- *Corresponding Author:
- Y. Li
Departments of orthopedics, Departments of Oncology
Taizhou People’s Hospital Affiliated to Medical College of The Nantong University
Taizhou, Jiangsu 225300, P. R. China
E-mail: fanggufan68024342@163.com
| This article was originally published in a special issue, “Biomedical Research in Healthcare Setting” |
| Indian J Pharm Sci 2020:82(3)Spl issue5;150-158 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
This study investigated the role of miR-182-3p in osteosarcoma. The effect of the miR-182-3p expression on cell function and the regulatory signal changes of Early B-cell Factor 2 targeted by miR-182-3p were studied in the osteosarcoma cell line in vitro. miR-182-3p was downregulated in both osteosarcoma patients and osteosarcoma cell line. The overexpression of miR-182-3p resulted in increased apoptosis of osteosarcoma cells and decreased migration and invasion of osteosarcoma cells. In addition, miR-182-3p regulated the messenger RNA stability of early B-cell factor 2 y directly binding to the 3 prime untranslated region of Early B-Cell Factor 2, which was a key regulator of osteosarcoma cell apoptosis. Overexpression of Early B-Cell Factor 2 could inhibit the apoptosis of osteosarcoma cells and promote the migration and invasion of osteosarcoma cells effectively. At the same time, overexpression of Early B-Cell Factor 2 could alleviate the phenotype induced by miR-182-3p. miR-182-3p was a tumor suppressing microRNA in osteosarcoma. Its function was realized by inhibiting Early B-Cell Factor 2. These results provided a new therapeutic target for metastatic osteosarcoma and a better understanding of the molecular regulation of Early B-Cell Factor 2.
Keywords
Osteosarcoma, miR-182-3p, Early B-Cell Factor 2, Cellular function
Osteosarcoma (OS) was a very common bone tumor, which mainly occurs in children and adolescents[1]. OS accounts for 3-5 % of the childhood cancers and the initial peak age of onset is between 10 to 14 y[2,3]. Because of its complex pathological process and the primary metabolic process, the mortality rate of OS is very high[4-6]. Since the application of chemotherapy, the 5 y survival rate of OS cases has increased to 60-75 %. To reduce the adverse reactions associated with chemotherapy and improve the clinical outcome of the OS patients, there is an urgent need to find new biomarkers or specific molecular-targeted therapy[7]. MicroRNA (miRNA) is an evolutionarily conservative small non-coding RNA (ncRNA), with a length of 22-24 nt. It was reported that the miRNA plays an important role in the pathological process of the disease, which was considered a new biomarker of cancer[8,9]. It has been shown that it regulates many physiological processes by regulating the expression of central genes, such as cell proliferation, differentiation, development and apoptosis[9-11]. The data shows that differential expression of miRNA may contribute to the initiation and progress of OS[12]. A study reported that miR-1284 as a new regulatory factor, inhibited the proliferation and migration of OS cells by targeting high-mobility group protein 1 (HMGB1)[13]. Huang et al. had shown that tumor inhibited the function of miR-124 in OS cells by targeting Snail2, which indicated that the miR-124 played a key role in the development of OS[14]. Other miRNAs (miR-143, miR-382 and miR-223) have also been confirmed to be disorder in OS and have been shown to have potential values in the prognosis, diagnosis and treatment of OS[12]. miR-182-3p was first discovered in the process of human posterior capsule opacity, it can regulate the transformation of epithelium to mesenchyma, which is the key process of cancer cell metastasis[11]. Later, researchers found that the miR-182-3p was involved in the migration and/or invasion of endometrial cancer cells[12], colorectal cancer cells[13], gliomas[14], oral squamous cell carcinoma[15] and laryngeal squamous cell carcinoma[16]. These evidences suggested that miR-182-3p played an anticancer role in various types of cancer cells by inhibiting the metastasis of cancer cells. Therefore, it is possible that miR-182-3p might also be involved in the metastasis of OS. In addition, it was reported that the miR-182-3p can also target SMAD4, RAB22A, CXCR4 and FOXC1. Early B-Cell Factor 2 (EBF2) was a transcription factor belonging to the COE (Collier/Olf/EBF) family, which was first found in the central nervous system[17]. Subsequently, EBF2 was also found to be a key regulator in plant development and lipogenesis[18]. Importantly, EBF2 is a key regulator of osteoblast-dependent differentiation through the RANK-RANKL signal transduction[19]. In OS, EBF2 was found to be overexpressed and activated as osteoprotegerin through transcriptional regulation. In addition, EBF2 can also inhibit the apoptosis of OS induced by tumor necrosis factor-related apoptosisinducing ligand (TRAIL)[20]. However, how the EBF2 was regulated is still unclear, especially in OS.
Therefore, in vitro experiments were used to study the role of miR-182-3p in OS. Firstly, the expression level of miR-182-3p in OS patients and cell line was detected. The function of miR-182-3p in OS cell line was analyzed by overexpressing miR-182-3p. The bioinformatics results showed that EBF2 was the target of miR-182-3p, which was then verified by the luciferase experiment. In addition, the data showed that the overexpression of EBF2 successfully verified the phenotype caused by miR-182-3p in OS, which indicated that the miR-182-3p played a role in OS through EBF2. The data of this study clarifed the anticancer effect of miR-182-3p in OS and revealed the regulatory role of EBF2, which provided a basis to further understand the key transcription factors in OS and other fields.
