- *Corresponding Author:
- M. Chen
Department of Pharmacy, The 309th Hospital of PLA, Beijing 100091, China
E-mail: lm19851229@126.com
| Date of Submission | 05 July 2016 |
| Date of Revision | 05 June 2017 |
| Date of Acceptance | 31 January 2018 |
| Indian J Pharm Sci 2018;80(2):291-297 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Sirolimus is metabolized by the CYP3A subfamily and is a substrate of the P-glycoprotein. CYP3A5 and MDR1 gene polymorphisms have been reported to influence plasma concentrations of this drug and dose requirements, but some recent studies reported conflicting results. The aim of this study was to develop population pharmacokinetics to identify the potential influencing factors that explain pharmacokinetic variability in healthy Chinese adults. Twenty seven healthy Chinese subjects were enrolled into this study and were genotyped for CYP3A5 (6986A>G), MDR1 (3435C>T). Blood samples at different time point were collected from each subject and blood concentrations were measured. The population pharmacokinetic model was established using the non-linear mixed-effects model method, CYP3A5, MDR1, gender and other factors were evaluated in the model as covariates. The results indicated that CYP3A5 polymorphisms significantly influenced the clearance, the central compartment distribution volume was larger in male than that in female subjects, body surface area showed remarkable impact on peripheral compartment volume. All parameters were estimated with acceptable precision. Visual predictive check and bootstrap indicated that the population model adequately captured the observed profiles. This model may be useful in guiding clinicians to adjust drug dosing regimen.
Keywords
Sirolimus, population pharmacokinetics, CYP3A5, MDR1, polymorphism
Sirolimus (rapamycin), a macrocyclic lactone and natural fermentation product of Streptomyces hygroscopicus, is widely used as immunosuppression drug to prevent organ rejection especially renal allograft rejection [1]. As one of the calcineurin inhibitors family, sirolimus exerts immunosuppressive effect through inhibition of the mammalian target of rapamycin protein by the sirolimus-FK binding protein complex [2-4].
Sirolimus is poorly absorbed, has a low bioavailability (14 % on average) and a long terminal elimination half-life of approximately 62 h [5,6]. The bioavailability, absorption and metabolism of sirolimus varies widely among individuals, so the blood concentrations reveal large differences among patients receiving the same dosage [1]. Since sirolimus has a narrow therapeutic index as well as large inter- and intra-individual pharmacokinetic variability, therapeutic drug monitoring and individualized immunosuppressive therapy are recommended [7,8] to reduce the risk of acute rejection, toxicity [9] and other adverse effects, such as thrombocytopenia, anaemia, leukopenia and hyperlipidaemia [10-12].
Like cyclosporine and tacrolimus, sirolimus is mainly metabolized by cytochrome P450 3A (CYP3A4 and 3A5) enzymes located at the gut mucosa and liver [13-15], and also is substrate of the multidrug efflux pump P-glycoprotein (P-gp, encoded by MDR1gene) [16,17]. Polymorphisms in the genes of these proteins could thus be associated with inter-individual variations in sirolimus bioavailability and metabolism [17-20]. It is reported that genetics may be accounts for 20-95 % of the variability in drug disposition and effects [21]. Several population pharmacokinetic studies [22–27] have been conducted to assess the pharmacokinetic characteristics of sirolimus in healthy volunteers or renal transplant recipients in White or African-American race. Among all those variants, the polymorphic CYP3A5 (6986A>G) plays a major role and has a significant association with trough levels standardized by the dosage (C0/dose) [28]. But for MDR1 (3435C>T), the impact of its polymorphisms to the inter-individual variability (IIV) in the blood concentration of sirolimus was controversial and incongruous [27,29,30].
Since ethnic differences in drug metabolism are fairly common [31], and the conclusion of different studies are controversial, the objective of this study was to investigate the population pharmacokinetics of sirolimus in healthy Chinese adults, and to identify potential factors that explain pharmacokinetic variability to facilitate individualized therapy.
