- *Corresponding Author:
- M. A. Zarka
Department of Pharmacognosy and Phytochemistry,
College of Pharmacy,
The Islamic University,
Al Najaf 54001,
Iraq
E-mail: Dr.m.abdulaal84@gmail.com
| Date of Received | 03 June 2020 |
| Date of Revision | 08 March 2021 |
| Date of Acceptance | 01 July 2021 |
| Indian J Pharm Sci 2021;83(4):648-662 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The hydro distilled essential oil obtained from Calothamnus quadrifidus leaves was analyzed by gas liquid chromatography-mass spectroscopy. Quantification of the oil constituents was carried out using the gas liquid chromatography-flame ionization detection device where 1,8-cineole was quantified as 525.62 μg/ml. The oil showed potential antioxidant activity when tested by 1,1-diphenyl-2-picrylhydrazine radical scavenging assay where the concentration required to scavenge 1,1-diphenyl-2-picrylhydrazine by 50 %=37.3 μg. The cytotoxic activity of the essential oil was evaluated using 3-(4,5-dimethylthiazol- 2-yl)-2,5-diphenyltetrazolium bromide assay compared with 1,8-cineole and doxorubicin. Calothamnus quadrifidus leaves essential oil cytotoxic activity was significant against human lung cell line A-549 (2.37 μg), human colon cell line HCT-116 (2.39 μg), human hepato-cellular cell line HepG-2 (2.55 μg) and intestinal epithelium carcinoma Caco-2 (5.80 μg) cell lines, respectively. The oil cytotoxicity exceeded 1,8-cineole except for intestinal epithelium carcinoma Caco-2. Additionally, the antimicrobial activity was evaluated using agar disc diffusion and minimum inhibitory concentration where the oil exhibited the best activity against Penicillium italicum and Geotrichum candidum with minimum inhibitory concentration=0.49 and 0.98 μg, respectively. Structure elucidation of the pure compounds isolated from Calothamnus quadrifidus leaves was done according to their chromatographic behavior, chemical and spectroscopic data. These compounds were isolated for the first time from Calothamnus quadrifidus leaves and identified as gallic acid, gallic acid methyl ester, 7,4`-dimethoxy-apigenin, 6,7-dimethoxy-apigenin, (Bis[3(2-methyl propan-1- one)-2, 6 dihydroxy-4- methoxy phenyl] methane, quercetin 3-O-beta-D-arbinopyranoside and quercetin 3-O-beta-D-4C1-xylopyranoside. In addition to the molecular docking of novel compound 3 was done in peroxiredoxins and the binding mode was found. Conformation 2 was found to be the most stable and may be responsible for the antioxidant effect as it showed the best free binding energy and affinity when compared to conformations.
Keywords
Anticancer, cytotoxicity, essential oils, 1,8-Cineole, phenolic, flavonoids, Calothamnus quadrifidus, molecular docking
Calothamnus in Greek means beautiful brush [1]. Calothamnus genus is a large group of mostly red-flowered shrubs indigenous to Western Australia. The genus Calothamnus comprises 25 species in Australia mainly and as an ornament in California [2]. Few reports about the biological activity of Calothamnus quadrifidus (C. quadrifidus) leaves are available. C. quadrifidus leaves aqueous methanolic extract (AME) reduced blood glucose levels when administered to streptozotocin-induced diabetic rats [3]. C. quadrifidus leaves AME also showed antioxidant potential when compared to ascorbic acid [3]. Moreover, it was proved that essential oil (EO) showed antifungal and antibacterial activities against different microorganisms [4]. The AME revealed significant inhibition of oedema formation as compared to saline-treated groups [3].
Few reports were conducted concerning the previously isolated compounds from Calothamnus species. Flavonoids and phenolic acids were isolated from C. quadrifidusleaves [4,5]. Kaempferol, kaempferol 3-O-beta(β)-D-glucuronide, kaempferol 3-O-alpha(α)-L-rhamnopyranoside, naringenin, quercetin, quercetin 3-O-β-D-glucopyranoside, quercetin 3-O-β-D-galactopyranoside, quercetin 3-O-α-L-rhamnopyranoside, quercetin 3-O-β-D-glucuronide, quercetin 3-O-β-D-glucuronide-3'-O-sulphate, luteolin and luteolin 7-O-β-D-glucopyranoside were reported from C. quadrifidus leaves [4,5].
Concerning phenolic acids which are identified in C. quadrifidus leaves were protocatechuic acid, ellagic acid, ellagic acid 3-O-methyl ether, ellagic acid 4-O-methyl ether, rosmarinic acid, gallic acid and ferulic acid were previously isolated [4,5]. Moreover, EO of C. quadrifidus leaves was reported to be rich in 1,8-cineole (79.95 %), terpinene-4-ol (1.13 %) with other minor constituents [6]. Antifungal and antibacterial activities of the EO against different microorganisms were previously estimated [6].
Materials and Methods
Instruments
The nuclear magnetic resonance (NMR) spectral data were measured using Bruker (400 and 100 MHz) for 1H and 13C NMR, heteronuclear multiple bond correlation and heteronuclear single quantum coherence spectroscopy respectively. The results were reported as δ ppm values relative to tetramethylsilane as an internal reference.
For column chromatography, polyamide S (FlukaChemie AG, Switzerland), Sephadex LH-20 (Sigma-Aldrich Steinheim, Germany) and microcrystalline cellulose (E. Merck-Darmstadt, Germany) were used. Whatman No.1 (Whatman Ltd., Maidstone, Kent, England) was used for paper chromatography. Ferric chloride (FeCl3) spray reagent was used for visualization of tannins compounds, while Naturstoff reagent and aluminum chloride were used for flavonoids under ultra-violet (UV) light. N-Butanol/acetic acid/water (n-BuOH/HOAc/H2O); 4:1:5 v/v/v, top layer (S1), 15 % aqueous HOAc (S2) and methylene chloride/methanol (MeOH); 9:1 (S3) were used as solvent systems for determination of Rf values.
Shimadzu, UV visible 1800 was used for UV spectroscopic analysis of the isolated compounds in methanol and different diagnostic shift reagents (200-450 nm). Compounds molecular weight was estimated using high pressure liquid chromatography coupled with photodiode array detector and electrospray ionization mass spectroscopic analysis (HPLC-PDA-ESI-MS) using a high-performance liquid chromatography (HPLC) Waters Corporation, Milford MA01757 U.S.A connected to Xevo family of tandem quadrupole (Xevo TQD) triple quadrupole mass spectrometry. Sample (10 μl) was injected into ACQUITY UPLC BEH C18 Column (130Å, 1.7 µm and 2.1 mm×50 mm). A binary gradient of 1 % formic acid in deionized water (solvent A) and methanol containing 0.1 % formic acid (solvent B) was as follows: 50 % solvent (A): 50 % solvent (B). The flow rate was 1 ml/min. The mass spectrometer was fitted to an atmospheric pressure electrospray ionization (ESI) source, operated in negative ion mode. The electrospray capillary voltage was set to 3250 V, with a nebulizing gas flow rate of 9 L/h and a drying gas temperature of 450°. Mass spectrometry data were acquired in the scan mode (mass range m/z 100-1450). The instrument was controlled by Xcalibur software (XcaliburTM2.0.7, Thermo Scientific). Microplate reader SunriseTM, TECAN, Inc, USA was used to determine the number of viable cells in the cytotoxicity assay.
Materials:
C. quadrifidus R.Br. leaves were collected from a local supplier at Saft El-Laban area during the flowering stage in September 2016. A Voucher specimen has been deposited at Orman botanical garden (No. 000105CC @ 05-01-05-01). Identification of the plant material was done by Trease Labib, former head specialist of plant taxonomy at Orman botanical garden, Giza, Egypt. The collected plant materials were air dried in the shade, powdered and it was kept in well closed containers till use.
Human cell lines of colon (HCT-116), hepato-cellular (HepG-2), lung (A-549) and intestinal epithelium carcinoma (Caco-2) were obtained from the American type culture collection (ATCC, Rockville, MD). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was obtained from Sigma-Aldrich, Germany. The cells were grown on Roswell Park Memorial Institute Medium (RPMI-1640) medium supplemented with 10 % inactivated fetal calf serum and 50 µg/ml gentamycin.
Staphylococcus aureus(RCMB 010028), Bacillus subtitles (RCMB 010067), Escherichia coli (RCMB 010052), Pseudomonas aeruginosa (RCMB 010043), Aspergillus fumigatus (RCMB 02568), Penicillium italicum (RCMB 03924) and Geotrichum candidum (RCMB 05097) strains were obtained from the Regional Center for Mycology and Biotechnology, Al-Azhar University, Cairo-Egypt. 1,1-diphenyl-2-picrylhydrazine (DPPH) were obtained from Sigma Chemical Company, USA. Ascorbic acid was obtained from El Nasr Pharmaceutical Co., Cairo, Egypt.