Materials and Methods
Patients and samples
A total of 20 OS primary tumor samples and adjacent tissues were collected from OS patients who underwent surgery in Taizhou People’s Hospital. All samples were divided into 2 parts, one for section staining and the other for RNA and protein extraction. All samples were used with the written informed consent of the patients and their families.
Cell culture
hFOB1.19, Saos-2, U2OS, and MG63 cells were purchased from ATCC and cultured according to the instructions of ATCC. Briefly, hFOB1.19 cells were cultured in a mixed medium (50 % Ham’s F12 medium and 50 % DMEM medium) supplemented with 2.5 mM glutamine, 0.3 mg/ml G418, and 10 % fetal bovine serum. Saos-2 and U2OS were cultured in McCoy’s 5a modified medium with 15 % fetal bovine serum. MG63 was cultured in the minimum essential medium with 10 % heat-inactivated fetal bovine serum. All cells were cultured at 37°, 95 % air and 5 % CO2.
In situ hybridization (ISH) of miR-182-3p
ISH was carried out according to the manufacturer’s instructions and the probe-target complex uses antidigoxin- alkaline phosphate conjugates, nitroblue tetrazolium, and 5-bromo-4-chloro-3-indole-indole phosphate as the staining targets. According to the intensity of cytoplasmic miR-182-3p, the cases were classified as follows, negative (no expression or weak expression in most cells), low expression (low expression in most cells or medium expression in <50 % cells), and high expression (medium-high expression in most cells).
Cell transfection with miRNA mimic, antimiR, and siRNA
The miRNA mimic and antimiR of miR-182-3p were purchased from Suzhou Ribo Life Science Co., Ltd. The EBF2 siRNA sequences were as follows, 5’-UGAAUAGCAAUGACUAACGA-3’. The cells were cultured in a 6-well plate for 24 h before transfection and transfected with a confluence degree of 60 %. Lipofectamine2000 was used for the transfection of miRNA mimic, anti-miR and siRNA according to the manufacturer’s instructions.
RNA extraction and real-time quantitative PCR (qPCR)
TRIzol reagent was used to separate the total RNA according to the manufacturer’s instructions. RNA was reverse transcribed with Prime Script TM RT kit. SYBR Green kit (RR420A, TaKaRa) was used for the real-time quantitative of the target gene. 2-ΔΔCt was used for the calculation of the fold, triplicate repeat. The primers used were as follows, miR-182-3p mimic: UUCCCUUGUCAUCCUAUGCCU; negative control (NC): 5’-UUCUCCGAACGUGUCACGUTT-3′; EBF2: 5’-ACCAACAACGGCACTCACTACAAG-3’, 5’-TTCGCAGCATCGACTACACATCAC-3’.
Cell viability analysis and apoptosis
Cell availability kit-8 (CCK8) was used for cell viability analysis. The cells were detected in the 96-well plate and processed according to the CCK8 kit manufacturer’s instructions. According to the manufacturer’s instructions, cell apoptosis was analyzed using the Annexin V-FITC apoptosis kit. After labeling apoptosis was analyzed using the Moflo Astrios Eq flow sorting system.
Transwell cell migration assay
The Transwell was placed in 24-well culture plates as the upper chamber and the lower chamber was filled with 10 % FBS DMEM. All cells were cultured in FBSfree DMEM overnight and then inoculated in the upper chamber (200 μl, 1×105) and cultured for 18 h. Then the cells were fixed with 4 % polyoxyethylene and stained with amethyst violet.
Transwell cell invasion assay
Transwell invasion analysis was performed according to the previously described method. In short, the Matrigel and the designated growth medium were mixed at 1:1 and then used to cover the Transwell chamber (Costar, Cambridge, MA, USA), the lower chamber was filled with 10 % FBS DMEM. Then the cells (5×104) were inoculated in the upper chamber and cultured for 24 h, with triplicate repeat. Then the cells were fixed with 4 % polyoxyethylene and stained with amethyst violet.
Protein extraction and Western blotting
Protein extraction and Western blotting experiments were carried out according to a previously described method. Each well of the 6-well plate was lysed with 60 μl RIPA lysis buffer and the cell lysate was centrifuged at 14 000 rpm for 15 min. The supernatant was collected and mixed with 2x SDS loading buffer for Western blotting analysis. Protein samples were analyzed by SDS-PAGE. Western blotting was performed using the standard procedures. Immunoreactive proteins were observed using the SuperSignal™ chemiluminescence assay.
In vivo analysis of tumor growth
For tumor growth assay, Saos-2 and MG63 cells were injected into the 8 w old nude mice. Once the tumor was formed, the tumor was measured every 2 d and the volume was calculated using the formula a*b2/2 [a:max, b:min]. After the mice were killed, the xenografted tumors were photographed and weighed and the RNA was extracted.
Luciferase reporter assay
The 3’UTR fragment of EBF2 containing miR-182- 3p binding sequence was cloned into TOP/FOP Flash reporter vector and the mutant plasmid was used as control. The cells were cultured in a 24-well plate and Fugene was used to the transfection of 250 ng TOP/ FOP Flash reporter plasmid and 50 ng Renilla plasmid as the internal reference. Two days after transfection, the cell lysate was collected to detect luciferase activity and TOP/FOP was used to measure the transcriptional activity.
