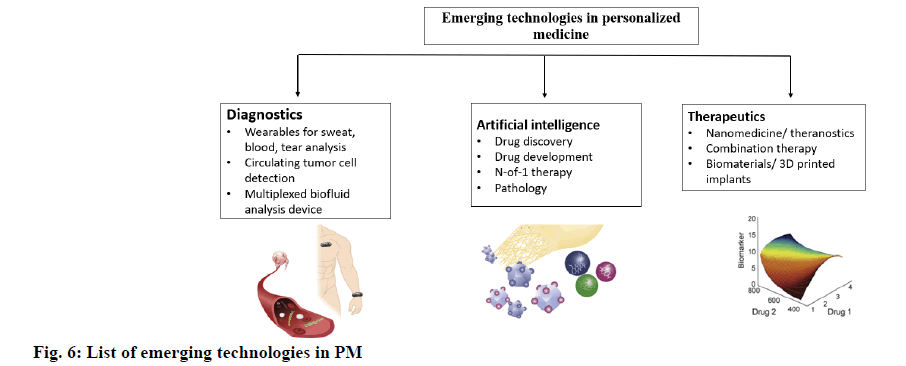
In pharmaceutical sector, developers have the opportunity to make PM a bigger reality as shown in fig. 6. Researchers have found that a dextran-dendrimer compound works differently as an adhesive not only in different organs but also in different environments in the same organ (e.g., colon cancer versus colitis) [20]. Such findings suggest that the application of most biomaterials cannot be generalized and that the identification of disease and organ condition is essential in the design of materials that are to remain temporarily within the body. Nonetheless, biomaterials can have a differential effect on cell fate, longevity and growth, but screening for the optimal material composition for a given application is missing in high-throughput approaches. One strategy is to use a mixture of small and large animal models to determine the best islet transplant polymer carrier. An approach that fully excludes animal models looks at how different formulations of the extracellular matrix (different tissues, different processing methods) influence the differentiation of stem cells, proliferation of cancer cells and apoptosis of cells. The creation of new platforms to narrow down the optimum formulation of biomaterials could improve personalized biocompatibility and therapeutic outcomes. The various emerging technologies used in the design of PM are given in Table 2.
| Technology platform | Status | Key details of study | Clinical trials. gov |
|---|---|---|---|
| Artificial intelligence | Phase II | Harnessing CURATE.AI to optimize N-of-1 combination therapy in multiple myeloma. | NCT03759093 |
| Artificial intelligence | Interventional | Developing N-of-1 training trajectories via CURATE.AI. | NCT03832101 |
| Artificial intelligence | Phase IV | HIV: Optimization of dose reduction of tenofovir (TDF) in antiretroviral therapy (ART). | NCT02632474 |
| Artificial intelligence | Phase II | Liver and Kidney transplant: Dynamic modulation of therapeutic dosing to optimize immunosuppression. | NCT03527238 |
| Artificial intelligence | Interventional | Radiotherapy: Image-guided adaptive radiotherapy is being studied to modulate intensity to reduce side effects and improve outcomes. | NCT04022018 |
| Genomic analysis/companion diagnostics | Feasibility | Non-small cell lung cancer (NSCLC): Co-clinical trials with genetic mouse models, NSCLC patient specimens (plasma, serum, tissue) and healthy patient samples to develop liquid biopsies. | NCT02597738 |
| Genome-guided therapy | Observational | Pain: Assessment of the cytochrome P450 2D6 (CYP2D6) and OPRM1 genotypes in guiding pain management. | NCT02664350 |
| Genome-guided therapy | Observational | Preterm birth: Genomic profiling to predict patient response to 17OHPC therapy to prevent preterm birth. | NCT02173210 |
| Genome-guide therapy | Phase II | Breast cancer: Biomarker analysis and genetic screening to develop individualized treatment regimens. | NCT02624973 |
| Genome-guided therapy | Phase II | Multiple tumor types: Matching of drugs to patients based on genomic alterations. | NCT02795156 |
| Genome-guided therapy | Phase III | Prostate cancer: Circulating tumor DNA is being used as a biomarker for genome-guide drug selection. | NCT03903835 |
| Microfluids/companion diagnostics | Observational | Multiple cancers types/circulating tumor cells: Correlation of titer with treatment response and progression for nasopharyngeal carcinoma, breast cancer, colorectal cancer, prostate cancer and gastric cancer. | NCT01022723 |