Materials and Methods
Study design
The study was approved by the Ethics Committee of the 309 Hospital and was accomplished in accordance with Good Clinical Practice guidelines (ethics approval number: 2014060501). Written informed consent was obtained before each volunteer was enrolled. The design and implementation of this clinical study conformed to the ethical principles of the Declaration of Helsinki. All the subjects were admitted to the hospital at 9:00 pm the day before the study. Breakfast was given 2 h after the drug was administered on the research day. The dose of sirolimus was 6 mg and whole blood was collected in EDTA tubes from each subject pre-dose, then at 0.5, 1, 2, 3, 4, 6, 8, 12, 16, 24, 48, 72, 96, 120 and 144 h post-dose. These samples were used to determine the sirolimus concentrations and were stored at –70°. All the subjects were administered drug in fasted state and then received a standard breakfast 30 min later.
Subjects
Subjects who were 18 to 45 y of age, had a body mass index between 18 and 25 kg/m2 and a body weight >50 kg (male) and >45 kg (female) were eligible for inclusion in the study. Subjects with active major infections, such as hepatitis B or C infection, human immunodeficiency virus were excluded. The demographic, biological and pharmacogenetic characteristics of the subjects were listed in Table 1.
| Characteristics | Median (range) values |
|---|---|
| Age (years) | 25.48 (20-36) |
| Sex (M/F; n) | 12/15 |
| Height (cm) | 168.78 (160-180) |
| Weight (kg) | 61.93 (45-75) |
| Body suface area (m2)# | 1.71 (1.45-1.93) |
| Haematocrit (%) | 38.63 (35.1-43.4) |
| Red blood cells (×1012 l-1) | 4.40 (3.78-5.06) |
| Haemoglobin (g l-1) | 130.04 (110-151) |
| Platelets (×109 l-1) | 234.15 (139-300) |
| Leukocytes (×109 l-1) | 5.86 (3.7-8.2) |
| Alanine transaminase (U l-1) | 16.93 (9-41) |
| Aspartate transaminase (U l-1) | 17.04 (12-24) |
| Cholesterol (mmol l-1) | 4.22 (2.76-5.59) |
| Triglycerides (mmol l-1) | 0.75 (0.4-1.4) |
| Proteins (g l-1) | 73.06 (65.9-81.6) |
| Albumin (g l-1) | 46.87 (44-50.6) |
| Blood urea nitrogen (mmol l-1) | 4.24 (2.81-8.15) |
| Serum creatinine (umol l-1) | 49.87 (40.4-63.2) |
| CYP3A5 genetype | |
| AA (*1/*1) | 3 |
| AG (*1/*3) | 10 |
| GG(*3/*3) | 14 |
| MDR1 genetype | |
| CC | 4 |
| CT | 16 |
| TT | 7 |
Table 1: Demographic, Biological and Pharmacogenetic Characteristics of Subjects Included in Study
Sirolimus assay
All sirolimus concentrations were measured by high performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) method using an Extend-C18 column (2.1×50 mm, 3.5 μm). The mobile phase consisted of methanol and 10 mM ammonium acetate and the flow rate was 0.4 ml·min-1 (gradient elution). Briefly, sample preparation consisted of adding 100 μl of a solution of ascomycin (internal standard, IS) in methanol, 200 μl acetonitrile, 200 μl zinc sulphate (2 M) and 100 μl, 50 % methanol to 100 μl whole blood, the mixture was centrifuged, and the supernatant was drawn off for test. The compounds of interest were separated by HPLC and detected by MS/MS in the positive ion, selected reaction monitoring mode using the following transitions: sirolimus (m/z 931.7→864.8) and IS (m/z 809.7→756.6; Figure 1). The calibration curve ranged from 0.5- 100.0 μg/l in whole blood. The within-day and betweenday coefficients of variation and bias values were <15 % over this range.