Immunohistochemical analysis
Sections of formalin-fixed paraffin-embedded tissues were prepared for immunohistochemical analysis. Paraffin was removed from the tissue and the sections were hydrated by gradient concentration of ethanol. The sections were blocked with 5 % sheep serum for 60 min and then incubated with the antiEBF2 antibody at 4° overnight. On the next d, the signal was observed with 3’-diaminobenzidine.
Statistical analysis
All experiments were repeated at least 3 times and all data were expressed as mean±SD. GraphPad Prism 8.0 was used for statistical analysis. The difference between the 2 groups was analyzed by the t-test, while the multiple comparisons was analyzed by the single factor analysis of variance. *p<0.05 means the difference was statistically significant.
Results and Discussion
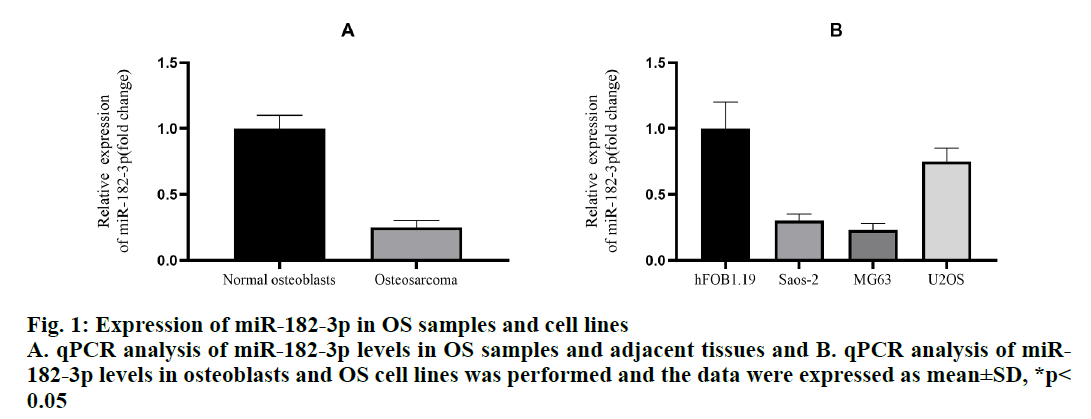
To evaluate the role of miR-182-3p in OS, the expression level of miR-182-3p in the clinical OS samples were determined. A total of 20 pairs of OS and adjacent tissues were examined and the results showed that the miR-182-3p decreased more than 75 % in OS (fig. 1A). In addition, the expression level of miR-182-3p in three OS cell lines and 1 human osteoclast line was detected. Compared to the human osteoblast cell line hFOB1.19, the expression of miR-182-3p in in the 3 OS cell lines decreased significantly (fig. 1B). In addition, ISH demonstrated that the expression of miR-182-3p was strong in normal osteoblasts, but hardly detectable in the OS samples. In OS cell lines, Saos-2 and MG63, miR-182-3p was downregulated more than 50 %, and in OS cell line U2OS, miR-182-3p was downregulated by about 25 %. Therefore, Saos-2 and MG63 were used to further study the role of miR-182-3p in OS.
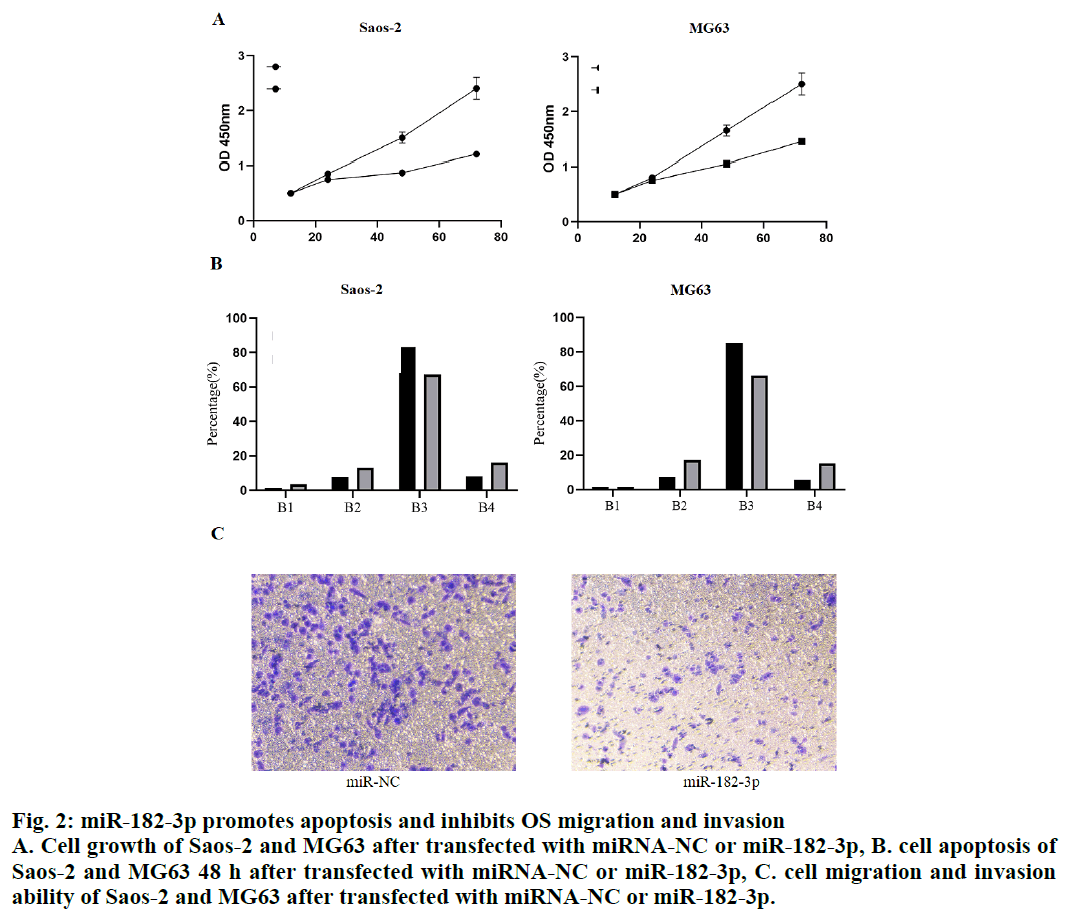
To understand the biological function of miR-182-3p in OS, miR-182-3p (50 nM) was transfected into Saos- 2 and MG63 cells. CCK8 assay was used to measure the number of cells at the specified time point after transfection. The results showed that the number of Saos-2 and MG63 cells decreased significantly after 48 h of transfection, indicating that the