- *Corresponding Author:
- K. Lu
School of Material and Chemical Engineering, Henan Institute of Engineering, Zhengzhou 451191, China
E-mail: lukui126@126.com
| Date of Submission | 21 Dec 2014 |
| Date of Revision | 18 Feb 2016 |
| Date of Acceptance | 18 Apr 2016 |
| Indian J Pharm Sci, 2016;78(2):278-282 |
Abstract
Peptide transporter 2 is a high affinity and low capacity transporter, that has recently been found in the mammalian kidney, brain, lung, and mammary glands, which is known to transport dipeptides and tripeptides and to reabsorb small peptides generated by luminal peptidases in the kidney. In addition, peptide transporter 2 can transport peptide-like drugs and play important roles in the transshipment and positioning of drugs. In this paper, an E.coli BL21(DE3) containing pET30a(+)-hPepT2(560-663) recombinant plasmid was obtained by using PCR, double restriction enzyme digestion, ligation, and transformation. Then, the effect of changing both the induction time and induction dose using Isopropyl β-D-1-Thiogalactopyranoside (IPTG) on the bacterial expression of hPepT2(560-663) was detected by SDS-PAGE electrophoresis, the optimum expression conditions (at 37°) were identified as: 3 h and a working concentration of 0.5 mM Isopropyl β-D-1-Thiogalactopyranoside. Finally, we purified the target protein hPepT2(560-663) via Sephadex G-75 gel chromatography. These results will facilitate in future studies on the nature and function of the protein.
Keywords
Prokaryotic expression, oligopeptide transport protein, hPepT2, bacterial induction
The human oligopeptide transporter (hPepT2) is primarily expressed in the kidney [1-5] and has been found to transport many dipeptides and tripeptides left behind by luminal peptidases, and to resorb some peptidomimetic drugs, such as β-lactam antibiotics, angiotensin converting enzyme inhibitor, renin inhibitor, and thrombin element inhibitor [6-8] due to its high-affinity transport characteristics. In addition, hPepT2 has a wide distribution throughout the body, such as the enteric nervous system, brain, lung,9 mammary glands, spleen, tumor cell, and bronchial epithelial cells [9-11], where it plays important roles in transporting and positioning of drugs [7].
The hPepT2 is an electrogenic transporter that can be energized by the Na+/H+ exchange system, thus allowing the co-transport of small peptides from the extracellular to the intracellular portions of the intestine and other tissues [12,13]. According to reports, hPepT2 has been predicted to contain 12 transmembrane domains (TMDS) with a large extracellular loop between TMDS 9 and 10 [14,15]. Furthermore, TMDS 1-6 are thought to decide the substrate affinity, while TMDS 7-9 are substrate binding domains, and the function of TMDS 10-12 are unknown [16]. Unfortunately, it has proven difficult to express the whole protein in vitro because hPepT2 contains a high content of rare codons (12%).
However, hPepT2(560-663) is a fragment isolated from TMDS 10-11 of hPepT2 that has a relatively low percentage of rare codons. As such, we constructed a prokaryotic expression plasmid pET30a(+)- hPepT2(560-663), and transformed it into E.coli BL21(DE3) for the expression of hPepT2(560-663). Through separation and purification, we obtained the target protein, which can now be used to study the nature and function of hPepT2.
The MyCycler PCR and UV gel imager were both purchased from BIO-RAD Company (USA), while the electrophoresis apparatus (DYY-8C), electrophoresis tank (DYCP-31C) and (DYCZ-28C) were obtained from Beijing Liuyi Instrument factory. The ultrasonic cell crusher was from Ningbo Scientz Biotechnology Co., Ltd (Ningbo, China).
The primers were synthesized by Sangon Biotech Co., Ltd (Shanghai, China) and the plasmid containing PepT2 gene was obtained from OriGene. Ampicillin, kanamycin, E. coli BL21(DE3), isopropyl β-D- 1-thiogalactopyranoside (IPTG), restriction endonucleases NdeI and BamH I, lysozyme, and T4 DNA ligase were all purchased from TaKaRa Biotechnology Co., Ltd (Dalian, China). DNA Marker I, 1 Kbp DNA Ladder Marker, and the blue prestained protein Marker (W) were purchased from Beijing Biomed Co., Ltd (Beijing, China). The TIANprep mini plasmid kit was from Tiangen Biotech Co., Ltd (Beijing, China) and the QIAquick PCR purification kit was purchased from QIAGEN (Germany). Finally, Sephadex G-75 medium was obtained from Solarbio (Beijing, China).