Genotyping
The CYP3A5 (6986A>G) and MDR1 (3435C>T) single-nucleotide polymorphisms were determined for each subject. Leucocytes were isolated from the whole blood samples, and then DNA was extracted from the leucocytes. The genomic DNA of each subject was used to characterize these genotypes, using a gene magnification fluorescence detector based on fluorescence in situ hybridization assay (TL988A gene magnification fluorescence detector, HuaXiaGene, China).
Pharmacokinetic model
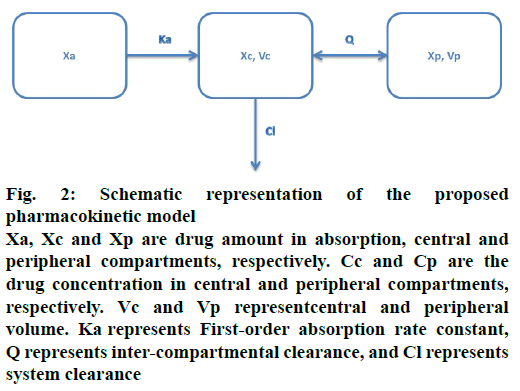
The movement of drug in healthy volunteers was described using a two-compartment model with firstorder absorption (Figure 2). The pharmacokinetic model was defined by the following differential Eqns. 1, 2, 3, 4 and 5, respectively: dXa/dt = –Ka×Xa; dXc/ dt = Ka×Xa+Q×Cp–(Q+Cl)×Cc; dXp/dt = Q×(Cc–Cp); Cc = Xc/Vc; Cp = Xp/Vp, where, Xa, Xc and Xp represent the drug amount in absorption, central and peripheral compartments, respectively. Cc and Cp are the drug concentrations in central and peripheral compartments, respectively. Vc and Vp represent central and peripheral volumes. Ka represents first-order absorption rate constant, Q represents inter-compartmental clearance, and Cl represents system clearance.
Figure 2: Schematic representation of the proposed pharmacokinetic model
Xa, Xc and Xp are drug amount in absorption, central and peripheral compartments, respectively. Cc and Cp are the drug concentration in central and peripheral compartments, respectively. Vc and Vp representcentral and peripheral volume. Ka represents First-order absorption rate constant, Q represents inter-compartmental clearance, and Cl represents system clearance
Data analysis
Data analysis was performed using Phoenix® NLME™ 1.2 (Certara, St Louis, MO, USA) software with first order conditional estimation with Lindstrom-Bates (FOCE L-B) method. The data from all volunteers were fitted simultaneously. IIV was characterized using an exponential variability model Eqn. 6: Pi = P×eηi, where, P is the population typical value and Pi represents the individual parameter. The random variable ηi is normally distributed with mean zero and variance of ω2, therefore, IIV follows a log-normal distribution. Residual error of pharmacokinetic model was calculated by proportional error model Eqn. 7: Cij = Cpij×(1+ε1ij), where, Cij, and Cpij are individual observation and prediction concentration, respectively. ε1ij is normally distributed with mean zero and variances of σ2.
Population covariate analysis
The effects of covariates such as gender, age, height, weight, body surface area (BSA), hematocrit, red blood cells, hemoglobin, platelets, leukocytes, alanine transaminase, aspartate transaminase, cholesterol, triglycerides, proteins, albumin, blood urea nitrogen, serum creatinine, CYP3A5 and MDR1 polymorphisms were evaluated for the population model. Each selected covariate was first tested by univariate non-linear mixedeffects modelling analysis to confirm its relevance. A decrease in the objective functional value (OFV) of at least 3.84 (p≤0.05) was first required to consider the covariate in the intermediate model. Classically, a covariate was not retained at this step in case of obvious over-parameter of the model. Secondly, all such significant covariates were added simultaneously into the intermediate model, and then each covariate was removed from the full (intermediate) model independently to confirm its relevance (backward deletion strategy), and the resulting variation in the OFV (compared with the full model) examined. An increase in the OFV of more than 6.64 was required to confirm the significance of the covariate and take it into account in the final model.