| Microfluids/companion diagnostics | Observational | Clear cell renal cancer/circulating tumor cells: Comparison of microfluidic technology that can detect CTCs under low epithelial cell adhesion molecule ( EpCAM) presence compared with EpCAM-based detection platform for clear cell renal cancer. | NCT02499458 |
| Microfluids/companion diagnostics | Observational | Metastatic breast cancer/circulating tumor cells: Applications of antibody cocktails to capture CTCs from metastatic breast cancer for microfluidic analysis. | NCT02904135 |
| Genome editing | Phase I | Mucopolysaccharidosis II: Intravenous deliver of SB-913, a zinc finger nuclease, to enable production of iduronate 2-sulfatase (IDS) enzyme from the liver | NCT03041324 |
| Genome editing | Phase I | Mucopolysaccharidosis I: Intravenous deliver of SB-318, a zinc finger nuclease, to insert α-L-iduronidase (IDUA) gene to enable liver-mediated enzyme production | NCT02702115 |
| Genome editing | Phase I | Severe hemophilia: Intravenous deliver of SB-FIX gene to enable factor IX clotting factor production. | NCT02695160 |
| Wearables | Observational | Perioperative risk assessment: Pairing wearable (Garmin Vivosmart HR+) with cardiopulmonary exercise testing indicators to develop broadly applicable risk assessment protocol to high risk elective surgery patient population. | NCT03328039 |
| Wearables | Observational | Chronic obstructive pulmonary disease (COPD): Application of wearable (on finger) for heart rate variability and oxygen saturation to detect and manage COPD in patients over 60 y in age. | NCT03268538 |
| Wearables | Observational | Cardiology: a shit-based electrocardiogram system is being used to monitor the following patient’s cohorts: Cardiac rehabilitation following myocardial infarct; pediatric super ventricular tachycardia; post pulmonary vein isolation; cardiac resynchronization therapy patients. | NCT03068169 |
| Digital health | Observational | Alzheimer’s disease: Cognitive battery software was used to predict dementia onset. | NCT03676881 |
| Digital health | Interventional | Health eBrain study: Application of neurocognitive assessment mobile health software to assess brain health combined with mindful intervention to address dementia and depression in Alzheimer’s disease care givers. | NCT02903862 |
| Biomaterials | Phase II | Wound healing: A nanodiamond based biomaterial platform to mediate periapical wound healing and prevention of reinfection following root canal therapy. | NCT03376984 |
| Biomaterials | Phase I | 3D printing : Personalized 3D printing of polycaprolactone (PCL) Is being explored for breast reconstruction after tumor removal. |
NCT03348293 |
| Electronic health records/infrastructure/genotyping | Observational | Pain: CYP2D6 genotyping for pain management/control and integration for pain questionnaire in electronic health record for physician guidance. | NCT02335307 |
Table 2: Emerging Technologies in PM