Extraction and isolation:
Essential oil: EO was obtained by hydrodistillation of fresh C. quadrifidus leaves (500 g) for 6 h using a Clevenger type apparatus. Both oils were dried over anhydrous sodium sulfate and kept in sealed vials at 2° for analyses [7].
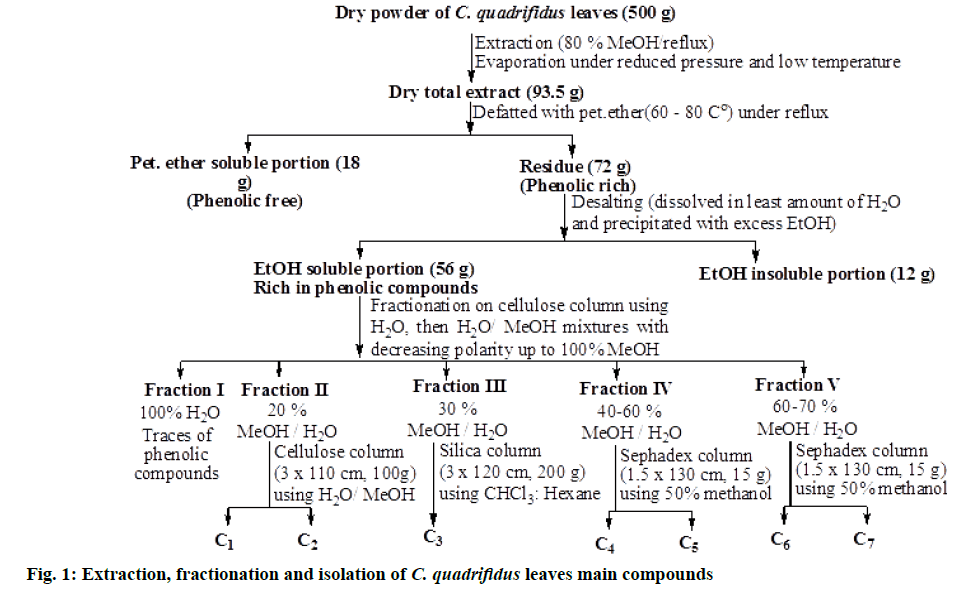
Extraction, fractionation and chromatographic investigation of C. quadrifidus R.Br. leaves: Air-dried C. quadrifidus leaves powder (500 g) was subjected to exhaustive extraction with hot 80 % aqueous methanol under reflux (2 L×6, 70°, 4 h). Evaporation of the solvent yielded a total dry extract of 93.5 g. It was defatted with petroleum ether (60-80°) under reflux (1 L×3, 60°) yielding 18 g of phenolic free residue based on 2D-PC using S1 and S2 solvent systems followed by spraying with different spray reagents (FeCl3 and Naturstoff reagent). Excess sugars and salts were desalted with excess ethanol to yield 56 g dry extract. About 19 g of the dry residue was suspended in water and fractionated on a cellulose column (100×3 cm, 150 g) packed with water, then gradient H2O/MeOH mixtures with decreasing polarity for elution. The bands on columns were traced using UV light and eluted fractions were examined by paper chromatography (PC) and thin layer chromatography (TLC) silica plates using different solvent systems and spray reagents. Similar fractions were combined to afford eight main collective fractions that were subjected separately to repeated columns chromatography on Sephadex LH-20, silica and/or cellulose and eluted with the suitable solvent system in each case for final purification.
Fraction I was eluted from the column by water, traces of polyphenols were found in this fraction on the basis of its 2D-PC and using different spray reagents.
Fraction II was eluted with 20 % MeOH in H2O. Its 2D-PC showed two shine violet spots under UV-light (λmax 254 nm) and faint blue colour with FeCl3 spray reagent. Separation of the two compounds was carried out on cellulose column (3×110 cm, 100 g) using H2O/MeOH mixtures to give two sub-fractions where each one was further purified on Sephadex column (1.5×130 cm, 15 g) using 50 % methanol which led to the isolation of chromatographically pure samples of compounds 1 and 2.
Fraction III was washed out from the column using 30 % MeOH in H2O. TLC was carried out using S3 solvent system and showed one major shiny blue spot along with other minor spots when visualized by UV light at λmax 254. Separation of 3 was carried out on silica column (3×120 cm, 200 g) using chloroform (CHCl3):hexane mixtures to give a pure sample of compound 3 (fig. 1).
TLC of fraction IV (eluted with 40-60 % MeOH/H2O) using CHCl3: MeOH (7:3 v/v) solvent system showed mainly two faint orange-yellow spots under UV light at λmax 254 which changed to yellow color on exposure to Ammonia (NH3) vapour. Further purification of the two compounds was carried out on a Sephadex column (1.5×130 cm, 15 g) using 50 % methanol. The crude of the two compounds mixture was concentrated and named compounds 4 and 5.
TLC of fraction V (eluted with 70-80 % MeOH/H2O) using S5 solvent system showed mainly two faint orange-yellow spots under UV light at λmax 254 which changed to yellow colour on exposure to NH3 vapour.
Further purification of the two compounds was carried out on a sephadex column (1.5×130 cm, 15 g) using 50 % methanol. The two compounds mixture was concentrated and named 6 and 7.
Biological activities:
Antioxidant activity using DPPH radical scavenging activity: Freshly prepared (0.004 % w/v) methanol solution of DPPH radical was used to evaluate antioxidant activity. 0.1 ml of EO in different concentrations (8-100 mg/ml) was added to 3 ml of 0.004 % methanol solution of DPPH [8]. The decrease in absorbance was measured at 515 nm [9]. EO of C. quadrifidus leaves (2-320 µg/ml) was treated similarly. The absorbance of the DPPH radical without antioxidant (control) and the reference compound ascorbic acid (5-80 µg/ml) were also measured. All the determinations were performed in three replicates and averaged. The percentage inhibition (PI) of the DPPH radical was calculated according to the formula:
PI=(AC-AT)/AC×100
Where AC=Absorbance of the control at t=0 min and AT=Absorbance of the sample+DPPH at t=16 min.
The percentage of DPPH radical-scavenging was plotted against each applied concentration of the threetested sample and ascorbic acid to determine SC50 (the concentration required to scavenge DPPH by 50 %).
Cytotoxic activity: EO of C. quadrifidus leaves, an authentic sample of 1,8-cineole and doxorubicin as a positive control (0-50 μg/ml) were evaluated for their cytotoxic activity against different human carcinoma cell lines including HCT-116, HepG-2, A-549 and Caco-2 using MTT assay [10,11]. The optical density was measured at 590 nm with the microplate reader (SunsiseTM, TECAN, Inc, USA) to determine the number of viable cells.
Percentage of viability= [1-(ODt/ODc)]×100 %
ODt is the mean optical density of wells treated with the tested oil; ODc is the mean optical density of untreated cells. The relation between cell viability percentage and concentration was plotted to get the survival curve for each tumor cell line. The 50 % inhibitory concentration (IC50) was estimated from graphic plots of the dose-response curve for each concentration.
Antimicrobial activity of C. quadrifidus R.Br. leaf oil: The antimicrobial activity was evaluated using the agar disc diffusion technique [8]. The used microorganism strains were Staphylococcus aureus (RCMB 010028) and Bacillus subtitles (RCMB 010067) as Gram-positive bacteria while Escherichia coli (RCMB 010052) and Pseudomonas aeruginosa (RCMB 010043) represented Gram-negative bacteria. The tested fungi were Aspergillus fumigatus (RCMB 02568), Penicillium italicum (RCMB 03924) and Geotrichum candidum (RCMB 05097). All organisms were obtained from the Regional Center for Mycology and Biotechnology, Al-Azhar University, Cairo, Egypt. C. quadrifidus leaves oil was dissolved in methanol at a concentration of 0.2 mg/ml where 100 µl was spotted on filter paper disc, air-dried and deposited on the surface of inoculated agar plates. The plates were kept at low temperature before incubation which favors the swelling and diffusion over microbial growth.
Experiments were performed in triplicate and diameters of inhibition zones were measured after 24 h for bacteria and 48 h for fungi. The antimicrobial activity was expressed as the inhibition zone diameter (mm). Amphotericin B was used as antifungal, while ampicillin and gentamycin were used as antibacteria controls (10 μg/ml). The percentage inhibition of diameter growth was calculated as following [12].
Percentage of inhibition=Inhibition zone diameter of sample/Inhibition zone diameter of standard×100
Molecular docking: Molecular docking studies were carried out using molecular operating environment ((MOE), 10.2008) software. All structure minimizations were performed with MOE until an root mean square deviation (RMSD) gradient of 0.1.kcal?mol-1Å-1 with Merck molecular force field (MMFF94x) force field and the partial charges were automatically calculated [13].
Results and Discussion
Both compound 1 (45 mg) and compound 2 (27 mg) are obtained as an off-white amorphous powder. They gave shine violet fluorescent spots under short UV light and blue colour with FeCl3 spray reagent. Rf values are 0.74 (S1) and 0.50 (S2) for compound 1 and 0.68 (S1) and 0.59 (S2) for compound 2. The identification was based upon cobalt-polycarbonate (Co-PC) with available authentic and comparison of spectroscopic data with the matched literature [14]. The two compounds named compound 1 and 2 were identified as gallic acid and its methyl ester, respectively.