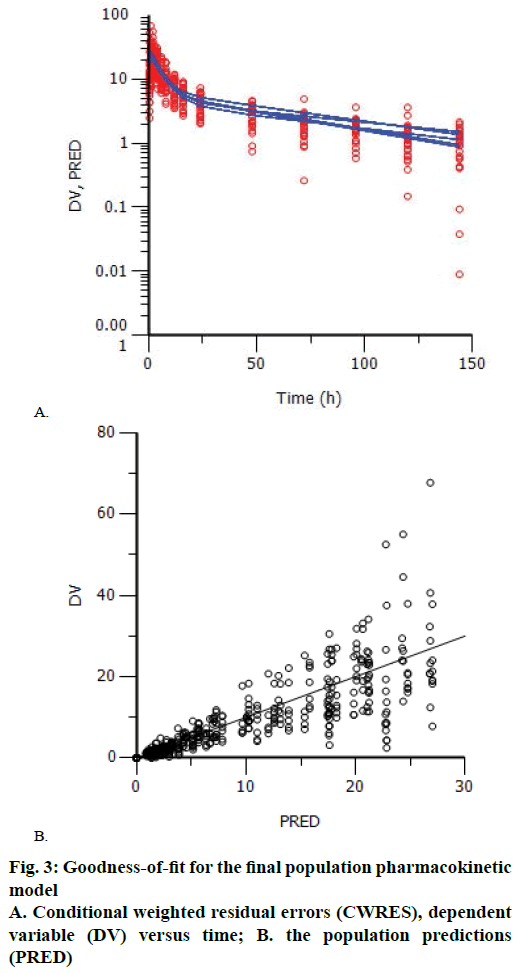
The plots of means of observation, prediction versus time were used to look for the goodness-of-fit of the final population model. According to OFV and goodness-of-fit, the best model was selected, and visual evaluation method was done by inspection of scatter plots of observation (dependent variable) versus prediction (PRED). The relative prediction errors were graphically described by conditional weighted residuals (CWRES) plotted against PRED and time after dose.
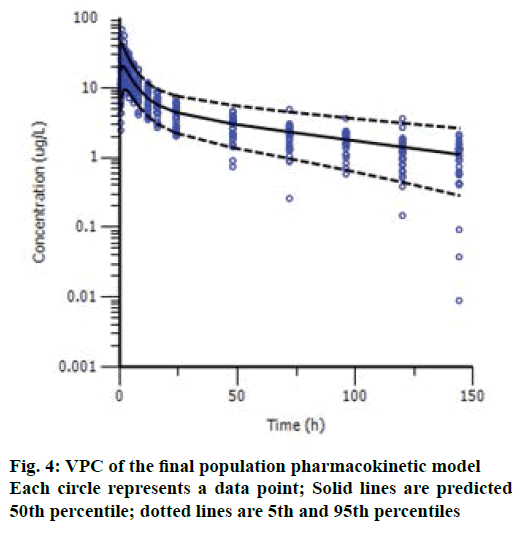
Model validation was performed using visual predictive check (VPC) for drug concentrations. 1000 simulations were generated using the final model and the simulated percentiles (2.5th, 50th, 97.5th) were calculated. The adequacy of the model was evaluated by comparing the distribution of observation with the distribution of simulated data. The final population model robustness was assessed with a bootstrap method. One thousand bootstrap datasets were generated by repeatedly sampling with replacements from the original dataset, and the final model was fitted to the new datasets. The median and 95 % confidence interval (95 % CI) of the parameters were compared with the final parameters estimates.
Results and Discussion
A two-compartment open model adequately characterized the drug concentration dataset. The stepwise forward addition showed that the age, body weight, MDR1 polymorphism and other factors did not have any influence on parameters to a statistically significant extent in this population. Gender and BSA were identified as covariates influencing central and peripheral volume, respectively: if gender= female, Vc= 184.461; if gender= male, Vc = 184.461×e0.266; Vp = 170.029×BSA2.165.