The main field of PM is pharmacokinetics which includes pharmacogenomics (PGx), it has provided some important information that is now extended to individual therapy. PGx is characterized as the genome-wide study of DMET genetic determinants, drug receptors and targets affecting phenotypes linked to therapeutic efficacy, health and medication. The term PGx is often used interchangeably with pharmacogenetics, which refers to the study of different single nucleotide polymorphisms (SNPs) in distinct genes with known functional plausibly linked to the drug reaction. The European Medicines Agency (EMA) and FDA have approved multiple pharmacogenomic biomarkers for clinical use. Approximately 15 % of EMA approved medical products and 138 FDA approved drugs bear pharmacogenomic marks [21,22]. Such biomarkers include variations of the germline or somatic gene (polymorphisms, mutations), functional defects with an inherited etiology, anomalies in gene expression and chromosome abnormalities. The most important biomarkers for PGx are genes that encode human leukocyte antigen (HLA) proteins, enzymes, transporters, drug targets and unique markers and mutations in somatic genome. Examination of a group of PGx markers is deemed even more important rather than providing evidence for particular drug gene pairs. Now a days, PGx analyzes are being conducted regularly during drug development process. Numerous tests are also available to evaluate the genetic makeup of a person and to predict drug reaction to facilitate the optimum range of medications and dosage. To date, numerous randomized controlled trials have given gold standard proof of the therapeutic utility of single drug gene PGx testing to direct the treatment of warfarin, acenocoumarol, phencopromone and thiopurine to guide the product collection of abacavir. In addition, many prospective cohort studies have been conducted showing clinical utility of single gene-drug PGx tests to guide the drug selection of carbamazepine and allopurinol. Therefore, cancer is one of the treatment fields where PGx is already used in clinical settings. Cetuximab, trastuzumab, imatinib and vemurafenib are just a few examples of PGx-labeled anticancer drugs [23-27].
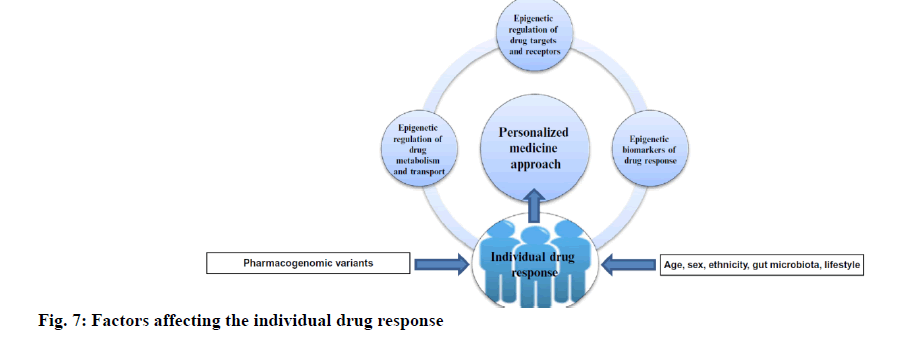
Genome-wide association studies (GWAS) in the last decade permitted the study of hundreds and thousands of genetic markers instead of candidate gene classes. It is now clear, that GWAS will not provide all the answers for any particular drug reaction phenotype and clarify all of the phenotypic differences in this regard. Currently, there are a much higher number of rare varieties of genes than previous. A research study revealed that 231 genes involved in drug metabolism and transportation were studied; it was found that each human carries over 18 000 variants and the actual number of SNPs in the gene family of CYP was more than double those reported in popular pharmacogenomic databases. Among the P450 genes, Cytochrome P450 1A2 (CYP1A2), CYP2A6, CYP2C19 and CYP3A4 have a large contribution of uncommon alleles to genetic variation. Ultimately, it can be calculated that unusual variants in ADME genes, not determined by common available PGx studies, account for 30-40 % of all genetically connected heterogeneity in drug response in a particular individual [28]. In addition, complexity/variations in phenotypes derive not only from knowledge about the genotype, but can also be affected by environmental factors and specific gene expression of the cell type. Apart from pharmacogenetics, novel biomarkers are required to predict individual responses to given drugs and develop new treatment strategies [29]. Epigenetics, which includes layers of regulatory mechanisms and information on environmental exposure in addition to the particular genome of each patient, allows better design of the personal drug regimen and better measurement of vulnerability and cure for diseases as shown in fig. 7.