miR-182-3p was the tumor inhibitory microRNA of OS (fig. 2A). To further understand the role of miR-182-3p in OS, the apoptosis of Saos-2 and MG63 cells 48 h after transfection was analyzed. The results showed that when miR-182- 3p was overexpressed in Saos-2 and MG63 cells, the positive cells of Annexin V, and the double positive cells of Annexin V and PI increased sharply, which showed that the miR-182-3p induced apoptosis. At the same time, a slight increase in the necrotic cells was observed when OS cells were transfected with miR-182- 3p (fig. 2B). The Transwell assay was used to evaluate the migration and invasion ability of OS cells in vitro. The results showed that the ability of cell migration and invasion was significantly impaired when miR-182-3p was overexpressed in Saos-2 and MG63 cells (fig. 2C). Therefore, these data revealed that the miR-182-3p played a tumor inhibitory role in OS.
Figure 2: miR-182-3p promotes apoptosis and inhibits OS migration and invasion
A. Cell growth of Saos-2 and MG63 after transfected with miRNA-NC or miR-182-3p, B. cell apoptosis of
Saos-2 and MG63 48 h after transfected with miRNA-NC or miR-182-3p, C. cell migration and invasion
ability of Saos-2 and MG63 after transfected with miRNA-NC or miR-182-3p.
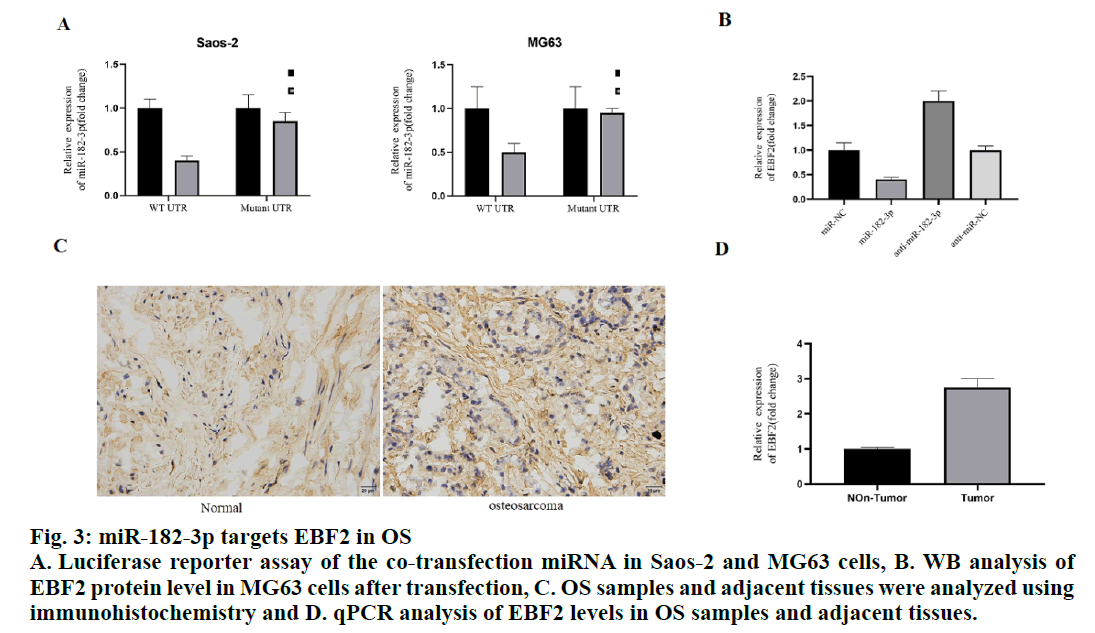
To study the mechanism of miR-182-3p inhibiting tumor growth, the online database for the target of miR-182-3p. TargetScan, miRanda, and PicTar were searched to predict the target of miR-182-3p. Among the predicted targets, EBF2 was found to be involved in the apoptosis of OS. Bioinformatic analysis showed that there was a strong binding between miR-182-3p and EBF2. To study whether miR-182-3p directly regulates EBF2, the wild-type EBF2 3 ‘UTR (WT UTR) and biding sites mutant EBF2 3’ UTR (mutant UTR) were co-transfected with miR-182-3p or control miRNA in Saos-2 and MG63 cells, respectively. Luciferase assay showed that the miR-182-3p inhibited WT UTR reporter activity in Saos-2 and MG63 cells. Importantly, miR-182-3p has no effect on mutant UTR, indicating that miR-182-3p binds EBF2 in vivo through the predicted binding site (fig. 3A).