The resulting population model with the covariates of CYP3A5 genotype is shown in Eqns. 11-13: if CYP3A5 = *1/*1, Cl = 10.813, if CYP3A5 = *1/*3, Cl = 10.813×e–0.034, if CYP3A5 = *3/*3, Cl = 10.813× e–0.250. All the pharmacokinetic parameters, IIV, and the residual errors are shown in Table 2. All the estimates showed an acceptable precision (relative standard error, RSE with the range from 7.650 to 49.6 %).
| Parameter (unit) | Model estimate | Bootstrap | ||||
|---|---|---|---|---|---|---|
| Estimate | RSE% | IIV (CV %)* |
95 % CI# | Estimate | 95 % CI# | |
| Ka (1/h) Cl (l/h) Vc (l) |
2.651 10.813 184.461 |
17.465 11.421 7.821 |
17.476 11.418 7.821 |
1.740-3.562 8.386-13.239 156.104-212.819 |
2.643 10.811 184.34 |
1.736-3.551 8.383-13.238 156.037-212.643 |
| Vp (l) | 170.029 | 38.701 | 38.701 | 40.681-299.377 | 168.217 | 41.271-295.163 |
| Q (l/h) | 23.596 | 7.650 | 7.648 | 20.049-27.144 | 23.592 | 20.048-27.136 |
| CYP3A5*1/*3-Cl□ | –0.034 | 38.235 | –0.059-0.010 | –0.034 | –0.059-0.010 | |
| CYP3A5*3/*3-Cl■ | –0.250 | 49.6 | –0.494-0.007 | –0.250 | –0.494-0.007 | |
| GNDR-Vc○ | 0.266 | 37.594 | 0.070-0.462 | 0.265 | 0.069-0.461 | |
| BSA-Vp◊ | 2.165 | 33.256 | 0.751-3.580 | 2.18468 | 0.781-3.588 | |
| σ※ | 17.452% | 6.078 | 0.154-0.195 | 0.175 | 0.154-0.195 | |
Table 2: Pharmacokinetic Parameters and Bootstrap Analysis of Final Population Model in Healthy Volunteers
Goodness-of-fit plots did not show systematic bias for the pharmacokinetic model predictions. The plots of CWRES versus time and PRED showed that the CWRES were scattered evenly around the zero line. There was no major trend in magnitude of CWRES (Figure 3). The plots of observed data (blood concentration) against PRED indicated that the data points distributed evenly around the line of identity (Figure 3). Since, random errors are ignored in the PRED, the deviations from the expected appearance are usually due to misspecifications in the model.
The VPC results are shown in Figure 4, and we can see an overall trend of fitted data. The plots of VPC suggested a suitable recapture of the central tendency in the concentrations (about 50 % of the observations fall above and 50 % below the median predicted values (solid line) at each time point). Ideally, 95 % of the observation should lie within the 95 % CI. As expected, VPC showed that 6.3 % (27/431) of drug concentrations fell outside the 95 % CI. Bootstrap results showed a success rate of 100.0 % (all the 1000 were successful). The 95 % CI of bootstrap analyses showed acceptable robustness of the final model. The data are presented together with the final estimates in Table 2. Bootstrapping of the population pharmacokinetic model yielded median parameter distributions to be similar to the estimates during the data fitting. Overall, suitable predictive performance was obtained with this two-compartment population model.
Many population pharmacokinetic models of sirolimus in healthy adults or renal transplant patients were reported, but most of them were focused on western populations [22–27]. Only one study was conducted on Chinese adult renal transplant patients [32], while a onecompartment model with first-order absorption and elimination was selected as the base model. In this study, we established a population pharmacokinetic model of sirolimus using a two-compartment model with firstorder absorption and elimination in healthy Chinese adults. We also tried one- and three-compartment model, and found sirolimus fits two-compartment model better, which was supported by several other studies [6,26,33]. This is the first population pharmacokinetic analysis of sirolimus performed in healthy Chinese adults to date. After screening and selecting potential covariates such as gender, weight, BSA, height, CYP3A5 (6986A>G), MDR1 (3435C>T) polymorphisms, only CYP3A5 (6986A>G) polymorphisms, gender and BSA were found to have influence on sirolimus pharmacokinetics and were taken into the final model as covariates.