Compound 3, isolated as a creamy white amorphous powder (15 mg). TLC properties: Rf-value is 0.84 (S3). It gave a shiny blue fluorescent spot under UV light (λmax 360 nm). The Electron ionization-mass spectrometry (EI-MS) spectrum of compound 3 showed the following fragments at m/z 432.08 (M+), 223.04 (M+-C11H13O4), 179 (M+-C13H17O5), 167.01 (M+-C15H21O4), 83 (M+-C4H4O2) and 43 (C3H4). UV λmax nm (MeOH): 286 and 350. The Proton nuclear magnetic resonance (1H-NMR), carbon-13 NMR Attached proton test (13C-NMR-APT) and two-dimensional NMR (2-D NMR) data are listed in Table 1.
| Carbon No. | 1H-NMR | 13C-NMR | HSQC | HMBC |
|---|---|---|---|---|
| 1/1` | - | 105.52 | - | - |
| 2/2` | - | 161.25 | - | - |
| 3/3` | 6.030 (2H, s, H-3/3`) | 92.53 | H-3/3`→ C-3/3` | C-2/C-4/C-5 |
| 4/4` | - | 165.13 | - | - |
| 5/5` | - | 104.95 | - | - |
| 6/6` | - | 158.49 | - | - |
| 7/7` | - | 211.47 | - | - |
| 8/8` | 3.852 (2H, m, H-8/8`) | 39.54 | H-8/8` → C-8/8` | CH3 |
| 2 OCH3 | 3.904 (6H, s, OCH3) | 56.29 | H-OCH3→ C-OCH3 | - |
| 4 CH3 | 1.095 (12H, d, CH3) | 19.22 | H-CH3→ C-CH3 | C-8/C-7/CH3 |
| 1a CH2 | 3.644 (2H, s, H-1a) | 15.75 | H-1a → C-1a | C-1/C-2 |
Table 1: 1H-NMR and 13C-NMR-APT (400 MHz, Deuterated Chloroform CDCL3, d1) of Compound 3
1H-NMR of compound 3 exhibited singlet at δ 13.273 for OH-6/6' and another signal at 9.099 for OH-2/2'. H-3 and 3' were established from the presence of singlet signal integrated for 2 protons at δ 6.030 as well as the CH2 at position 1a appeared as singlet signal at 3.644. CH group at C-8 and C-8? were indicative of the presence of a multiplate signal at 3.852 (2H, m, H-8/8'). The four CH3 groups were indicated from the presence of doublet signal at 1.07, besides the two methoxy group was established from the presence of the singlet signal at δ 3.904 ppm [15,16].
In the 13C-NMR-APT spectra; the upfield value at 92.53 ppm assigned for unsubstituted C-3. The carbon at 15.75 ppm was assigned to CH2 at position 1a. The presence of two CH and CH3 group in the side chain group was indicative of the presence of carbon signal at 39.54 and 19.22 respectively as well as the two methoxy appeared at 56.29. Moreover, the carbon signal at 92.53 gave evidence for the carbon at C-3' and the carbonyl carbon was established from the carbon signal at 211.47.
Heteronuclear single quantum coherence (HSQC) spectrum showed that each proton of H-3, H-8, H of OCH3, H of CH3 and H of 1a-CH2 at δ 6.030, 3.852, 3.904, 1.095 and 3.644 respectively is in correlation with the directly attached carbons. Heteronuclear multiple bond correlation (HMBC) spectrum confirmed the hypothesis of the structure where the methylene bridge protons at δ 3.644 (1a) was correlated with carbons at 161.21 (C-2), carbon at 158.49 (C-6) and C-1 at (104.94). CH3 group at δ 1.07 was correlated with the signal of CH-8 at δ 39.54 and C=O (C-7) at 211.74. The C-8 at δ 3.85 was correlated with CH3 (19.22). H-3 at δ 6.030 was correlated with C-4 (165.13) and C-5 at (105.52).
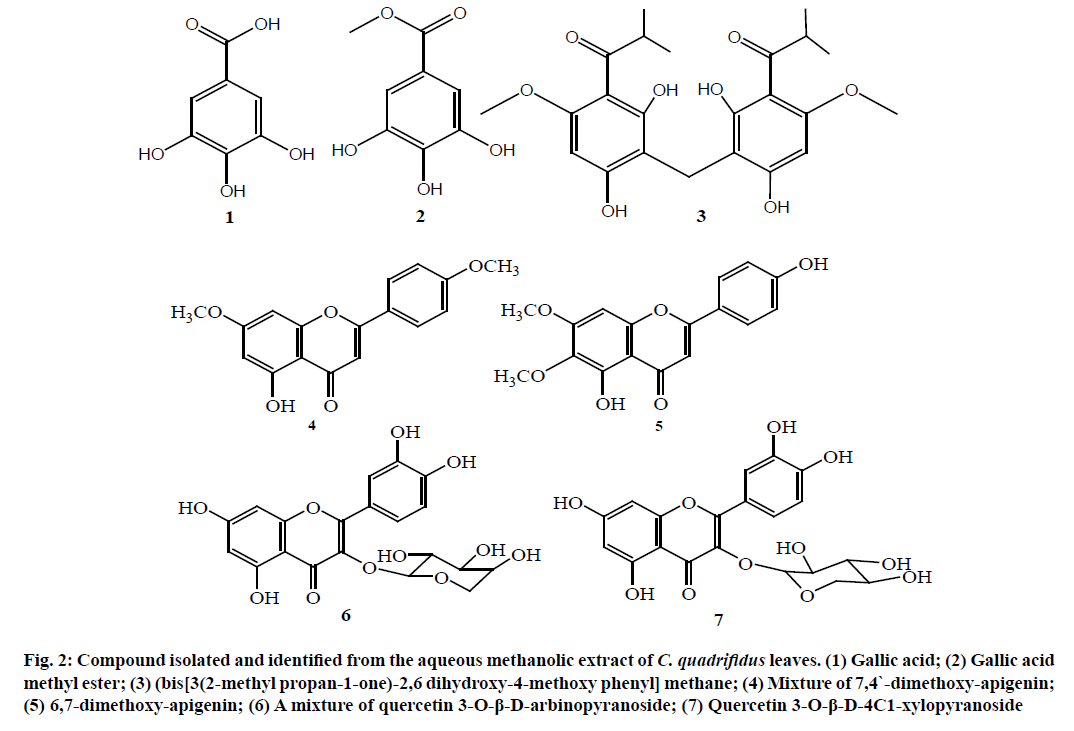
From the above spectral data, the structure of compound 3 was established as (bis [3(2-methyl propan-1-one)-2,6 dihydroxy-4-methoxy phenyl] methane [15,16]. This is the first report for compound 3 in nature (fig. 2) and in C. quadrifidus leaves.
Figure 2: Compound isolated and identified from the aqueous methanolic extract of C. quadrifidus leaves. (1) Gallic acid; (2) Gallic acid methyl ester; (3) (bis[3(2-methyl propan-1-one)-2,6 dihydroxy-4-methoxy phenyl] methane; (4) Mixture of 7,4`-dimethoxy-apigenin; (5) 6,7-dimethoxy-apigenin; (6) A mixture of quercetin 3-O-β-D-arbinopyranoside; (7) Quercetin 3-O-β-D-4C1-xylopyranoside
A mixture of compounds 4 and 5 was obtained as white amorphous powder (12 mg). TLC chromatographic properties: Rf-value is 0.54 (S3). It gave a dark purple fluorescent spot under UV light (λmax 360 nm) which turned to orange colour with Naturstoff spray reagent and bluish-green color with FeCl3. The liquid chromatography electrospray ionisation mass spectrometry (LC-ESI-MS) positive mode spectrum of compound 4 showed the pseudo-molecular ion peak at (M+H)+ at m/z 299 while compound 5 ESI-MS spectrum showed (M+H)+ at m/z 315. The LC-ESI-MS spectrum revealed the presence of two compounds one of which is a compound 4 while the other is a compound 5 with a pseudo-molecular ion peak (M+H)+ at m/z 299 and 315 respectively [17,18]. The difference between the m/z of these compounds can be attributed to the hydroxyl group.
The 1H-NMR spectrum revealed the presence of two compounds through the presence of two protons at δ 12.803 and 12.789 which are identical for the hydroxyl at C-5 of compound 4 and 5 respectively. Four methoxy groups appeared as four singlets at δ 3.849, 3.723, 3.821 and 3.816. There are four protons grouped appeared as two multiplets due to superimposed signals with the A2B2 spin system at δ 6.924 (H-3'and H-5') and 7.773 (H-2' and H-6') which are identical for B ring protons of both compounds. Two protons singlet at δ 6.407 revealed the presence of two superimposed H-3 protons, one proton for each compound. Two protons singlet at δ 6.500 indicated the presence of two superimposed H-8 protons. H-6 proton of compound 4 was assigned to δ 6.532.