Figure 3: miR-182-3p targets EBF2 in OS
A. Luciferase reporter assay of the co-transfection miRNA in Saos-2 and MG63 cells, B. WB analysis of
EBF2 protein level in MG63 cells after transfection, C. OS samples and adjacent tissues were analyzed using
immunohistochemistry and D. qPCR analysis of EBF2 levels in OS samples and adjacent tissues.
Then, the miR-182-3p and Antagomir were transfected into MG63 cells, respectively and the level of EBF2 mRNA was detected to further analyze the regulatory relationship between miR-182-3p and EBF2. The data showed that the EBF2 mRNA and protein levels were significantly inhibited in MG63 cells overexpressing miR-182-3p, while EBF2 mRNA and protein levels were significantly upregulated in MG63 cells with low expression of miR-182-3p (fig. 3B). These results suggested that the expression of EBF2 in OS patients needed to be studied. The immunohistochemical results showed that EBF2 was upregulated in OS samples compared to the adjacent tissues (fig. 3C). In addition, the level of EBF2 mRNA in OS samples with low expression of miR-182-3p was analyzed, which revealed that the level of EBF2 mRNA in this sample was significantly increased (fig. 3D), indicating that the miR-182-3p regulated the expression of EBF2 in human osteoblasts.
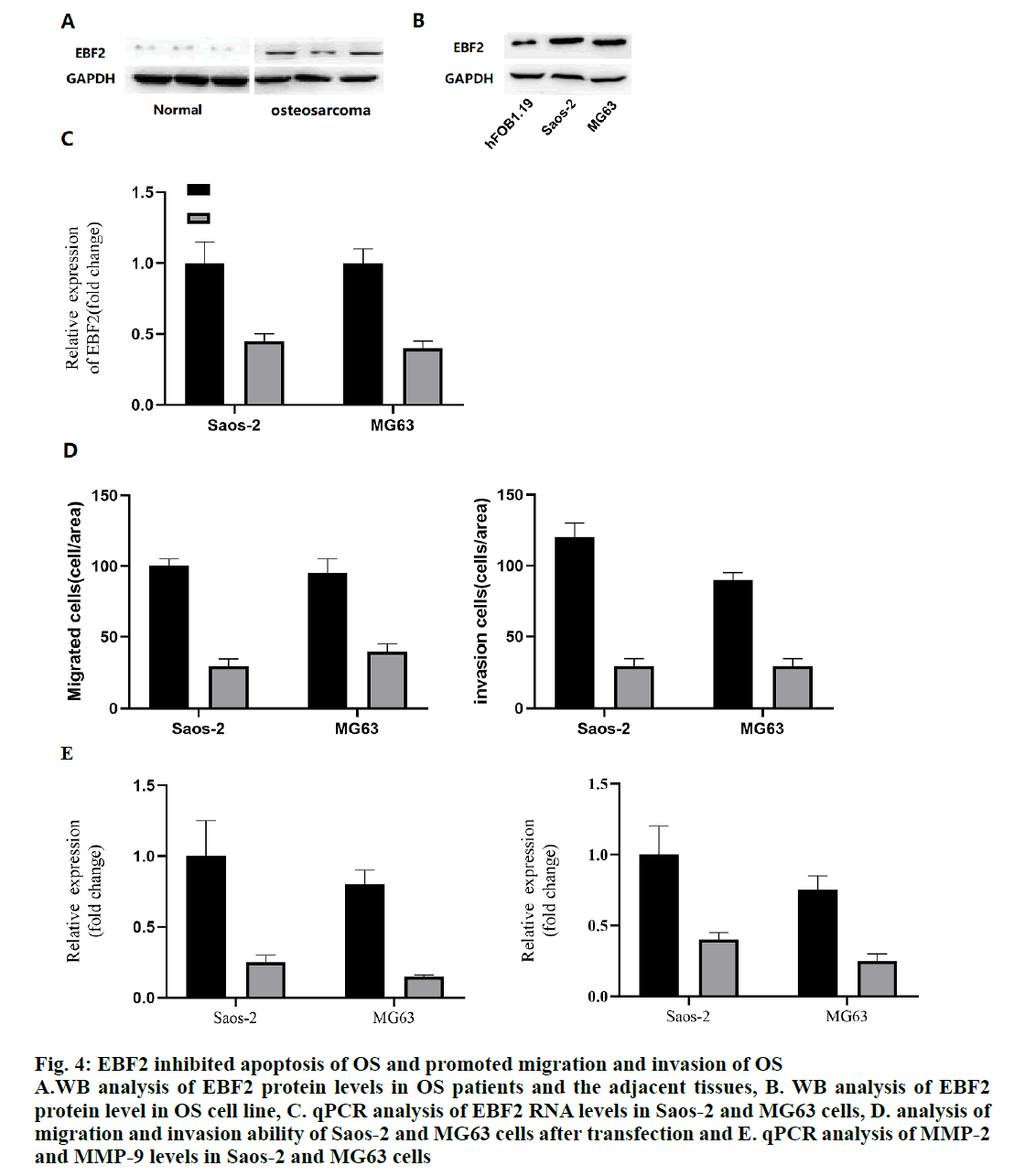
Because of the significant increase of EBF2 mRNA in OS patients, the protein level of EBF2 in OS samples were measured. As expected, the level of EBF2 protein in OS samples was significantly higher than that in the normal tissues (fig. 4A). In addition, these data also showed that the EBF2 protein levels were significantly upregulated in OS cell lines Saos-2 and MG63 (fig. 4B). Therefore, the expression of EBF2 in Saos-2 and MG63 cells was knocked down with siRNA and the results showed that EBF2 successfully interfered at both mRNA and protein levels (fig. 4C). The migration and invasion of OS cells transfected with EBF2 siRNA was analyzed to find that reduction in EBF2 significantly