According to the selected hPepT2(560-663) gene fragment, appropriate upstream 5'-GGAATTCCATATGCACTGTAGAACAGAAGAT- 3' and downstream 5'-CGCGGATCCCTATGCCACAACAAGCACGATGA- 3' were designed according to the design principles of the primer: amplified by PCR, verified by 2% agarose gel electrophoresis, and purified through DNA purification kit.
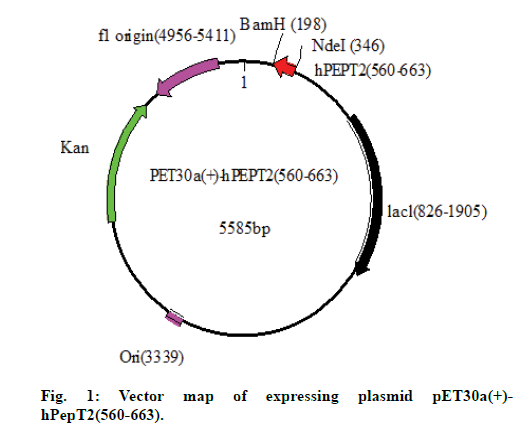
The purified PCR product and pET30a(+) plasmid were double enzyme digested by NdeI and BamHI, respectively, at 37° for 10 h, then the hPepT2(560-663) gene and the large fragment of pET30a(+) were extracted using a gel extraction kit and ligated by T4 ligase at 16° for 18 h. Subsequently, 5 μl of the ligation mixture was transformed into competent cells of E. coli BL21(DE3), which were then seeded onto well plates and cultured under standard conditions of 37° for 24 h. A single colony was selected and inoculated into a 5 ml LB liquid medium containing Kan for minipreparation and sequencing (figs. 1 and 2).
The competent cells of BL21(DE3)/pET30a(+)- hPepT2(560-663) were inoculated into a liquid LB medium (50 μg/ml kan), cultured for 12 h, then 100 μl of the culture medium was seeded into 10 tubes containing 5 ml LB (50 μg/ml kan) and shaken until the 600 nm absorbance value reached 0.5. Finally, the induced dose and induced time were used as the variables for optimizing the expression condition of hPepT2(560-663).
We added 1000 μl E.coli BL21(DE3)/pET30a(+)- hPepT2(560-663) to 50 ml LB (50 μg/ml kan), and allowed it to shake for 12 h at 37° to activate the bacterium, before it was added to 1 l LB of liquid medium, based on a percentage of 100 μl/5 ml. After shaking the subsequent culture for 3 h, we next added IPTG at a working concentration of 0.4 mM to induce the bacteria, and shook the culture at 37° for another 3 h. This solution was then centrifuged (5000 r/min for 5 min at 4°), the cells were collected, washed three times by 4-5 times volume of PBS (phosphate buffered saline) buffer (50 mM), and suspended in 60 ml PBS solution. Protease inhibitors and lysozyme were added to the solution and then the cells were immediately ruptured by supersonic wave in an ice bath. Supernatant and precipitate were detected by SDS-PAGE electrophoresis after centrifugation. Samples containing the protein were purified through sephadex G-75 medium column. The eluent was collected by automatic fraction collector and detected by UV spectrophotometer and SDS-PAGE electrophoresis.
Results revealed that the size of the purified PCR product was between 300 bp and 400 bp (fig. 3, Lane 2); the length of this purified PCR product was consistent with the hPepT2(560-663) gene. The band in lane 3 of fig. 3 showed the result of the double digestion of the PCR product. There are no obvious differences between the bands of lane 2 and lane 3.