The 13C-NMR-APT spectrum showed the signals of two compounds. Compound 4 showed 15 carbon resonances characteristic for apigenin among which the key signals at δ ppm 183.26 (C-4), 127.99 (C-2'/6'), 114.46 (C-3'/5'), 91.80 (C-8) and 104.49 (C-3/6) [17,18]. While compound 5 showed 15 carbon resonances characteristic for apigenin among which the key signals at δ ppm 182.41 (C-4), 127.95 (C-2'/6'), 89.34 (C-8) and 104.03 (C-3), 102.79 (C-6) [17,18]. Two methoxy were assigned for compound 4 at 55.90 (7-OCH3) and 55. 53 (4`-OCH3) while compound 5 methoxy signals were assigned at 60.50 (6-OCH3) and 55.90 (7-OCH3) [17,18]. 1H-NMR and 13C-NMR data for compound 4 and 5 along with the reported literature [17,18] are listed in Table 2. These data suggested the tentative structure of the compounds; compound 4 and 5 to be 7,4'-dimethoxy-apigenin and 7,6-dimethoxy-apigenin, respectively. Both of these compounds are identified for the first time from C. quadrifidus leaves. Characterization and identification of compounds 6 and 7 is a yellow amorphous powder (25 mg) with Rf values 0.45 (S3) and 0.42 (S3) showing dark purple spot under UV(λmax 360 nm) which changed to orange fluorescence and pale green colour upon spraying with Naturstoff and FeCl3 reagents respectively. Acid hydrolysis afforded quercetin aglycone (Co-TLC with authentic samples) in the organic phase and xylose and arabinose in the aqueous phase (Co-PC with authentic samples) which suggested the presence of a mixture of two compounds named compounds 6 and 7.
| Carbon No. | Compound 4 | Compound 5 | ||||||
|---|---|---|---|---|---|---|---|---|
| δH | δH[13B] | δC | δC[13B] | δH | δH[13A] | δC | δC [13A] | |
| 2 | - | - | 163.61 | 163.9 | - | 163.38 | 163.6 | |
| 3 | 6.407 (1H, s) | 6.54 (s) | 104.49 | 104.3 | 6.407 (1H, s) | 6.65 (1H, s, H-3) | 104.03 | 103.3 |
| 4 | - | - | 183.26 | 182.4 | - | - | 182.41 | 182.3 |
| 5 | 12.65 (1H, s) | - | 155.93 | 157.6 | - | - | 155.93 | 152.6 |
| 6 | 6.532 (1H, brs) | 6.49 (d, J=2.5) | 104.49 | 98 | - | - | 102.79 | 104.49 |
| 7 | - | - | 167.48 | 165.3 | -- | - | 157.28 | 158.6 |
| 8 | 6.500 (1H, brs) | 6.37 (d, J=2.5) | 91.8 | 92.5 | 6.500 (1H, s) | 6.66 (1H, s) | 89.34 | 91.6 |
| 9 | - | - | 158.47 | 162.5 | - | - | 162.47 | 153.6 |
| 10 | - | - | 105.31 | 105 | - | - | 105.31 | 106.6 |
| 1` | - | - | 123.74 | 123.5 | - | - | 123.86 | 122.7 |
| 2` | 7.773 (1H, m) | 7.85 (d, J=9) | 127.99 | 128 | 7.773 (2H, m) | 7.38 (d, J=2Hz, H-2') | 127.95 | 128.4 |
| 3` | 6.924 (1H, m) | 7.02 (d, J=9) | 114.46 | 114.4 | 6.924 (2H, m) | 7.02 (d, J=9) | 114.46 | 114.6 |
| 4` | - | 164.4 | 164 | - | - | 162.6 | 162.4 | |
| 5` | 6.924 (1H, m) | 7.02 (d, J=9) | 114.46 | 114.4 | 6.924 (2H, m) | 7.02 (brd, J=8.5 Hz) | 114.46 | 114.5 |
| 6` | 7.773 (1H, m) | 7.85 (d, J=9) | 127.99 | 128 | 7.773 (2H, m) | 7.57 (dd, J=8.5, 2 Hz) | 127.95 | 128.4 |
| 5-OH | 12.803 (1H) | 12.8 | -- | -- | 12.789 (1H, s) | - | - | |
| 7-OCH3 | 3.849 (3H,s) | 3.89 (s) | 55.9 | 55.7 | 3.816 (3H, s) | 3.82 (s) | 55.9 | 56.5 |
| 4`- OCH3 | 3.723 (3H,s) | 3.88 (s) | 55.53 | 55.5 | - | - | - | - |
| 6-OCH3 | - | - | -- | 3.821 (3H, s) | 3.84 (s) | 60.5 | 60.1 | |
Table 2: 1H-NMR (400 MHz, CDCL3-d1) and 13C-NMR-APT (100 MHz, CDCL3-d1) Spectral Data for Compounds 4 and 5 Mixture
UV λmax nm (MeOH): 254, 290 (sh), 354; (+NaOMe): 261, 300 (sh), 405; (+NaOAc): 265, 323(sh), 372; (+NaOAc/H3BO3): 265, 310, 406 (+AlCl3): 275, 300 (sh), 337, 431; (+AlCl3/HCl): 270, 290 (sh), 397.
The compound 7 gives the same NMR data of 6 except δ ppm 12.65 (1H, s, H-bonded OH-5), 5.37 (1H, d, J=7.2 Hz, H-1'') as well as in case of 13C-NMR-APT data, compound 5 showing the same carbons of aglycone and differ only in these carbon resonances of sugar moiety being 76.17 (C-3''), 73.90 (C-2''), 69.61 (C-4''), 65.82 (C-5'') [19].
The presence of two signals at 12.62 and 12.65 ppm in 1H -NMR spectrum characteristic for the proton of OH at C5 indicated the presence of quercetin aglycone for both compounds through a comparison with the previous quercetin compounds isolated [20]. Moreover, compound 6 is finally identified as quercetin 3-O-β-D-arbinopyranoside (Gvaijaverin) based on comparison of its 1H-NMR and 13C-NMR-APT [20]. 1H-NMR and 13C-NMR-APT of compound 7 showed that the sugar moiety is xylose due to the presence of doublet signal at δ 5.37 (J=7.2 Hz) and it is confirmed from 13C-NMR-APT (Table 3) which showed the five-carbon signal characteristic for xylose especially C-3''(76.17) and C-5''(65.62) [19,20]. The Final identification of compound 7 is quercetin 3-O-β-D-xylopyranoside. From the above data, the mixture is composed of two compounds, one major as quercetin 3-O-β-D-arbinopyranoside [6] and the minor one as quercetin 3-O-β-D-4C1-xylopyranoside [7] in ratio 2.5:1. Both compounds 6 and 7 were identified in C. quadrifidus leaves for the first time.
| Carbon No. | Compound 6 | Compound 7 | ||
|---|---|---|---|---|
| δH | δC | δH | δC | |
| 2 | ----- | 157.26 | ----- | 157.26 |
| 3 | ----- | 134.26 | ----- | 134.26 |
| 4 | ----- | 178.03 | ----- | 178.03 |
| 5 | ----- | 161.57 | ----- | 161.57 |
| 6 | 6.26 (1H, brs) | 98.48 | 6.26 (1H, brs) | 98.48 |
| 7 | ----- | 164.6 | ----- | 164.6 |
| 8 | 6.482 (1H, d, J=1.2 Hz) | 93.33 | 6.482 (1H, d, J=1.2 Hz) | 93.33 |
| 9 | ----- | 156.97 | ----- | 156.97 |
| 10 | ----- | 104.21 | ----- | 104.21 |
| 1` | ----- | 121.64 | ----- | 121.64 |
| 2` | 7.599 (1H, d, J=1.96 Hz) | 114.76 | 7.599 (1H, d, J=1.96 Hz) | 114.76 |
| 3` | ----- | 144.53 | ----- | 144.53 |
| 4` | ----- | 148.54 | ----- | 148.54 |
| 5` | 6.933 (1H, m) | 116.09 | 6.933 (1H, m) | 116.09 |
| 6` | 7.68 (1H, dd, J=1.96, 8.8 Hz) | 121.47 | 7.68 (1H, dd, J=1.96, 8.8 Hz) | 121.47 |
| 5-OH | 12.622 (1H, s) | 12.65 (1H, s) | ||
| 1`` | 5.3 (1H, d, J=5.6 Hz) | 103.32 | 5.37 (1H, d, J=7.2 Hz) | 103.32 |
| 2`` | 3.80-3.23 (m, remaining sugar protons). | 72.77 | 3.80-3.23 (m, remaining sugar protons). | 73.9 |
| 3`` | 71.52 | 76.17 | ||
| 4`` | 67.78 | 69.61 | ||
| 5`` | 65.62 | 65.82 | ||
Table 3: 1H-NMR (400 MHz, CD3OD-d4) and 13C-NMR-APT (100 MHz, CD3OD-d4) Spectral Data for Compounds 6 and 7 Mixture
C. quadrifidus leaves EO yield was 0.5 % v/w and was injected in GLC-MS device for identification of the EO constituents. The identification of the oil components was based upon the comparison of mass spectral data and KI with Wiley registry of mass spectral data 8th Edition, NIST Mass Spectral Library (December 2005) and the available literature [6,21]. A total of 32 compounds composing 94.26 % of the oil composition were identified. Merely, oxygenated monoterpenes represented by 1,8-Cineole (53.98 %) comprises 68.96 % of the EO. Monoterpenes represented 0.55 % while non-identified components were found to be less than 6 %. C. quadrifidus EO leaves chemical composition is presented in Table 4.