It is now clear that epigenetic changes in phase I and II drug-metabolizing enzymes (DME) expressions are significant contributors to human drug response. Many epigenetic modifications affecting the expression of the DME gene include changes in deoxyribonucleic acid (DNA) methylation in 5’−C−phosphate−G−3’ (CpG) islands. In most cases the changes in DNA methylation leads to the histone modifications. In addition to these mechanisms the expression of CYP genes and phase II enzymes was found to be directly regulated by micro ribonucleic acids (miRNAs) or indirectly regulated by nuclear receptors through the binding of miRNAs. In this game the new players are long noncoding RNAs (lncRNAs). These are involved in the epigenetic regulation of coding genes by controlling up or down messenger RNAs (mRNAs), methylation and transcription of specific gene polymorphisms [30-32].
Its aberrant levels are likely to cause protein related dysregulatory disorders. However, its relationship with most diseases and DMEs has not yet been elucidated and needs further research. While DME gene expression is regulated by a variety of epigenetic modifications, including DNA methylation, histone modifications, miRNAs and lncRNAs, all of which can significantly alter drug effects [33-35].
Pharmacoepigentics in PM
Epigenetic biomarkers of drug response:
DNA methylation: It is an epigenetic biomarker with the largest translational ability in transition to PM. An irregular DNA methylation pattern is one of the hallmarks of many tumors, since changes in methylation occur early during carcinogenesis. It is stable in fixed samples over time and there are robust identification methods recently introduced by highthroughput profiling techniques such as whole genome bisulfite sequencing and CpG methylation-specific array technology. In turn, next-generation sequencing (NGS) opens the way for routine testing of biomarker panels for DNA methylation. Its profiling can be done not only in fresh frozen tissue but also in formalin-fixed or improperly preserved material. Methylguanine DNA methyltransferase (MGMT) methylation was one of the first DNA methylation biomarkers to be detected, clinically validated and reviewed. The patients taking part in clinical trials that benefit from MDxHealth’s MGMT assay, which evaluates the methylation status of the MGMT assay. This test is used in the evaluation of patients who are likely to respond to alkylating agents as a laboratory developed method, or as a research only device. This proprietary methylated gene assay is also beneficial for new developers of brain cancer drugs as they can focus their new drugs more effectively on patients who do not usually respond to the conventional treatment regime of alkylating agents [36-38]. Methylation of adenosine triphosphate (ATP)-binding cassette (ABC) drug transporters and genes encoding the proteins involved in apoptosis and signal transduction may theoretically be useful biomarkers to predict drug resistance growth, but so far no studies are known to use this method. The biomarkers of DNA methylation have tremendous potential to contribute to personalized/ precision medicine. The biomarkers with diagnostic/ prognostic capacity for DNA methylation are already being used in clinical trials or in oncology clinical applications. Much more needs to be done to develop pharmacoepigenetic biomarkers suitable for monitoring pharmacotherapy [39,40].
MicroRNA markers: For addition to DNA methylation inhibitors there are other epigenetic genes that can be used in the transition of PM. It has been shown that miRNA markers exhibit great potential as novel diagnostic and/or prognostic biomarkers, especially in clinical oncology. Different miRNAs alter [ the expression of several CYP genes and nuclear receptors. Both paraffin parts and body fluids are distinguished by temporal and spatial precision, flexibility and stability. It is claimed that automated polymerase chain reaction (PCR), miRNA microarrays and high throughput deep sequencing techniques enhance their penetration into clinics. Despite the growing number of potential miRNA biomarkers published in the literature, the transition from bench to bedside of miRNA-based biomarkers is still relatively slow, close to the DNA methylation markers. None of the suggested issues discuss how to control pharmacotherapy [41-45].
Epigenetic drugs:
It is now clear that not only the epigenetic status affect drug reaction, but also this can be modulated by medicines. Epigenetic therapy is defined as the use of drugs to treat or prevent disease-related epigenetic deficiencies and can be a step forward in the treatment of cancer and other diseases in which epigenetic regulation plays an important role. Regional hypermethylation of CpGs is accompanied by a general decrease in methylated cytosine levels (genomic hypomethylation) in cancer cells. Both hypo and hyper-methylation can promote cancer growth and are therefore ‘natural’ drug targets interfering with epigenetic machinery. In addition to cancer, epigenetics is suspected to play a major role in the pathogenesis of many other multifactorial disorders such as schizophrenia and bipolar disorder, insomnia, cardiac hypertrophy, heart failure several neurological diseases. To date, the most widely studied epidrugs have been DNA methyl transferase inhibitors (DNMTi), histone acetyl transferase inhibitors (HATi/KATi) and histone methyme [46-49].