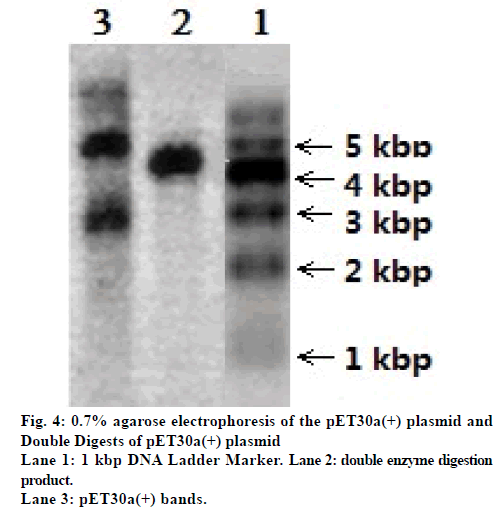
Fig. 4 shows the 0.7% agarose electrophoresis of the pET30a(+) plasmid and its double digestion product. The length of the pET30 was 5448 bp. The three bands in lane 3 of fig. 4 reveal the three conformational states of the pET30a(+) plasmid: supercoiled plasmids, linear plasmids, and ringopened plasmids. The results of the double digestion of pET30a(+) are shown in fig. 4, lane 2; only one band compared to the three bands in lane 3, revealing that the double digestion of the linear pET30a(+) plasmid was completed.
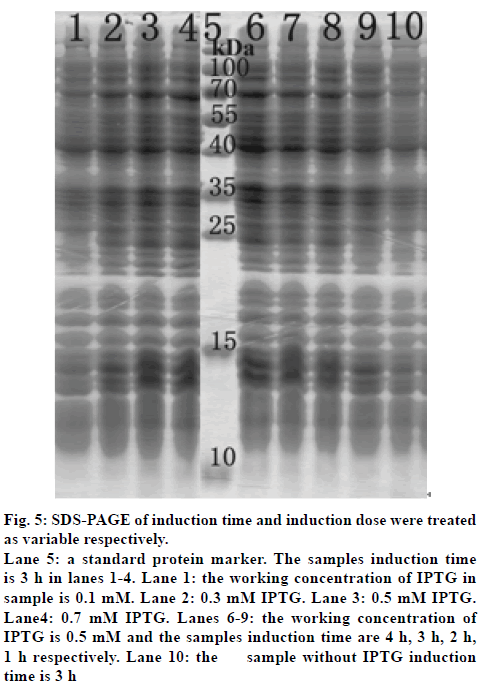
Polyacrylamide gel electrophoresis (SDS-PAGE) was then used to detect the influence of the induction dose and induction time on the expression of hPepT2(560-663) (fig. 5). The bacteria were induced for 3 h using working concentrations of 0.1, 0.3, 0.5 and 0.7 mM IPTG in lanes 1-4, respectively. For the determination of the optimal induction time, we used a working concentration of IPTG of 0.5 mM, and the induction times of the samples were 4, 3, 2 and 1 h in lanes 6-9, respectively. Lane 10 was the control lane that lacked an induction dose or time.
Figure 5: SDS-PAGE of induction time and induction dose were treated
as variable respectively.
Lane 5: a standard protein marker. The samples induction time
is 3 h in lanes 1-4. Lane 1: the working concentration of IPTG in
sample is 0.1 mM. Lane 2: 0.3 mM IPTG. Lane 3: 0.5 mM IPTG.
Lane4: 0.7 mM IPTG. Lanes 6-9: the working concentration of
IPTG is 0.5 mM and the samples induction time are 4 h, 3 h, 2 h,
1 h respectively. Lane 10: the sample without IPTG induction
time is 3 h
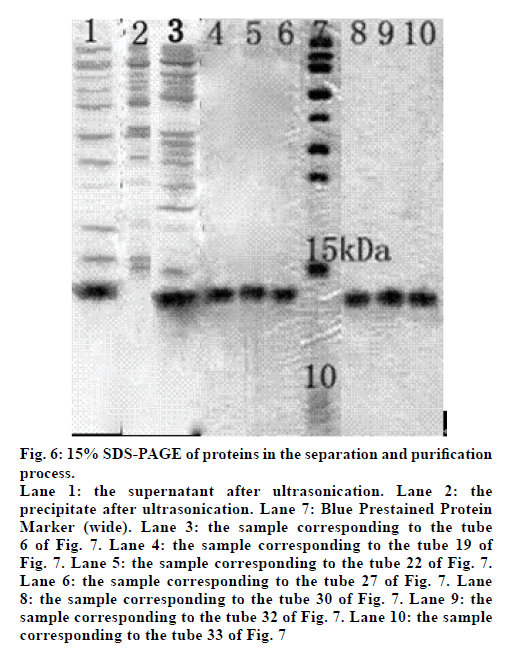
We next induced the expression of the E. coli BL21(DE3) containing recombinant plasmid pET30a(+)-hPepT2(560-663). The cells were collected and suspended in PBS solution, and then the soluble protein in the supernatant was separated with the precipitate after ultrasonic cytolysis and centrifugation. Lanes 1 and 2 of fig. 6 show the bands of the supernatant and precipitate, respectively. The supernatant band was 10-15 kDa, corresponding to the target protein with a theoretical molecular weight was 11.5 kDa, while the precipitate did not contain the target protein (fig. 6).