The sirolimus CL was significantly decreased when the CYP3A5*1 wild type mutated to *3, the CL typical value of CYP3A5*1 wild type, CYP3A5*1/*3 type and CYP3A5*3/*3 type were 10.813 l/h, 10.445 l/h, and 8.110 l/h, respectively, which means those patients bearing at least one active CYP3A5*1 may require higher doses of sirolimus to achieve therapeutic plasma concentrations than CYP3A5*3/*3. CYP3A5*3 is located on intron three, its mutation results in a splicing defect of the mRNA and produces an unstable and non-functional protein, which induces the decrease of enzymatic activity and drug clearance. The data observed in this study coincide with prediction, and also with previous reports [34-36]. From the population pharmacokinetics model, we also found that gender showed remarkable impact on central volume, and BSA influenced peripheral volume of sirolimus. The typical value of sirolimus central volume observed in this study was 184.461 l in female and 240.537 l in male, which was reported ranged from 187-249 l [27]. The typical value of sirolimus peripheral volume was 170.029 l. The central volume in compartment model refers to organs and tissues with abundant blood flow, such as liver, kidney and heart, while the peripheral volume refers to organs and tissues with poor blood flow, such as muscle, fat, skin and skeleton. For one drug, its distribution volume is usually affected by the volume of the organs and tissues itself. As the males generally have bigger body configuration than females, so the central volume of distribution are bigger in males than in females. This conclusion was similar to that of a previous study [37]. For BSA, those with bigger body configuration may have larger BSA, and also have larger peripheral volume. BSA was mentioned to be correlated with apparent volume of distribution of a peripheral compartment in a previous study [6].
A previous study has reported that MDR1 (3435C>T) were significantly associated with long-term sirolimus dose requirements [30], which means its polymorphisms may influence pharmacokinetics of sirolimus. But in this study, no significant influence of MDR1 (3435C>T) polymorphisms on the pharmacokinetics of sirolimus were found, this conclusion was supported by several other studies [38-40].
In summary, estimates of sirolimus population pharmacokinetics were obtained in this study. The influence of CYP3A5 (6986A>G) polymorphisms, gender, BSA were identified as important covariates for sirolimus CL, central and peripheral volume respectively. However, it is a relatively small-scale study, and the finding need to be confirmed by research of larger samples.
Acknowledgements
This work was supported by the Capital Health Development project (2011-5007-04) from Science and Technology Commission of Beijing.
Conflicts of interest
There are no conflicts of interest.
Financial support and sponsorship
Nil.
References
- Mahalati K, Kahan BD. Clinical pharmacokinetics of sirolimus. Clin Pharmacokinet 2001;40:573-85.
- Sehgal SN. Sirolimus: its discovery, biological properties, and mechanism of action. Transplant Proc 2003;35:7S-14S.
- Dumont FJ, Staruch MJ, Koprak SL, Melino MR, Sigal NH. Distinct mechanisms of suppression of murine T cell activation by the related macrolides FK-506 and rapamycin. J Immunol 1990;144:251-8.
- Kahan BD, Chang JY, Sehgal SN. Preclinical evaluation of a new potent immunosuppressive agent, rapamycin. Transplantation 1991;52:185-91.
- MacDonald A, Scarola J, Burke JT, Zimmerman JJ. Clinical pharmacokinetics and therapeutic drug monitoring of sirolimus. Clin Ther 2000;22:B101-21.
- Ferron GM, Mishina EV, Zimmerman JJ, Jusko WJ. Population pharmacokinetics of sirolimus in kidney transplant patients. Clin Pharmacol Ther 1997;61:416-28.