reduced the migration and invasion ability of Saos-2 and MG63 cells (fig. 4D), indicating that EBF2 is the downstream target of miR-182-3p. It also indicated that targeting miR-182-3p will certainly block the activity of EBF2 in OS cell lines, thus making miR- 182-3p a key regulator. In addition, the mRNA levels of MMP-2 (matrix metalloproteinase) and MMP-9 were also analyzed. The results showed that MMP-2 and MMP-9 in EBF2 knockout cells decreased significantly, indicating that the migration ability of the OS cell line changed (fig. 4E).
Figure 4: EBF2 inhibited apoptosis of OS and promoted migration and invasion of OS
A.WB analysis of EBF2 protein levels in OS patients and the adjacent tissues, B. WB analysis of EBF2
protein level in OS cell line, C. qPCR analysis of EBF2 RNA levels in Saos-2 and MG63 cells, D. analysis of
migration and invasion ability of Saos-2 and MG63 cells after transfection and E. qPCR analysis of MMP-2
and MMP-9 levels in Saos-2 and MG63 cells
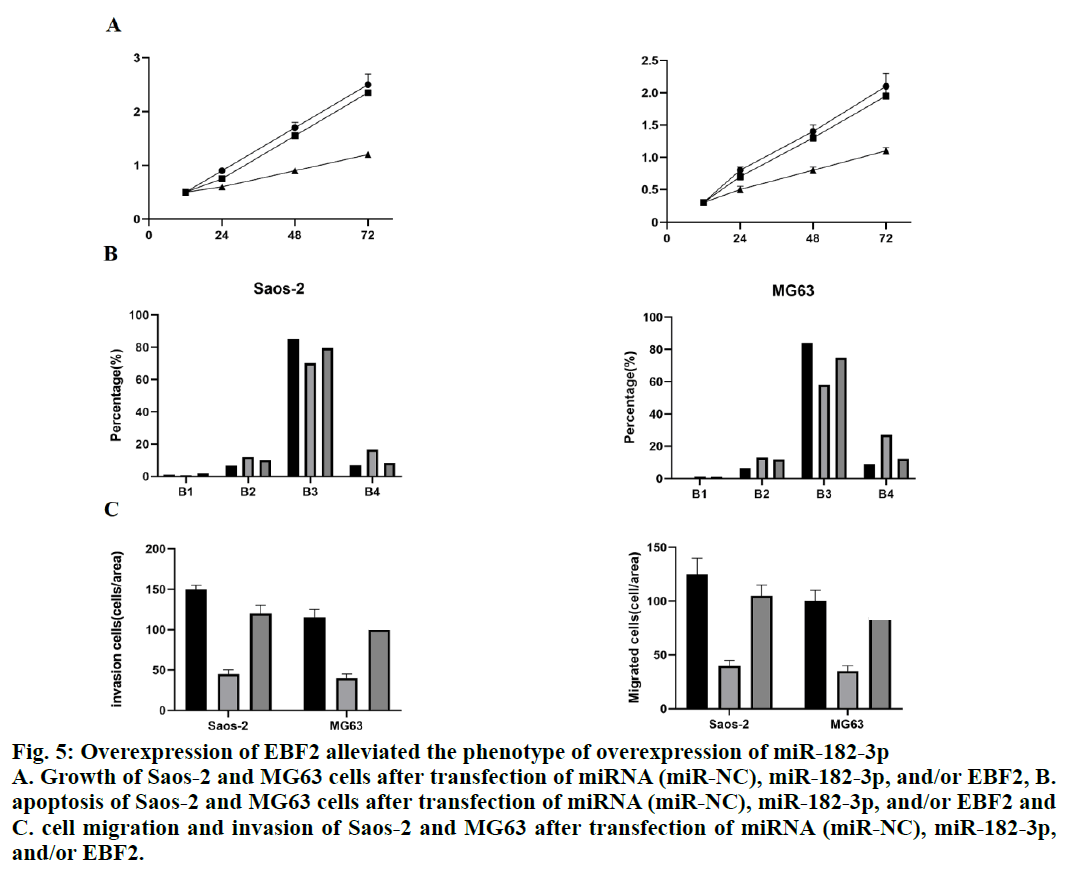
To verify the regulation between miR-182-3p and EBF2, miR-182-3p and the plasmid expressing EBF2 were co-transfected into Saos-2 and MG63 cells. The number of cells was analyzed by CCK8 assay. The results showed that the number of Saos- 2 and MG63 cells with overexpressing miR-182-3p decreased significantly, while the overexpression of EBF2 decreased the loss of cell number caused by the overexpression of miR-182-3p (fig. 5A), which showed that the miR-182-3p regulated cell growth by targeting EBF2. In addition, the co-expression of miR- 182-3p and EBF2 partially decreased the apoptosis of Saos-2 and MG63 cells induced by miR-182-3p alone (fig. 5B). The migration and invasion of Saos-2 and MG63 cells co-expressing miR-182-3p and EBF2 was also analyzed. The co-expression of miR-182-3p and EBF2 partially decreased the reduction of migration and invasion caused by the overexpression of miR- 182-3p alone. (fig. 5C), which showed that miR-182-3p played a role by targeting EBF2.