| No. | Compound name | KI index | Area % | M+ | B.P. | Mass fragments |
|---|---|---|---|---|---|---|
| Monoterpene hydrocarbons | ||||||
| 1 | α-Pinene | 939 | 0.06 | 136 | 93 | 121, 105, 77, 67, 53, 41 |
| 2 | β-Pinene | 979 | 0.1 | 136 | 93 | 121, 107, 79, 69, 53, 41 |
| 3 | Myrcene | 991 | 0.05 | 136 | 93 | 121, 79, 69, 53, 41 |
| 4 | Para-cymene | 1025 | 0.17 | 134 | 119 | 103, 91,77,65, 51 |
| 5 | γ- Terpinene | 1060 | 0.17 | 136 | 93 | 121, 105, 77, 65, 41 |
| Oxygenated monoterpenes | ||||||
| 6 | 1,8-Cineole | 1031 | 53.98 | 154 | 43 | 139, 108, 93, 84, 81, 69, 56, 41 |
| 7 | cis-β-Terpineol | 1144 | 5.39 | 136 | 71 | 55, 69, 80, 93, 107, 121 |
| 8 | Terpinene-4-ol | 1177 | 1.25 | 154 | 71 | 136, 121, 111, 93, 71, 69 |
| 9 | α-terpineol | 1189 | 2.41 | 154 | 59 | 136, 121, 93, 81, 41, 41 |
| 10 | Neral | 1238 | 0.66 | 152 | 41 | 137, 119, 94, 81, 69, 53 |
| 11 | Geraniol | 1253 | 0.24 | 150 | 69 | 139, 123, 111, 93, 53, 41 |
| 12 | Geranial | 1267 | 1.02 | 152 | 69 | 137, 123, 109, 84, 53, 41 |
| 13 | Gernyl acetate | 1381 | 4.11 | 196 | 69 | 136, 121, 93, 43, 41 |
| Others | ||||||
| 14 | n-Octane | * | 0.33 | 114 | 70 | 99, 85, 71, 57 |
| 15 | 1,4- dimethyl cyclohexane | * | 0.05 | 112 | 97 | 70, 56, 55, 41 |
| 16 | Ethyl-cyclohexane | * | 0.06 | 112 | 83 | 41, 55, 67, 82 |
| 17 | Ethyl-benzene | * | 0.1 | 106 | 91 | 40, 51, 65, 77 |
| 18 | Heptane | 700 | 4.81 | 100 | 43 | 41, 57, 70, 71 |
| 19 | Capylene | 792 | 0.31 | 98 | 69 | 41, 55, 70,97 |
| 20 | I-octene | 792 | 1.85 | 98 | 70 | 41, 55, 56, 84, 98 |
| 21 | Octene | 802 | 5.26 | 112 | 70 | 43, 55, 85 |
| 22 | (2Z)-octene | 812 | 0.11 | 112 | 70 | 41, 55, 84, 98 |
| 23 | Methyl-valerate | 828 | 0.06 | 114 | 43 | 57, 70, 71 |
| 24 | Isopropyl butrate | 845 | 0.33 | 114 | 57 | 43, 70, 99 |
| 25 | Ethyl isovalerate | 858 | 0.21 | 111 | 43 | 41, 57, 70, 84, 85 |
| 26 | n-Nonone | 900 | 0.05 | 128 | 43 | 57, 71, 85, 99 |
| 27 | Decane | 1000 | 4.24 | 100 | 43 | 41, 57, 71, 85 |
| 28 | n-Decane | 1000 | 0.1 | 142 | 57 | 41, 43, 71, 85, 99 |
| 29 | n-Undecane | 1100 | 0.08 | 156 | 57 | 43, 71, 84, 98, 113 |
| 30 | Eugenol | 1359 | 3.1 | 164 | 164 | 149, 131, 121, 103, 91, 77 |
| 31 | Caryophyllene acetate | 1701 | 2.4 | 264 | 43 | 69, 81, 93, 109, 121 |
| 32 | Hexadecanoic acid | 2861 | 1.2 | 256 | 73 | 43, 60, 115, 129, 213 |
| Monoterpene hydrocarbons | 0.55 | |||||
| Oxygenated monoterpenes | 68.96 | |||||
| Others | 24.75 | |||||
| Total identified % | 94.26 | |||||
| Total unidentified % | 5.74 | |||||
Note: *Identified by mass fragment from NIST database
Table 4: Chemical Constituents of Hydrodistilled Essential Oil of C. quadrifidus Leaves
Upon comparison between EO composition, it was found that our results were in accordance with literature where 1,8 cineole was identified as the major component. However, the EO was reported to include 30 components including 1,8-cineole (80 %), α-pinene (2.34 %), β-pinene (3.37 %), myrcene (3.37 %) and the rest of the components were stated to be less that 1 % [6]. The concentration of major EO component of C. quadrifidus leaves was calculated via gas liquid chromatography-flame ionization detection (GLC-FID) analysis using 1,8-cineole as an external standard since it represented the major identified component in the tested oil and oxygenated monoterpenes. The calibration curve of cineole exhibited high linearity with the coefficient of determination (R2) of 0.9999 at the used concentrations.
1,8-cineole represented the major component of C. quadrifidus leaves oil (525.62 µg/ml) followed by α-terpineol (12 µg/ml), terpinen-4-ol (4.21 µg/ml), β-pinene (5.36 µg/ml), myrcene (3.13 µg/ml), α-pinene (2.68 µg/ml). Quantitative determination via GLC-FID analysis using the calibration curve of 1,8-cineole of C. quadrifidus leaves EO was estimated for the first time in this study
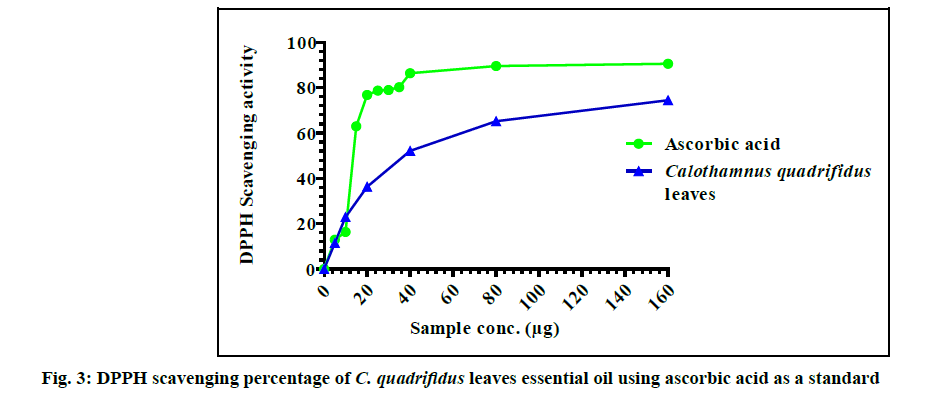
The anti-oxidant activity of C. quadrifidus leaves EO and ascorbic acid against DPPH radicals were expressed in fig. 3. The tested oil showed antioxidant capacity in a dose-dependent manner. The best scavenging capacity for C. quadrifidus EO (74.49 %) were observed at 160 µg. C. quadrifidus leaves oil showed an antioxidant capacity with SC50=37.3 µg that may be attributed to its oxygenated hydrocarbons content.
In previous reports, 1,8-cineole, linalool, α-pinene and p-cymene showed weak DPPH scavenging activity percentage [22]. According to the available literature C. quadrifidus AME revealed a marked significant scavenging activity at different concentrations (2-100 mg/ml) showed. The scavenging capacity of the AME of C. quadrifidus leaves was found to be SC50=8 mg/ml [3]. Comparing the scavenging activity of both the EO and the AME of C. quadrifidus it is obvious that the antioxidant potential of the EO is much greater than that of AME. This free radical potential may be attributed to the oxygenated contents of the EO.