Figure 6: 15% SDS-PAGE of proteins in the separation and purification
process.
Lane 1: the supernatant after ultrasonication. Lane 2: the
precipitate after ultrasonication. Lane 7: Blue Prestained Protein
Marker (wide). Lane 3: the sample corresponding to the tube
6 of Fig. 7. Lane 4: the sample corresponding to the tube 19 of
Fig. 7. Lane 5: the sample corresponding to the tube 22 of Fig. 7.
Lane 6: the sample corresponding to the tube 27 of Fig. 7. Lane
8: the sample corresponding to the tube 30 of Fig. 7. Lane 9: the
sample corresponding to the tube 32 of Fig. 7. Lane 10: the sample
corresponding to the tube 33 of Fig. 7
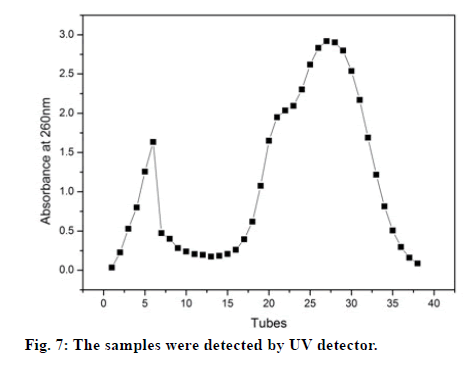
The supernatant containing the target protein was purified by using sephadex G-75 gel filtration chromatography. The samples were collected in 1 ml/min/tube via an automatic fraction collector, and the absorbance values of the samples at 260 nm were detected by using UV detector. The percent of amino acids containing phenyl in the hPepT2(560-663) sequence was found to only be 0.11%, leading to the very low absorbance of protein observed at 280 nm. The fig. 7 shows the results of the samples initially detected by the UV detector. The samples in tubes 3-7 and 17-35 contained many proteins due to the high absorbance value observed at 260 nm. Therefore, the samples in tubes 6, 19, 22, 27, 30, 32 and 33 were further evaluated by SDS-PAGE, the results correspond to lanes 3, 4, 5, 6, 8, 9 and 10 in fig. 6. Furthermore, this data revealed that the samples in the first peak were a mixture of the whole protein while the samples collected from the second peak were the target protein.
The target gene fragment and pET30a(+) large fragment with the same cohesive ends were obtained by restriction enzyme digestion, and ligated together using T4 ligase. The products of the ligation reaction were transformed into E. coli BL21(DE3) competent cells. The recombined plasmid from E.coli BL21(DE3) was sequenced, and the result revealed that the gene fragment between the restriction sites was consistent with the hPepT2(560-663) gene. As such, this was determined to be the target protein and expressed further.
We next determined optimal conditions for producing a target protein with the proper theoretical molecular weight of hPepT2 (560-663) of 11.5 kDa. Fig. 5 shows the results of the SDS-PAGE, in which the induction dose (lanes 1-4) and induction time (lanes 6-9) were treated as the variables. We found that the production of hPepT2(560-663) increased as the working concentration of IPTG increased from 0.1 mM to 0.7 mM. Lighter bands in lane 1 indicated that the expression of the protein was incomplete when IPTG was 0.1 mM. Meanwhile, the smeared bands in lane 4 (working concentration of IPTG was 0.7 mM) made it difficult to separate the target protein. As such, the bands in lane 3, produced using 0.5 mM of IPTG, were determined to yield the highest proportion of the target protein. As for the optimal induction time, the production of hPepT2(560-663) decreased with a reduction of the induction time from 4 h to 1 h. However, with an induction time of 4 h, the protein bands were heavier and blurred, indicating that this lengthy induction time resulted in the poor selective expression of hPepT2(560-663). After comprehensively analyzing the bands in lanes 6-9, we determined that the optimal induction time was 3 h. Interestingly, the light bands found in the control lane 10 suggested that the target protein could be expressed at a very low level even without IPTG induction, but not to an optimal degree. Taken together, the optimal induction condition of hPepT2 (560-663) was determined to be 0.5 mM IPTG for 3 h at 37°.