Figure 5: Overexpression of EBF2 alleviated the phenotype of overexpression of miR-182-3p
A. Growth of Saos-2 and MG63 cells after transfection of miRNA (miR-NC), miR-182-3p, and/or EBF2, B.
apoptosis of Saos-2 and MG63 cells after transfection of miRNA (miR-NC), miR-182-3p, and/or EBF2 and
C. cell migration and invasion of Saos-2 and MG63 after transfection of miRNA (miR-NC), miR-182-3p,
and/or EBF2.
Exploring the pathogenesis of OS, especially the mechanism of OS metastasis, is the basis for improving the survival rate of adolescent OS. The identification of differentially expressed genes was a feasible way to crack the molecular mechanism of OS. It was found that miR-182-3p was significantly downregulated in OS patients and OS cell lines. Previous studies have shown that miR-182-3p was low expressed in renal clear cell carcinoma[21], colorectal cancer[13], gastric cancer[22], thyroid papillary carcinoma[23], glioma[14], oral squamous cell carcinoma[15], prostate cancer[24], hepatocellular carcinoma[25], and laryngeal squamous cell carcinoma[16]. Combined with these results in OS, miR-182-3p showed a strong anticancer effect in many kinds of cancers. In addition, these data showed that overexpression of miR-182-3p in both OS cell lines inhibited the cell viability in vitro, further confirming the role of miR-182-3p in inhibiting the growth of cancer cells, which was consistent with the findings on other types of cancer[12-15,21-25]. Therefore, all these data showed that miR-182-3p could inhibit tumors. Importantly, apoptosis increased significantly when miR-182-3p was overexpressed in Saos-2 and MG63 cells, indicating that the presence of miR-182-3p could induce apoptosis in OS cells. Studies on prostate cancer, liver cancer, and thyroid papillary carcinoma have also shown that miR-182-3p can promote apoptosis.
The main threat to OS was the distant metastasis. Therefore, it is important to distinguish the genetic changes in OS metastasis. The present data showed that the overexpression of miR-182-3p suppresses the migration and invasion of OS. Transwell assays results showed that miR-182-3p can be used as a predictor to prevent the local OS from developing into the metastatic OS, which was consistent with the existing results. That is, miR-182-3p inhibits the migration and invasion of OS[12-16,25]. However, it is not clear whether knockout of miR-182-3p in osteoblasts will lead to OS, because miR-182-3p may not be the root cause of OS. It is meaningful to detect the cellular changes in normal cells when miR-182-3p was knocked out.
Most importantly, it was found EBF2 was a new target of miR-182-3p. It was reported that EBF2 plays an important role in osteoclast differentiation and OS apoptosis. These data showed that in the OS cell line, miR-182-3p directly bind to the untranslated region of EBF2 and regulated the mRNA level of EBF2, indicating that miR-182-3p regulated OS through EBF2 protein. In addition, these data also showed that EBF2 was overexpressed in OS tissues, while the level of miR-182-3p was suppressed, indicating that EBF2 has a carcinogenic effect. The Immunohistochemistry and western blot results showed an increase in EBF2 in OS patients and cell lines. Elevated mRNA and protein levels of EBF2 were also observed in a group of OS patients who received chemotherapeutic[18]. In addition, present data also showed that EBF2 was required for OS migration. Previous studies have shown that EBF2 activates the osteoprotegerin promoter, which was synergistic with the Wnt/beta-catenin pathway. Current data showed that the overexpression of miR- 182-3p promoted apoptosis, while the co-expression of miR-182-3p and EBF2 weakened this pro-apoptotic effect, indicating that miR-182-3p and EBF2 were functionally related. In addition, the supplement of EBF2 also reduced the inhibition of migration and invasion caused by the overexpression of miR-182-3p, indicating that EBF2 was also involved in the transition from epithelial cells to mesenchymal cells mediated by miR-182-3p.