Today the FDA has licensed two groups of epigenetic drugs for therapeutic use in the US: DNMTi and HDACi. First-class members, azacitidine (5-azacytidine or Vidaza) and its deoxyderivative decitabine (5-aza- 20-deoxycytidine), have been suggested for the diagnosis of chronic myelomonocytic leukemia and myelodysplastic syndrome. Both drugs cause large hypomethylation leading to cellular dysregulation, which impacts, in fact, rapidly dividing cells. Such drugs were essentially designed to induce genes that have been silenced in cancer, but they may also enable the expression of oncogenes and prometastatic genes, because their action is not strongly locus specific. The pluripotent stem cells (iPSCs) produced in recent years have become a highly important method of regenerative medicine [50-53]. Somatic cell reprogramming entails erasing somatic memories and creating a pluripotent state similar to that of embryonic stem cells (ESCs). iPSCs produced by ectopic expression of key transcriptional factors exhibit similar characteristics to ESCs with remarkable developmental plasticity and capacity for indefinite self-renewal, offering significant prospects for disease modeling and future clinical therapy/application. To summarize, the development of iPSC reprogramming strategies including epigenetic modifiers provides a robust forum for stem cell research and regenerative medicine studies [54]. It is now clear that during the reprogramming process, epigenetic modification and remodelling play key roles. In addition to previously defined epigenetic modifications, the recently discovered N6-methyladenosine, a retained epitranscriptomic alteration of eukaryotic mRNAs, has been shown to have a positive effect on reprogramming to pluripotence. Hence, further studies are needed for the crosstalk between RNA methylation and DNA methylation during reprogramming. With the application of small molecules/epidrugs in this process, deeper knowledge of epigenetic modification in iPSC induction and elaboration of protocols will pave the way for personal regenerative medicine/therapy [55-57]. In 2018, FDA approved 25 new molecular entities as PMs out of 59 approvals [58]. The list of approved PMs is given in Table 3.
| S. No | Brand name | Drug name | Purpose/Use |
|---|---|---|---|
| 1 | Lutathera | Lutetium Lu 77 dotatate | Gastroenteropancreatic neuroendocrine tumors |
| 2 | Biktarvy | Bictegravir/emtricitabine/tenofovir alafenamide | HIV 1 infection |
| 3 | Symdeko | Tezacaftor, ivacaftor | Cystic fibrosis |
| 4 | Trogarzo | Ibalizumab-uiyk | HIV 1 infection |
| 5 | Crysvita | Burosumab-twza | x-linked hypophosphatemia |
| 6 | Doptelet | Avatrombopag and lusutrombopag | Thrombocytopenia in patients with chronic liver diseases |
| 7 | Palynziq | Pegvaliase-pqpz | Phenylketonuria |
| 8 | Mektovi | Binimetinib | Metastatic melanoma |
| 9 | Braftovi | Encorafenib | Metastatic melanoma |
| 10 | Tibsova | Ivosidenib | Refractory acute myeloid leukemia |
| 11 | Krintafel | Tafenoquine | Plasmodium vivax malaria |
| 12 | Mulpletaf | Lusutrombopag | Thromobocytopenia in chronic liver disease |
| 13 | Onpattro | Patisiran | Polyneuropathy of hereditary transthyretin-mediated amyloidosis |
| 14 | Galafold | Migalastat | Fabry disease |
| 15 | Takhzyro | Lanadelumab-flyo | Type I and II hereditary angioedema |
| 16 | Pifeltro | Doravirine | HIV 1 infection |
| 17 | Vizimpro | Dacomitinib | Advanced non-small cell lung cancer (NSCLC) |
| 18 | Libtayo | Cemiplimab-rwlc | Cutaneous squamous cell carcinoma (CSCC) |