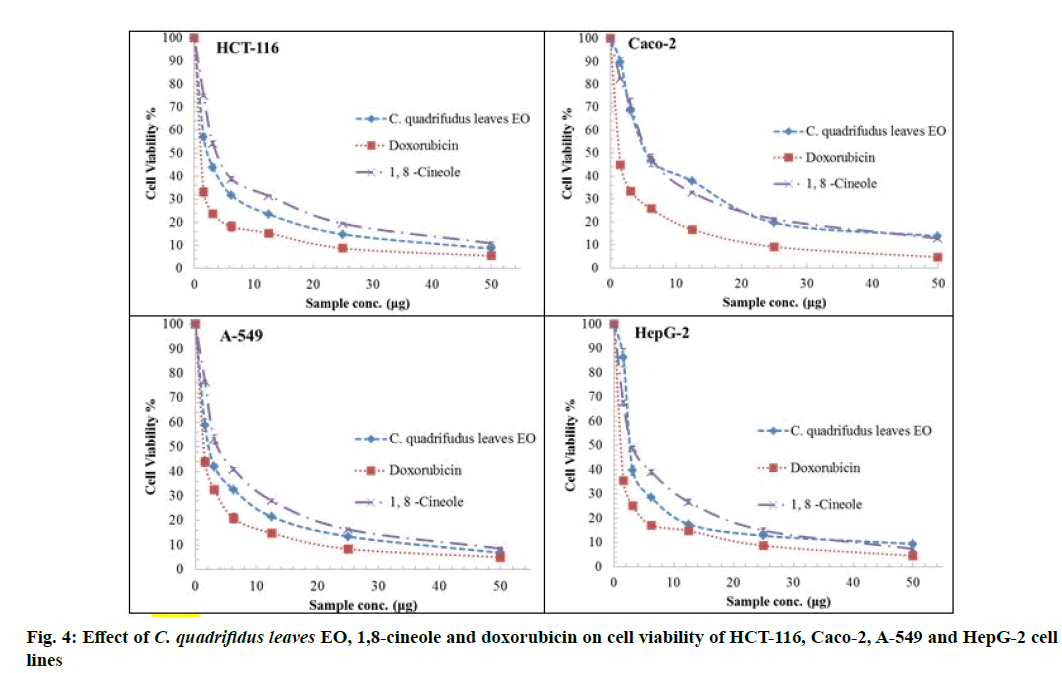
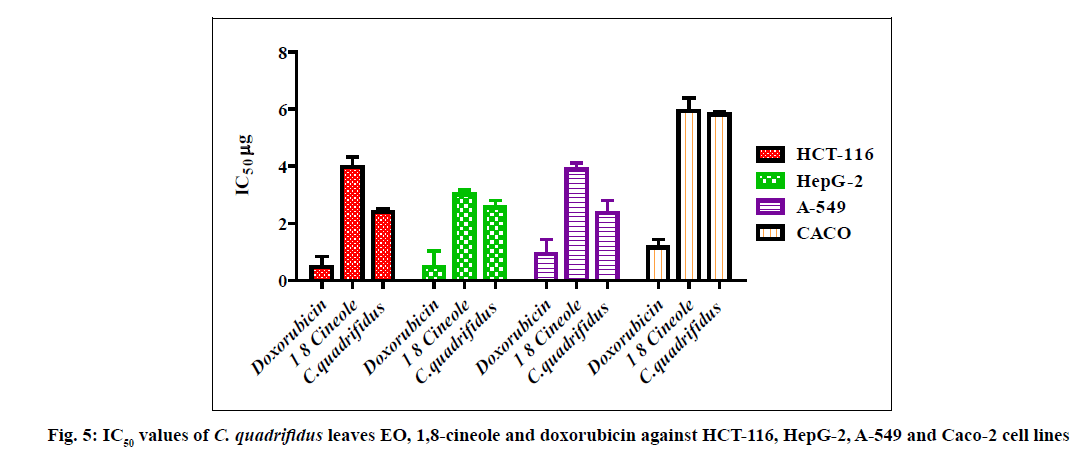
The cytotoxic activity of tested oils, 1,8-cineole and standard doxorubicin against HCT-116, HepG-2, A-549 and Caco-2 cell lines are represented in fig. 4 and fig. 5. IC50 values were used to assess the activity of the EO and 1,8-cineole against cell lines under investigation based on the protocol of the National Cancer Institute (NCI) which recommended that crude extracts of plant origin should be considered significant for IC50 values≤30 μg/ml as well as IC50 values≤4 μg/ml for pure substances [23]. Moreover, IC50 values were classified as follows: IC50≤20 μg/ml are highly active, IC50 21-200 μg/ml are moderately active, IC50 201-500 μg/ml are weakly active and IC50>501 μg/ml are inactive [24].
The role of the major component in the EO was assessed using 1,8-cineole [6] where it showed comparable potencies against all tumour cell lines. IC50 ranged from 3.02 to 5.92 µg/ml. 1,8-Cineole cytotoxicity was significant against HepG-2 (3.02 μg/ml) followed by A-549 (3.89 μg/ml), HCT-116 (3.97 μg/ml) and Caco-2 (5.92 μg/ml) cell lines. In this study, the synergistic cytotoxic effect of the various oil components is clear as the cytotoxic effect of the major constituent 1,8-cineole is less than that of the EO except against HepG-2 [24]. EO of C. quadrifidus leaves showed variable potencies against all tumour cell lines used with IC50 ranged from 2.37-5.8 μg/ml (fig. 5). The cytotoxicity of EO of C. quadrifidus leaves was significant against A-549 (2.37 μg), HCT-116 (2.39 μg), HepG-2 (2.55 μg) and Caco-2 (5.80 μg) cell lines. The role of the major component in the EO was assessed using 1,8-cineole.
This is the first report for cytotoxic activity of C. quadrifidus leaves EO.
In vitro cytotoxic activities against liver and colon carcinoma were reported for EO of different Myrtaceae plants containing 1,8-cineole, linalool and α-terpineol with IC50 values 0.36-0.69 µg [25], which are in accordance with our results. Terpinen-4-ol induced a significant in vitro inhibition of colorectal cells growth in a dose-dependent manner [26]. Additionally, myrcene exhibited strong cytotoxic activities against HCT-116 and HepG-2 (IC50=1.27 and 0.93 µg respectively) in a dose-dependent manner when evaluated by MTT assay [8]. Geraniol (a minor component of the two EO tested) was reported to inhibit 70 % of cell growth of Caco-2 cell line with cells accumulating in the S transition phase of the cell cycle and inhibition of DNA synthesis [27]. It also inhibited colon cancer cell proliferation by inducing membrane depolarisation and interfering with ionic canals and signalling pathways [28].
The results indicated that the two oils showed selective activities against different microorganisms as indicated by inhibition zone diameters, percentage of standard activity and minimum inhibitory concentration (MIC) values. P. aeruginosa was resistant to the oils (Table 5 and Table 6).
| Microorganism | Inhibition zone diameter in mm (% of standard activity) | |||
|---|---|---|---|---|
| C. quadrifidus | Ampicillin | Gentamycin | Amphotericin B | |
| G +ve bacteria | ||||
| S. aureus | 21.3±0.44 (77.7) | 27.4±0.18 (100) | N/A | N/A |
| B. subtilis | 21.9±0.37 (67.6) | 32.4±0.10 (100) | N/A | N/A |
| G -ve bacteria | ||||
| P. aeruginosa | - | N/A | 17.3±0.15 (100) | N/A |
| E. coli | 9.2±0.25 (41.3) | N/A | 22.3±0.18 (100) | N/A |
| Fungi | ||||
| A. fumigatus | 17.2±0.58 (72.5) | N/A | N/A | 23.7±0.10 (100) |
| P. italicum | 20.9±0.44 (95.4) | N/A | N/A | 21.9±0.12 (100) |
| G. candidum | 20.4±0.58 (70.7) | N/A | N/A | 28.7±0.22 (100) |
(-): No activity and N/A is non-applicable
Table 5: Antimicrobial Activity Evaluation of C. quadrifidus EO
| Microorganism | Inhibition zone diameter in mm (% of standard activity) | |||
|---|---|---|---|---|
| C. quadrifidus | Ampicillin | Gentamycin | Amphotericin B | |
| G +ve bacteria | ||||
| S. aureus | 21.3±0.44 (77.7) | 27.4±0.18 (100) | N/A | N/A |
| B. subtilis | 21.9±0.37 (67.6) | 32.4±0.10 (100) | N/A | N/A |
| G -ve bacteria | ||||
| P. aeruginosa | - | N/A | 17.3±0.15 (100) | N/A |
| E. coli | 9.2±0.25 (41.3) | N/A | 22.3±0.18 (100) | N/A |
| Fungi | ||||
| A. fumigatus | 17.2±0.58 (72.5) | N/A | N/A | 23.7±0.10 (100) |
| P. italicum | 20.9±0.44 (95.4) | N/A | N/A | 21.9±0.12 (100) |
| G. candidum | 20.4±0.58 (70.7) | N/A | N/A | 28.7±0.22 (100) |
(--): No activity
Table 6: MIC of Tested C. quadrifidus EO
MIC is generally considered more accurate than disc diffusion assay for EO. The limitation of the oils activity can be attributed to the low water solubility of the oil and its components, which limits their diffusion into the agar medium. Only the more water-soluble components can diffuse into the medium. The hydrocarbon components may remain on the surface of the medium or evaporate [29]. MIC of the C. quadrifidus leaves oil against the tested organisms ranged from 0.98 to 15.63 μg/ml. The least MIC values were detected against B. subtilis (0.98 μg) and E. coli (1.25 μg). Concerning S. aureus and P. italicum there are significant antimicrobial activities observed as indicated by their MIC values compared with the corresponding standards. Additionally, C. quadrifidusleaves EO showed significant antifungal activity against P. italicum and to less extent against G. candidum which is in accordance with the literature [6].