- Holt DW, Denny K, Lee TD, Johnston A. Therapeutic monitoring of sirolimus: its contribution to optimal prescription. Transplant Proc 2003;35:157S-61S.
- Kahan BD, Keown P, Levy GA, Johnston A. Therapeutic drug monitoring of immunosuppressant drugs in clinical practice. Clin Ther 2002;24:330-50.
- Lopez-Montenegro Soria MA, Kanter Berga J, Beltran Catalan S, Milara Paya J, et al. Genetic polymorphisms and individualized tacrolimus dosing. Transplant Proc 2010;42:3031-3.
- Hong JC, Kahan BD. Sirolimus-induced thrombocytopenia and leukopenia in renal transplant recipients: risk factors, incidence, progression, and management. Transplantation 2000;69:2085-90.
- Brattstrom C, Wilczek H, Tyden G, Bottiger Y, Sawe J, Groth CG. Hyperlipidemia in renal transplant recipients treated with sirolimus (rapamycin). Transplantation 1998;65:1272-4.
- Kahan BD, Napoli KL, Kelly PA, Podbielski J, Hussein I, Urbauer DL, et al. Therapeutic drug monitoring of sirolimus: correlations with efficacy and toxicity. Clin Transplant 2000;14:97-109.
- Lampen A, Zhang Y, Hackbarth I, Benet LZ, Sewing KF, Christians U. Metabolism and transport of the macrolide immunosuppressant sirolimus in the small intestine. J Pharmacol Exp Ther 1998;285:1104-12.
- Sattler M, Guengerich FP, Yun CH, Christians U, Sewing KF. Cytochrome P-450 3A enzymes are responsible for biotransformation of FK506 and rapamycin in man and rat. Drug Metab Dispos 1992;20:753-61.
- Arceci RJ, Stieglitz K, Bierer BE. Immunosuppressants FK506 and rapamycin function as reversal agents of the multidrug resistance phenotype. Blood 1992;80:1528-36.
- Crowe A, Lemaire M. In vitro and in situ absorption of SDZ-RAD using a human intestinal cell line (Caco-2) and a single pass perfusion model in rats: comparison with rapamycin. Pharm Res 1998;15:1666-72.
- Sakaeda T, Nakamura T, Okumura K. Pharmacogenetics of MDR1 and its impact on the pharmacokinetics and pharmacodynamics of drugs. Pharmacogenomics 2003;4:397-410.
- Mourad M, Wallemacq P, De Meyer M, Brandt D, Van Kerkhove V, Malaise J, et al. The influence of genetic polymorphisms of cytochrome P450 3A5 and ABCB1 on starting dose- and weight-standardized tacrolimus trough concentrations after kidney transplantation in relation to renal function. Clin Chem Lab Med 2006;44:1192-8.
- Li D, Gui R, Li J, Huang Z, Nie X. Tacrolimus dosing in Chinese renal transplant patients is related to MDR1 gene C3435T polymorphisms. Transplant Proc 2006;38:2850-2.
- Lukas JC, Calvo R, Zografidis A, Ortega I, Suarez E. Simulation of sirolimus exposures and population variability immediately post renal transplantation: importance of the patient's CYP3A5 genotype in tailoring treatment. Biopharm Drug Dispos 2010;31:129-37.
- Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. New Engl J Med 2003;348:538-49.
- Wu FL, Tsai MK, Chen RR, Sun SW, Huang JD, Hu RH, et al. Effects of calcineurin inhibitors on sirolimus pharmacokinetics during staggered administration in renal transplant recipients. Pharmacotherapy 2005;25:646-53.
- Tejani A, Alexander S, Ettenger R, Lerner G, Zimmerman J, Kohaut E, et al. Safety and pharmacokinetics of ascending single doses of sirolimus (Rapamune, rapamycin) in pediatric patients with stable chronic renal failure undergoing dialysis. Pediatr Transplant 2004;8:151-60.