| 19 | Revcovi | Elapegademase-lvlr | Adenosine deaminase severe combined immune deficiency (ADA-SCID) |
| 20 | Tegsedi | Inotersen | Polyneuropathy of hereditary transthyretin-mediated amyloidosis |
| 21 | Talzenna | Talazoparib | Advanced breast cancer |
| 22 | Lorbrena | Lorlatinib | NSCLC |
| 23 | Vitrakvi | Larotrectinib | Solid tumor cancer |
| 24 | Firdapse | Amifampridine | Lambert-Eaton Myasthenic |
| 25 | Xospata | Gilteritinib | Refractory acute myeloid leukemia (AML) |
Table 3: FDA Approved PMs
Conclusion
PM has the ability to fulfill the patient benefit demand by reducing healthcare costs, drug development costs and time. At a time when initiatives such as the HGP started and the technology to personalize medication would continue to advance in unimaginable ways, the word PM got new definition. Growth in the new era of pharmacoepigenic biomarkers and drug target will certainly be important to optimize a true personalized/ precision medicine for many diseases by genetically assisted drug therapy. The concept of PM is using a person’s combined knowledge to determine illness, risk prognosis or patient reaction and thereby improving the health of people. Using the customized method in the future, each patient should obtain their full genetic details on the day of their birth to keep them in an electronic medical record. Such knowledge will allow physicians and practitioners to introduce some more efficient solutions to health care focused on patient sensitivity to various diseases.
Acknowledgements:
The authors are thankful to Vignan’s foundation for science technology and research, Guntur for providing the necessary facilities.
Conflicts of Interest:
The authors declared no conflict of interest.
References
- Lonie IM. Cos versus Cnidus and the historians: Part I. Hist Sci 1978;16(1):42-75.
- Lonie IM. Cos versus Cnidus and the historians: Part 2. Hist Sci 1978;16(2):77-92.
- Collins FS. Personalized medicine: a new approach to staying well. The Boston Globe; 2005.
- Naylor S. What’s in a name? The evolution of ‘P-Medicine’. J Precis Med 2015;1:15-29.
- Hood L, Flores M. A personal view on systems medicine and the emergence of proactive P4 medicine: predictive, preventive, personalized and participatory. N Biotechnol 2012;29(6):613-24.
- U.S. Food and Drug Administration. Paving the way for personalized medicine: FDA’s role in a new era of medical product development; 2013.
- Redekop WK, Mladsi D. The faces of personalized medicine: a framework for understanding its meaning and scope. Value Health 2013;16(6):S4-9.
- Personalized Medicine 101: The Story. Personalized medicine Coalition; 2015.
- Jain KK. Personalized medicine. Curr Opin Mol Ther 2002;4(6):548-58.
- Personalized Medicine Timeline. http://www.personalizedmedicinetimeline.com
- Heyn H, Mendez-Gonzalez J, Esteller M. Epigenetic profiling joins personalized cancer medicine. Expert Rev Mol Diagn 2013;13(5):473-9.
- Sykiotis GP, Kalliolias GD, Papavassiliou AG. Pharmacogenetic principles in the Hippocratic writings. J Clin Pharmacol 2005;45(11):1218-20.
- March R, Schott C. Personalized/Precision Medicine/Personalised Healthcare: the art of giving different names to the same thing. Per Med 2017;14(6):463-6.
- US National Research Council. Toward precision medicine: Building a knowledge network for biomedical research and a new taxonomy of disease. Washington, DC: National Academic Press; 2011.
- Collins FS, Varmus HA. New initiative on precision medicine N Engl J Med 372:793-5.
- Perez B. China’s ‘precision medicine’ initiative gets lift from latest genomics company funding. South China Morning Post; 2017.