Authors’ Contributions
Yaxing Li conceived and designed the experiments. Haitao Jiang and Fangyong Lou performed the experiments. Wen Guo analyzed the data and wrote the paper.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81302336).
Conflict of Interests
The authors declared no conflicts of interest.
References
- Gianferante DM, Mirabello L, Savage SA. Germline and somatic genetics of osteosarcoma- connecting aetiology, biology and therapy. Nat Rev Endocrinol 2017:480-91.
- Ji Q, Xu X, Li L, Goodman BS, Bi W, Xu M, et al. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis 2017;8:e3103.
- Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis 2016;7:e2389.
- Zhou W, Hao M, Du X, Chen K, Wang G, Yang J. Advances in targeted therapy for osteosarcoma. Discov Med 2014;17:301-7.
- Kushlinskii N E, Fridman M V, Braga E A. Molecular Mechanisms and microRNAs in Osteosarcoma Pathogenesis. Biochemistry 2016;81:315-28.
- Blakaj A, Lin H. Piecing together the mosaic of early mammalian development through microRNAs. J boil Chem 2008;283:9505-08.
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008;18: 997-1006.
- Wang C, Jing Q. Non-coding RNAs as biomarkers for acute myocardial infarction. Acta pharmacol Sin 2018;39:1110-19.
- Yao ZS, Li C, Liang D, Xiang X B, Tang JJ, Ye L Q, et al. Diagnostic and prognostic implications of serum miR-101 in osteosarcoma. Cancer Biomark 2018;22:127-133.
- Kim YM, Jin JJ, Lee SJ, Seo TB, Ji E. Treadmill exercise with bone marrow stromal cells transplantation facilitates neuroprotective effect through BDNF-ERK1/2 pathway in spinal cord injury rats. J Exerc Rehabil 2018;14:335-340.
- Goradel NH, Mohammadi N, Haghi‐Aminjan H, Farhood B, Negahdari B, Sahebkar A. Regulation of tumor angiogenesis by microRNAs: State of the art. J Cellular Physiol 2019;23:1099-110.
- Wang C, Jing J, and Cheng L. Emerging roles of non-coding RNAs in the pathogenesis, diagnosis and prognosis of osteosarcoma. Invest New Drugs 2018;36:1116-32.
- Lv S, Guan M. miRNA-1284, a regulator of HMGB1, inhibits cell proliferation and migration in osteosarcoma. Biosci Rep 2018;38.
- Huang J, Liang Y, Xu M, Xiong J, Wang D, Ding Q. MicroRNA‑124 acts as a tumor‑suppressive miRNA by inhibiting the expression of Snail2 in osteosarcoma. Oncol lett 2018;15:4979-87.
- Clough E, Barrett T. The gene expression omnibus database. InStatistical genomics 2016;1418:93-110.
- Pathan M, Keerthikumar S, Ang CS, Gangoda L, Quek CYJ, Williamson AN, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015;15:2597-601.
- Su WL, Kleinhanz RR, Schadt EE. Characterizing the role of miRNAs within gene regulatory networks using integrative genomics techniques. Mol Syst Biol 2011;7:490.
- Chen G, Wang Q, Yang Q, Li Z, Du Z, Ren M, et al. Circular RNAs hsa_circ_0032462, hsa_circ_0028173, hsa_circ_0005909 are predicted to promote CADM1 expression by functioning as miRNAs sponge in human osteosarcoma. PLoS One 2018;13:e0202896.
- Garcia AP, Zalacain M, Folio C, Zandueta C, Sierrasesúmaga L, San Julián M, et al. Profiling of Chemonaive Osteosarcoma and Paired-Normal Cells Identifies EBF2 as a Mediator of Osteoprotegerin Inhibition to Tumor Necrosis Factor–Related Apoptosis-Inducing Ligand–Induced Apoptosis. Clin Cancer Res 2009;15:5082-91.
- Xu J, Li D, Cai Z, Zhang Y, Huang Y, Su B, et al. An integrative analysis of DNA methylation in osteosarcoma. J Bone Oncol 2017;9:34-40.
- Yang Z, Chen Y, Fu Y, Yang Y, Zhang Y, Chen Y, et al. Meta-analysis of differentially expressed genes in osteosarcoma based on gene expression data. BMC Med Genet 2014;15:80.
- Gu HJ, Zhou B. Focal adhesion kinase promotes progression and predicts poor clinical outcomes in patients with osteosarcoma. Oncol lett 2018;15:6225-32.
- Chen J, Liu C, Yang QQ, Ma RB, Ke Y, Dong F, et al. Isoliquiritigenin suppresses osteosarcoma U2OS cell proliferation and invasion by regulating the PI3K/Akt signalling pathway. Chemotherapy 2018;63:155-61.
- Bulla SC, Badial PR, Silva RC, Lunsford K, Bulla C. Platelets inhibit migration of canine osteosarcoma cells. J Comp Pathol 2017;156:3-13.
- Shi Z, Zhou H, Pan B, Lu L, Wei Z, Shi L, et al. Exploring the key genes and pathways of osteosarcoma with pulmonary metastasis using a gene expression microarray. Mol Med Rep 2017;16:7423-31.