- Thomas PP, Manivannan J, John GT, Jacob CK. Sirolimus and ketoconazole co-prescription in renal transplant recipients. Transplantation 2004;77:474-5.
- Zahir H, Keogh A, Akhlaghi F. Apparent clearance of sirolimus in heart transplant recipients: impact of primary diagnosis and serum lipids. Ther Drug Monit 2006;28:818-26.
- Dansirikul C, Morris RG, Tett SE, Duffull SB. A Bayesian approach for population pharmacokinetic modelling of sirolimus. Br J Clin Pharmacol 2006;62:420-34.
- Djebli N, Rousseau A, Hoizey G, Rerolle JP, Toupance O, Le Meur Y, et al. Sirolimus population pharmacokinetic/pharmacogenetic analysis and bayesian modelling in kidney transplant recipients. Clin Pharmacokinet 2006;45:1135-48.
- Anglicheau D, Le Corre D, Lechaton S, Laurent-Puig P, Kreis H, Beaune P, et al. Consequences of genetic polymorphisms for sirolimus requirements after renal transplant in patients on primary sirolimus therapy. Am J Transplant 2005;5:595-603.
- Li Y, Yan L, Shi Y, Bai Y, Tang J, Wang L. CYP3A5 and ABCB1 genotype influence tacrolimus and sirolimus pharmacokinetics in renal transplant recipients. Springerplus 2015;4:637.
- Sam WJ, Chamberlain CE, Lee SJ, Goldstein JA, Hale DA, Mannon RB, et al. Associations of ABCB1 3435C>T and IL-10-1082G>A polymorphisms with long-term sirolimus dose requirements in renal transplant patients. Transplantation 2011;92:1342-7.
- Johnson JA. Influence of race or ethnicity on pharmacokinetics of drugs. J Pharm Sci 1997;86:1328-33.
- Ritter JM. Communicating about potential drug harms: safety implications for patients. Br J Clin Pharmacol 2009;68:147-8.
- Brattstrom C, Sawe J, Tyden G, Herlenius G, Claesson K, Zimmerman J, et al. Kinetics and dynamics of single oral doses of sirolimus in sixteen renal transplant recipients. Ther Drug Monit 1997;19:397-406.
- Saeki M, Saito Y, Nakamura T, Murayama N, Kim SR, Ozawa S, et al. Single nucleotide polymorphisms and haplotype frequencies of CYP3A5 in a Japanese population. Hum Mutat 2003;21:653.
- Josephson F, Allqvist A, Janabi M, Sayi J, Aklillu E, Jande M, et al. CYP3A5 genotype has an impact on the metabolism of the HIV protease inhibitor saquinavir. Clin Pharmacol Ther 2007;81:708-12.
- Wong M, Balleine RL, Collins M, Liddle C, Clarke CL, Gurney H. CYP3A5 genotype and midazolam clearance in Australian patients receiving chemotherapy. Clin Pharmacol Ther 2004;75:529-38.
- Zimmerman JJ. Exposure-response relationships and drug interactions of sirolimus. AAPS J 2004;6:e28.
- Elens L, Bouamar R, Shuker N, Hesselink DA, van Gelder T, van Schaik RH. Clinical implementation of pharmacogenetics in kidney transplantation: calcineurin inhibitors in the starting blocks. Br J Clin Pharmacol 2014;77:715-28.
- Hesselink DA, van Gelder T, van Schaik RH, Balk AH, van der Heiden IP, van Dam T, et al. Population pharmacokinetics of cyclosporine in kidney and heart transplant recipients and the influence of ethnicity and genetic polymorphisms in the MDR-1, CYP3A4, and CYP3A5 genes. Clin Pharmacol Ther 2004;76:545-56.
- Hesselink DA, Bouamar R, Elens L, van Schaik RH, van Gelder T. The role of pharmacogenetics in the disposition of and response to tacrolimus in solid organ transplantation. Clin Pharmacokinet 2014;53:123-39.