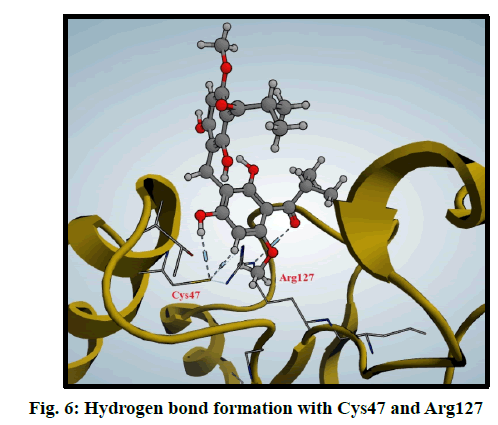
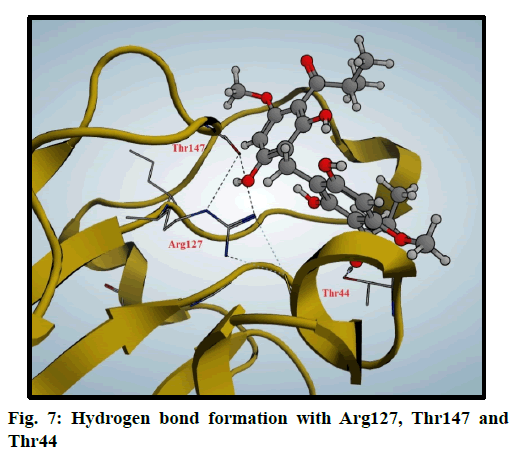
Peroxiredoxins are a family of antioxidant enzymes which also control cytokine-induced peroxide levels which control signal transduction in mammalian cells. The active site structure contains cysteine 47 (Cys47) that is known as peroxidase Cys which is involved in the reduction of peroxide and also, Arginine 127 (Arg127) is an important residue in the interactions and fixation of the substrate [30]. Molecular docking of compound 3 was done in this peroxiredoxins and the binding mode was found to be in two poses (fig. 6 and fig. 7)
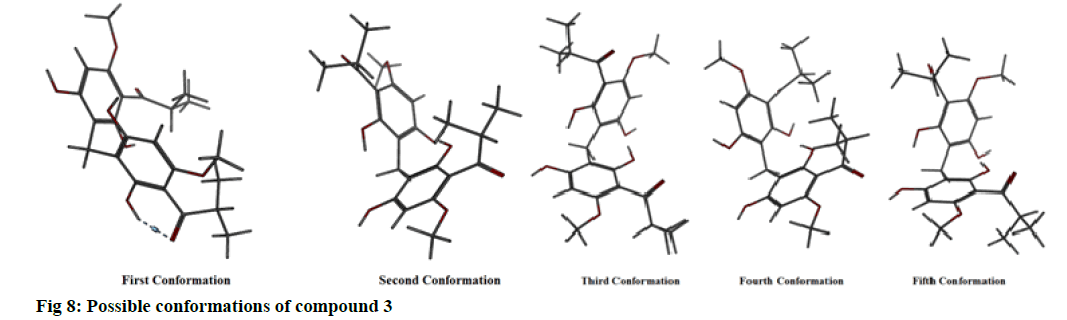
The possible conformations of this compound were found to be five (fig. 8). The energy profile for all of them was computed (Table 7). It was found that confirmation 2 showed the least total energy (43.85) and electrostatic energy as well (-16.80). The energy profile for all conformations was also computed when they are in the bound form (in complex with the enzyme) (Table 8).
| Conformation | Total Energy | Stretching Energy | Angle Energy | Torsional Energy | Vdw Energy | Electrostatic Energy | Solvation Energy |
|---|---|---|---|---|---|---|---|
| 1 | 51.74 | 2.88 | 11.81 | 29.11 | 15.17 | -7.23 | 0 |
| 2 | 43.85 | 2.19 | 12.25 | 33.89 | 12.32 | -16.8 | 0 |
| 3 | 51.65 | 2.28 | 13.23 | 36.54 | 13.5 | -13.9 | 0 |
| 4 | 49.14 | 2.93 | 13.06 | 27.5 | 16.16 | -10.51 | 0 |
| 5 | 47.17 | 1.99 | 12.88 | 33.84 | 10.41 | -11.95 | 0 |
Table 7: Energy Profile for All Possible Conformations
| Conformation | Total Energy | Stretching Energy | Angle Energy | Torsional Energy | Electrostatic Energy | Solvation Energy |
|---|---|---|---|---|---|---|
| 1 | 24047.77 | 103.23 | 339.01 | 1129.13 | -7.23 | 0 |
| 2 | 19685.58 | 102.6 | 340.4 | 1136.6 | -12.44 | 0 |
| 3 | 63733.51 | 103.28 | 340.26 | 1127.52 | -10.51 | 0 |
| 4 | 20437.88 | 102.35 | 340.08 | 1133.86 | -14.29 | 0 |
| 5 | 32000.92 | 102.5 | 339.45 | 1132.91 | -10.8 | 0 |
Table 8: Energy Profile for all Possible Conformations in the Bound Form
In the bound form, almost all the conformations showed the same values of both stretching and angle energies. Confirmation 2 was the best in the electrostatic energy and the total energy as it demonstrated the least values. We could not find any correlation with Van der Waals energy. The free binding energy of the binding of each conformation to the protein was calculated in kcal/mol together with the in silico affinity (pki) (Table 9).
| Conformation | Free binding Energy ΔG | Affinity pki |
|---|---|---|
| 1 | -23.08 | 7.25 |
| 2 | -33.58 | 10.92 |
| 3 | -30.71 | 8.59 |
| 4 | -30.17 | 8.39 |
| 5 | -30.98 | 9.27 |
Table 9: Free Binding Energy of the Binding of Each Conformation to the Protein
Another proof that conformation 2 is the most stable and may be responsible for the antioxidant effect it showed the best free binding energy and affinity when compared to others. Structure elucidation of the pure compounds isolated from C. quadrifidus leaves was done according to their chromatographic behaviour, chemical and spectroscopic data. These compounds were isolated for the first time from C. quadrifidus leaves and identified as gallic acid [1], gallic acid methyl ester [2], (bis [3(2-methyl propan-1-one)-2,6 dihydroxy-4-methoxy phenyl] methane [3], a mixture of 7,4'-dimethoxy-apigenin [4] and 6,7-dimethoxy-apigenin [5] and a mixture of quercetin 3-O-β-D-arbinopyranoside [6] with quercetin 3-O-β-D-4C1-xylopyranoside [7]. High phenolic content indicate the presence of high antioxidant potential that requires further investigation.
The EO obtained from C. quadrifidus fresh leaves revealed the presence of 32 compounds composing 94.26 % of the whole oil were identified. 1,8-Cineole was estimated to be the major component of the oil (525.62 µg/ml) followed by α-terpineol (12 µg/ml) and terpinen-4-ol (4.21 µg/ml) using GLC-FID analysis. The oil best free radical scavenging capacity (86.86 %) was observed at 640 µg.
The EO exhibited potential antioxidant activity in a dose-dependent manner when tested by DPPH radical scavenging assay compared with ascorbic acid. The oil also showed significant activity against human carcinoma cell lines including colon, hepatic, lung and intestinal. Additionally, the antimicrobial activity was evaluated using agar disc diffusion and determination of MIC against ampicillin, gentamycin and amphotericin B as positive controls. According to MIC values, the most sensitive strain to C. quadrifidusleaves EO was B. subtilis. Generally, the antimicrobial activity of EO is mostly due to the presence of phenols, aldehydes and alcohols [31]. Moreover, as typical lipophiles, the EO can penetrate the cell wall and cytoplasmic disrupting the structure of polysaccharides, fatty acids and phospholipids and permeabilize them, causing loss of ions and reduction of membrane potential leading to depletion of the Adenosine triphosphate (ATP) pool [32] in addition to cytoplasm coagulation [33] leading to cell death by apoptosis and necrosis [34]. Most of the EO components reported in this study were reported to have antimicrobial action. 1,8-Cineole, terpinen-4-ol, linalool and α-terpineol exhibited significant inhibition of S. aureus and E. coli [35]. Also, 1,8-cineole showed fungicidal activity against A. flavus and A. parasiticus [36]. The different antibacterial activity of the EO, compared with those of their major components, can be attributed to the synergistic effect of the different components in the oil and/or by the presence of other active constituents in small concentrations [37].