The bands in lane 3 of fig. 6 suggest that the first absorption peak seen in fig. 7 is a mixture of the whole protein. Meanwhile, the bands in lane 4-10 of fig. 6 suggest that the second absorption peak seen in fig. 7 was actually the target protein. As such, we were able to obtain a pure protein following sephadex G-75 gel filtration chromatography.
In conclusion, we constructed a recombinant plasmid pET30a(+)-hPepT2 (560-663) and transformed it into E.coli BL21(DE3) to obtain the expression system of BL21(DE3)/pET30a(+)-hPepT2 (560- 663). The expression condition of hPepT2 (560- 663) was optimized by altering the induction time and induction dose. Under optimum conditions, hPepT2 (560-663) was purified via sephadex G-75 gel chromatography for use in further studies related to the nature and function of protein.
Financial Assistance
This work was supported by National Natural Science Foundation of China (No.21172054, No.21301050), Innovation Scientists and Technicians Troop Construction Projects of Henan province (No.104200510022) and Innovation Scientists and Technicians Troop construction projects of Zhengzhou city (No.10LJRC174).
Conflict of Interests
None declared.
References
- Ramamoorthy S, Liu W,Ma YY,Yang-Feng TL,Ganapathy V, Leibach FH. Proton/peptide cotransporter (PEPT2) from humankidney: functional characterization and chromosomallocalization. Biochim Biophys Acta 1995;1240:1-4.
- Shen H, Smith DE, Brosius FC. Developmental Expression of PEPT1and PEPT2 inRat Small Intestine, Colon, and Kidney. Pediatr Res 2001;49:789-95.
- Li J, Wang YJ. Significant role of transporters in drug renal transport. Chin J Clin Pharm 2006;102:303-6.
- Cang J, Liu KX. The role of oligopeptide transporter in drug transport in kidney and its activity regulation. Drugs and Clinlc 2010;7:49-52.
- Xu Q, Wang C,Meng Q, Liu Q, Sun P, Sun H, et al. The oligopeptide transporter 2-mediated reabsorption of entecavir in rat kidney. Eur J Pharm Sci 2014;52:41-7.
- Biegel A, Knutter I,Hartrodt B, Gebauer S, Theis S, Luckner P, et al.The renal type H+/peptide symporterPEPT2:structure-affinity relationships. Amino Acids 2006;31:137-56.
- Zhao DX, Xue YL, Lu K, Wu JZ. Role of Oligopeptide Transporter PepT2 in Drug Transportation. Chin J Biochem Mol Biol 2010;26:1-8.
- Guo X, Meng Q, Liu Q, Wang C, Sun H, Kaku T, et al.Construction, identification and application of HeLa cells stably transfected with human PEPT1 and PEPT2. Peptides 2012;34:395-403 .
- Groneberg DA, Doring F, Theis S, Nickotaus M, Fischer A, Daniel H. Peptide transport in the mammary gland: expression and distribution of PEPT2 mRNA and protein. Am J Physiol Endocrinol Metab 2002;282:E1172-9.
- Daniel H, Rubio-Aliaga I. An update on renal peptide transporters. Am J Physiol Renal Physiol 2003;284:F885-92.
- Takano M, Horiuchi T, Sasaki Y, Kato Y, Nagai J, Yumoto R. Expression and function of PEPT2 during transdifferentiation of alveolar epithelial cells. Life Sci 2013;93:630-6.
- Daniel H. Function and molecular structure of brush border membrane peptide/H+ symporters. J Membrane Biol 1996;154:197-203.
- Pedretti A, De Luca L, Marconi C, Regazzoni L, Aldini G, Vistoli G. Fragmental modeling of hPepT2 and analysis of its binding features by docking studies and pharmacophore mapping. Bioorg Med Chem 2011;19:4544-51.
- Saito H, Terada T, Okuda M, Sasaki S, Inui K. Molecular cloning and tissue distribution of rat peptide transporter PEPT2. Biochim Biophys Acta 1996;1280:173-7.
- Rubio-Aliaqa I, Boll M, Daniel H. Cloning and characterization of the gene encoding the mouse peptide transporter PEPT2. Biochem Biophys Res Commun 2000;276:734-41.
- Daniel H, Kottra G. The proton oligopeptide cotransporter family SLC15 in physiology and pharmacology. Pflugers Arch 2004;447:610-8.