- Yu AM, Ingelman-Sundberg M, Cherrington NJ, Aleksunes LM, Zanger UM, Xie W, et al. Regulation of drug metabolism and toxicity by multiple factors of genetics, epigenetics, lncRNAs, gut microbiota and diseases: a meeting report of the 21st International Symposium on Microsomes and Drug Oxidations (MDO). Acta Pharm Sin B 2017;7(2):241-8.
- Aronson SJ, Rehm HL. Building the foundation for genomics in precision medicine. Nature 2015;526:336-42.
- Rehm HL, Berg JS, Brooks LD, Bustamante CD, Evans JP, Landrum MJ, et al. ClinGen-The clinical genome resource. N Engl J Med 2015;372:2235-42.
- Oliva N, Carcole M, Beckerman M, Seliktar S, Hayward A, Stanley J, et al. Regulation of dendrimer/dextran material performance by altered tissue microenvironment in inflammation and neoplasia. Sci Transl Med 2015;7:272-91.
- Kronfol MM, Dozmorov MG, Huang R, Slattum PW, McClay JL. The role of epigenomics in personalized medicine. Expert Rev Precis Med Drug Dev 2017;2(1):33-45.
- Van der Wouden CH, Cambon‐Thomsen A, Cecchin E, Cheung KC, Davila‐Fajardo CL, Deneer VH, et al. Implementing pharmacogenomics in Europe: design and implementation strategy of the ubiquitous pharmacogenomics consortium. Clin Pharmacol Ther 2017;101(3):341-58.
- Ventola CL. Role of pharmacogenomic biomarkers in predicting and improving drug response: part 1: the clinical significance of pharmacogenetic variants. Pharm Ther 2013;38(9):545-60.
- Liou SY, Stringer F, Hirayama M. The impact of pharmacogenomics research on drug development. Drug Metab Pharmacokinet 2011;27(1):2-8.
- Moaddeb J, Haga SB. Pharmacogenetic testing: current evidence of clinical utility. Ther Adv Drug Saf 2013;4(4):155-69.
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature 2007;447:433-40.
- Ivanov M, Barragan I, Ingelman-Sundberg M. Epigenetic mechanisms of importance for drug treatment. Trends Pharmacol Sci 2014;35(8):384-96.
- Kim IW, Han N, Burckart GJ, Oh JM. Epigenetic changes in gene expression for drug‐metabolizing enzymes and transporters. Pharmacotherapy 2014;34(2):140-50.
- He JH, Han ZP, Li YG. Association between long non‑coding RNA and human rare diseases. Biomed Rep 2014;2(1):19-23.
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009;10(3):155-9.
- Majchrzak-Celinska A, Paluszczak J, Kleszcz R, Magiera M, Barciszewska AM, Nowak S, et al. Detection of MGMT, RASSF1A, p15INK4B and p14ARF promoter methylation in circulating tumor-derived DNA of central nervous system cancer patients. J Appl Genet 2013;54(3):335-44.
- Majchrzak-Celinska A, Kosicka K, Paczkowska J, Glowka FK, Breborowicz GH, Krzyscin M, et al. HSD11B2, RUNX3 and LINE-1 methylation in placental DNA of hypertensive disorders of pregnancy patients. Reprod Sci 2017;24(11):1520-31.
- Zeng H, He B, Yi C, Peng J. Liquid biopsies: DNA methylation analyses in circulating cell-free DNA. J Genet Genomics 2018;45(4):185-92.
- Noehammer C, Pulverer W, Hassler MR, Hofner M, Wielscher M, Vierlinger K, et al. Strategies for validation and testing of DNA methylation biomarkers. Epigenomics 2014;6(6):603-22.
- Mikeska T, Craig JM. DNA methylation biomarkers: cancer and beyond. Genes 2014;5(3):821-64.
- Mikeska T, Bock C, Do H, Dobrovic A. DNA methylation biomarkers in cancer: progress towards clinical implementation. Expert Rev Mol Diagn 2012;12(5):473-87.