EO of other Myrtaceae plants containing α-and β-pinene, 1,8-cineole, linalool and α-terpineol (which were reported in the EO of our study) showed variable antimicrobial activities against S. aureus, B. subtilis, E. coli and P. aeruginosa [25]. To sum up, the C. quadrifidusleaves EO biological properties that can attributed to the complex mixtures of numerous molecules, compared to 1,8-cineole alone. This emphasizes the theory that natural products action is potentiated by synergism; in other words, the EO variation of concentrations and compositions is necessary for both synergistic and antagonistic activities on the major constituents.
Peroxiredoxins are a family of dominant antioxidant enzymes in mammalian cells. Molecular docking of compound 3 was done in peroxiredoxins and the binding mode was found. Five binding conformations were found to compound 3. The energy profile for all compound 3 confirmation was computed. It was found that confirmation 2 showed the least total energy and electrostatic energy as well. In the bound form almost all the conformations showed the same values of both stretching and angle energies still conformation 2 was the best in the electrostatic energy and the total energy as it demonstrated the least values. Conformation 2 of compound 3 was found to be the most stable and may be responsible for the antioxidant effect as it showed the best free binding energy and affinity when compared to conformations.
References
- Baily L. Manual of cultivated plants, Revised edition. The Macmillan Company 1958:724-6.
- Hawkeswood TJ. Studies in the genus Calothamnus (Myrtaceae: Leptospermoideae): Redescription of Calothamnus chrysantherus F. Muell., with notes on distribution, habitat and conservation and a list of collection records 2007.
- Abd Elazeem R. A Pharmacognostical Study of Calothamnus quadrifidus Family Myrtaceae. Faculty of Pharmacy: Helwan University 2009.
- Ayoub NA, El-Ahmady SH, Singab AN, Al-Azizi MM. Phytotherapeutic studies on Calothamnus quadrifidus R. Br (Myrtaceae). Nat Prod Ind J 2007;3(1):30-7.
- El Dib RA, Marzouk MS, Moharram FA, El-Shenawy SM, Elazeem RM. Chemical and biological investigation of Calothamnus quadrifidus R. Br. Bull Fac Pharm (Cairo University) 2009;47:193-202.
- El-Ahmady SA, Ayoub NA, Singab AN, Al-Azizi MM, Kubeczka KH. Chemical composition and antimicrobial activity of the essential oils from Eucalyptus cinerea, Callistemon viminalis and Calothamnus quadrifidus (Myrtaceae). Nat Prod Ind J 2007;3:23-9.
- Chemical composition and anti-inflammatory activity of the essential oils of Psidium guajava fruits and leaves. J Essent Oil Res 2013;25(6):475-81.
- Al-Gendy A, Moharram F, Zarka M. Chemical composition, antioxidant, cytotoxic and antimicrobial activities of Pimenta racemosa (Mill.) JW Moore flower essential oil. J Pharmacogn Phytochem 2017;6(2):312-9.
- Yen GC, Duh PD. Scavenging effect of methanolic extracts of peanut hulls on free-radical and active-oxygen species. J Agric Food Chem 1994;42(3):629-32.
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Method 1983;65(1-2):55-63.
- Elaasser M, Abdel-Aziz M, El-Kassas R. Antioxidant, antimicrobial, antiviral and antitumor activities of pyranone derivative obtained from Aspergillus candidus. J Microbiol Biotechnol Res 2011;1(4):5-17.
- Himratul-Aznita WH, Mohd-Al-Faisal N, Fathilah AR. Determination of the percentage inhibition of diameter growth (PIDG) of Piper betle crude aqueous extract against oral Candida species. J Med Plant Res 2011;5(6):878-84.
- Ghorab MM, Alsaid MS, Al-Dosary MS, Nissan YM, Attia SM. Design, synthesis and anticancer activity of some novel thioureido-benzenesulfonamides incorporated biologically active moieties. Chem Central J 2016;10(1):1-3.
- Barakat HH, Hussein SA, Marzouk MS, Merfort I, Linscheid M, Nawwar MA. Polyphenolic metabolites of Epilobium hirsutum. Phytochemistry 1997;46(5):935-41.
- Arisawa M, Fujita A, Suzuki R, Hayashi T, Morita N, Kawano N, et al. Studies on cytotoxic constituents in pericarps of Mallotus japonicus, Part I. J Nat Prod 1985;48(3):455-9.
- Kabran FA, Okpekon TA, Roblot F, Séon-Méniel B, Leblanc K, Bories C, et al. Bioactive phloroglucinols from Mallotus oppositifolius. Fitoterapia 2015;107:100-4.
- Gohari AR, Ebrahimi H, Saeidnia S, Foruzani M, Ebrahimi P, Ajani Y. Flavones and flavone glycosides from Salvia macrosiphon Boiss. Iran J Pharma Res 2011;10(2):247.
- Saeidnia S, Moradi-Afrapoli F, Gohari A, Malmir M. Cytotoxic Flavonoid from Achillea talagonica Bioss. J Med Plants 2009;1(29):52-6.
- Agrawal PK, Bansal MC, Porter LJ, Foo LY. Carbon-13NMR of Flavonoids (Studies in Inorganic Chemistry). BV Amsterdam: Elsevier Science Publishers 1989;39:XVI-564.
- Harborne JB, Williams CA. Flavone and flavonol glycosides. In: The flavonoids 1975:376-441.
- Adams RP. Identification of Essential Oils by Gas Chromatography/ Mass spectroscopy 4th edition: Academic Press; 2007.
- Aazza S, Lyoussi B, Miguel MG. Antioxidant and antiacetylcholinesterase activities of some commercial essential oils and their major compounds. Molecules 2011;16(9):7672-90.
- Geran R. Protocols for screening chemical agents and natural products against animal tumors and other biological systems. Cancer Treat Rep 1972;3:51-61.
- Srisawat T, Chumkaew P, Heed-Chim W, Sukpondma Y, Kanokwiroon K. Phytochemical screening and cytotoxicity of crude extracts of Vatica diospyroides symington type LS. Tropical J Pharm Res 2013;12(1):71-6.
- Aboutabl E, Meselhy K, Elkhreisy E, Nassar M, Fawzi R. Composition and bioactivity of essential oils from leaves and fruits of Myrtus communis and Eugenia supraxillaris (Myrtaceae) grown in Egypt. J Essent Oil Bear Plant 2011;14(2):192-200.
- Shapira S, Pleban S, Kazanov D, Tirosh P, Arber N. Terpinen-4-ol: A novel and promising therapeutic agent for human gastrointestinal cancers. PloS one 2016;11(6):e0156540.
- Carnesecchi S, Schneider Y, Ceraline J, Duranton B, Gosse F, Seiler N, et al. Geraniol, a component of plant essential oils, inhibits growth and polyamine biosynthesis in human colon cancer cells. J Pharmacol Exp Ther 2001;298(1):197-200.
- Bakkali F, Averbeck S, Averbeck D, Idaomar M. Biological effects of essential oils: A review. Food Chem Toxicol 2008;46(2):446-75.
- Griffin S, Markham J, Leach D. An agar dilution method for the determination of the minimum inhibitory concentration of essential oils. J Essent Oil Res 2000;12(2):249-55.
- Wood Z, Schröder E, Harris JR, Poole L. Structure, mechanism and regulation of peroxiredoxins. Trends Biochem Sci 2003;28(1):32-40.
- Sacchetti G, Maietti S, Muzzoli M, Scaglianti M, Manfredini S, Radice M, et al. Comparative evaluation of 11 essential oils of different origin as functional antioxidants, antiradicals and antimicrobials in foods. Food Chem 2005;91(4):621-32.
- Turina A, Nolan M, Zygadlo J, Perillo M. Natural terpenes: self-assembly and membrane partitioning. Biophys Chem 2006;122(2):101-13.
- Gustafson J, Liew Y, Chew S, Markham J, Bell H, Wyllie S, et al. Effects of tea tree oil on Escherichia coli Lett. Appl Microbiol 1998;26(3):194-8.
- Armstrong J. Mitochondrial membrane permeabilization: the sine qua non for cell death. Bioessays 2006;28(3):253-60.
- Carson C, Mee B, Riley T. Mechanism of action of Melaleuca alternifolia (tea tree) oil on Staphylococcus aureus determined by time-kill, lysis, leakage and salt tolerance assays and electron microscopy. Antimicrob Agents Chemother 2002;46(6):1914-20.
- Vilela G, De Almeida G, D'Arce M, Moraes M, Brito J, da Silva M, et al. Activity of essential oil and its major compound, 1, 8-cineole, from Eucalyptus globulus Labill., against the storage fungi Aspergillus flavus Link and Aspergillus parasiticus Speare. J Stored Prod Res 2009;45(2):108-11.
- Suzuki É, Baptista E, Resende Do Carmo A, Chaves M, Afonso M, Chicourel E, et al. Potential of the essential oil from Pimenta pseudocaryophyllus as an antimicrobial agent. Acta Pharmaceutica. 2014;64(3):379-85.