- Rieger JK, Klein K, Winter S, Zanger UM. Expression variability of absorption, distribution, metabolism, excretion–related microRNAs in human liver: influence of nongenetic factors and association with gene expression. Drug Metab Dispos 2013;41(10):1752-62.
- Lithwick-Yanai G, Dromi N, Shtabsky A, Morgenstern S, Strenov Y, Feinmesser M, et al. Multicentre validation of a microRNA-based assay for diagnosing indeterminate thyroid nodules utilising fine needle aspirate smears. J Clin Pathol 2017;70(6):500-7.
- Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int 2015;2015.
- Baer-Dubowska W, Majchrzak-Celinska A, Cichocki M. Pharmocoepigenetics: a new approach to predicting individual drug responses and targeting new drugs. Pharmacol Rep 2011;63(2):293-304.
- Salarinia R, Sahebkar A, Peyvandi M, Reza Mirzaei H, Reza Jaafari M, Matbou Riahi M, et al. Epi-drugs and Epi-miRs: moving beyond current cancer therapies. Curr Cancer Drug Targets 2016;16(9):773-88.
- Broderick JA, Zamore PD. MicroRNA therapeutics. Gene Ther 2011;18(12):1104-10.
- Ginsburg GS, Willard HF. Genomic and personalized medicine: foundations and applications. Transl Res 2009;154(6):277-87.
- Hood L. A personal journey of discovery: developing technology and changing biology. Annu Rev Anal Chem 2008;1:1-43.
- Ross JS, Ginsburg GS. The integration of molecular diagnostics with therapeutics: implications for drug development and pathology practice. Am J Clin Pathol 2003;119(1):26-36.
- Tobon GJ, Pers JO, Canas CA, Rojas-Villarraga A, Youinou P, Anaya JM. Are autoimmune diseases predictable? Autoimmun Rev 2012;11(4):259-66.
- Auffray C, Charron D, Hood L. Predictive, preventive, personalized and participatory medicine: back to the future. Genome Med 2010;2:57.
- Khoury MJ, Gwinn M, Glasgow RE, Kramer BS. A population perspective on how personalized medicine can improve health. Am J Prev Med 2012;42(6):639-45.
- Pirmohamed M. Acceptance of biomarker‐based tests for application in clinical practice: criteria and obstacles. Clin Pharmacol Ther 2010;88(6):862-6.
- Parikh NI, Vasan RS. Assessing the clinical utility of biomarkers in medicine. Biomark Med 2007;1(3):419-36.
- Tektonidou MG, Ward MM. Validity of clinical associations of biomarkers in translational research studies: the case of systemic autoimmune diseases. Arthritis Res Ther 2010;12(5):1-10.
- Hawgood S, Hook-Barnard IG, O’Brien TC, Yamamoto KR. Precision medicine: beyond the inflection point. Sci Transl Med 2015;7(300):300-17.
- Personalized medicine at FDA. A progress and outlook report by personalized medicine coalition; 2018.
- Zhao Z, Ukidve A, Dasgupta A, Mitragotri S. Transdermal immunomodulation: Principles, advances and perspectives. Adv Drug Deliv Rev 2018;127:3-19.
- Ramirez JE, Sharpe LA, Peppas NA. Current state and challenges in developing oral vaccines. Adv Drug Deliv Rev 2017;114:116-31.
- Yeo DC, Wiraja C, Paller AS, Mirkin CA, Xu C. Abnormal scar identification with spherical-nucleic-acid technology. Nat Biomed Eng 2018;2(4):227-38.
- Artzi N, Shazly T, Baker AB, Bon A, Edelman ER. Aldehyde‐amine chemistry enables modulated biosealants with tissue‐specific adhesion. Adv Mater 2009;21:3399-403.
- Kordou Z, Siamoglou S, Patrinos GP. Legal aspects of genomic and personalized medicine. Applied Genomics and Public Health. Academic Press; 2020. p. 259-74.