- *Corresponding Author:
- N. Roy
Centre of Pharmacoinformatics, National Institute of Pharmaceutical Education and Research, Sector 67, S. A. S. Nagar, Punjab - 160 062, India
E-mail: nilanjanroy@niper.ac.in
| Date of Submission | 14 June 2006 |
| Date of Revision | 9 July 2007 |
| Date of Acceptance | 2 September 2007 |
| Indian J Pharm Sci, 2007, 69 (5): 609-615 |
Abstract
The low success rate of converting lead compounds into drugs owing to unfavorable pharmacokinetic parameters has evoked a renewed interest in understanding more clearly what makes a compound drug-like. This article reviews a number of computational techniques for identifying drug-like molecules, ranging from simple counting schemes to sophisticated machine learning techniques such as neural networks, along with their application and challenges.
Keywords
Drug-like, non-drug likes, comprehensive medicinal chemistry (CMC), available chemical databases (ACD) and modern drug data report (MDDR)
The phrase ‘drug–like’ is becoming more widespread. According to Walters and Murcko, drug-like compounds are molecule which contain functional groups and/or have physical properties consistent with the majority of known drugs, and hence can be inferred as compounds which might be active biologically or might show therapeutic potential [1,2]. Lipinski defines those compounds as ‘drug-like’, which have sufficiently acceptable ADME/T properties to survive through the Phase I clinical trials [3]. However, drugs as well as drug-like compounds are distributed extremely meagerly through chemical space, which is estimated to contain 1040 to 10100 molecules. For a drug, properties like synthetic ease, stability, oral availability, good pharmacokinetic properties, lack of toxicity and minimum addictive potential are of utmost importance. Many of these properties depend on the inherent biological and physicochemical parameters of the molecule; however the complex structure of the whole drug molecule makes correlating attempts difficult. One interesting approach is to study the parameters of the fragments of whole drug molecule. The present review explores what makes a molecule drug like, the methods for prediction of drug likeness, along with notes on currently available drug like and non drug like databases.
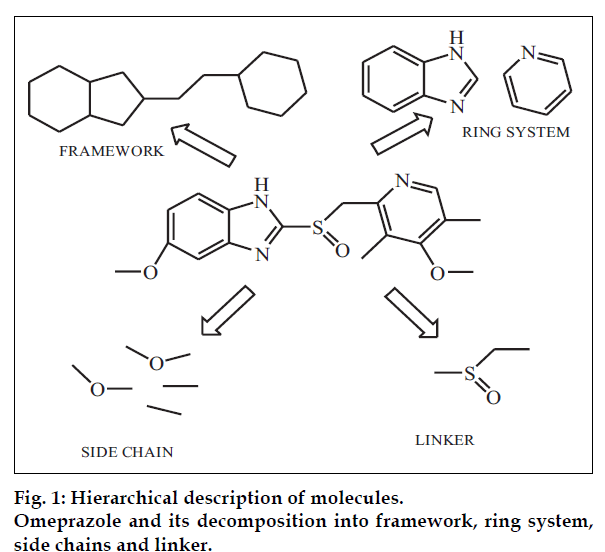
In order to understand the concept of ‘drug-likeness’, it is necessary to understand the common features present in a drug molecule. Bemis and Murcko have performed a general analysis of the shapes of molecules with the help of a simple graph approach that considers only two-dimensional structures [4,5]. According to this approach any molecule can be dissected into four units: ring, linker, side chain and finally framework (fig. 1). Ring system is the cyclic part within the graph representation of molecule and sharing an edge (a connection between two atoms or a bond). E.g. omeprazole has pyridine and benzimidazoline ring systems. Linker atoms form the direct path connecting the two rings. Side chain atoms are any non-ring, non-linker atoms such as the four side chains in omeprazole; two single-atom side chains and two, two-atom side chains. Finally framework is defined as the union of ring systems and linkers in a wire-frame.
In order to determine what makes a molecule drug-like one must begin by examining the molecules in hand with sets of known drugs and with sets of non-drug (or molecules assumed to be non-drug). Methods to identify drug-like molecules are based on their ability to distinguish known drugs from non-drugs in the set of compounds by comparing with one or more of the following widely available drug databases.
The comprehensive medicinal chemistry (CMC) database
CMC is derived from the drug compendium in Pergamon’s Comprehensive Medicinal Chemistry. The database contains more than 7000 compounds, used or tested as medicinal agents in humans.
The modern drug data report (MDDR)
MDDR contain more than 100 000 drugs launched or under development. These compounds are referenced in the patent literature, conference proceeding and other sources.
The world drug index (WDI)
The WDI 1997 contains 51 596 compounds, of these, 7570 have been assigned a United State Adapted Name (USAN) and 6307 have been assigned an International Nonproprietary Name (INN) combining these gives 8323 unique compounds, of which 3515 have an entry in the indication and usage (IU) field.
Available chemical databases (ACD)
The ACD is a collection of more than 300 000 commercially available compounds. The set of non-drugs is typically created by selecting random compounds from the available chemical databases [4].
Drug-likeness is mostly a statistics of descriptors derived from databases of other compounds. It can therefore, be used to evaluate the drug-likeness of other compounds and selection from screening libraries such as combinatorial libraries or virtual libraries rather than that of a single compound. For a broader view on the subject of drug-likeness and its use in library design the reader is suggested to go through recent review articles [6-11].
Methods Of Drug-Likeness Prediction
Simple counting methods
Simple counting method involved correlation of molecular descriptors or properties implicit to drug likeness. Properties such as oral bioavailability or membrane permeability have often been correlated to log P, molecular weight (MW) and number of hydrogen bond acceptors and donors in a molecule. Simple counting methods include “Lipinski’s rule of 5” and its implementation in prediction of the drug likeness, along with extended concept of Ghose and Opera.
The “rule of 5” (RO5) provides a heuristic guide for determining, if a compound will be orally bioavailable. The rules were derived from the analysis of 2245 compounds with a USAN or INN and the entries in the indication and usage field of the database were included in the analysis. The assumption was that compounds meeting these criteria had entered human clinical trials and therefore must have possessed many of the desirable characteristics of drugs [12]. The RO5 states that molecules showing poor absorption or permeation are more likely to have- more than 5 H-bond donors, MWT over 500, log P over 5 and, more than 10 H–bond acceptors. However there are plenty of examples available for RO5 violation amongst the existing drugs. Majority of violations come from antibiotics, antifungals, vitamins, and cardiac glycosides. Still these classes of compound are orally bioavailable because they possess groups which act as substrates for transporters. If a compound fails the RO5 there is a high probability that oral activity problems will be encountered. However, passing the RO5 is no guarantee that a compound is drug-like. Moreover, the “RO5” says nothing about specific chemistry or structural features found in drugs or non-drugs. Ghose et al., extended this work by characterizing 6304 compounds (taken from the CMC Database) based on computed physicochemical properties. They established qualifying ranges which cover more than 80% of the compounds in the set. Ranges were established for A log P (20.4 to 5.6) [13,14], molar refractivity (40 to 130), molecular weight (160 to 480), and number of atoms (20 to 70). A similar study was performed by Oprea [15], who carried out a Pareto analysis of compounds from MDDR, CMC, Current Patents Fast-alert, New Chemical Entities and ACD. The Pareto analysis was used to determine property ranges covering 80% of the compounds in a particular database. In addition to the properties discussed above, Oprea also considered counts of ring bonds, rigid bonds and rotatable bonds. Rotatable bond count has been widely used filter following the observation that greater than 10 rotatable bonds correlates with decreased oral bioavailability in rat. An analysis of small drug-like molecules suggests a filter of log D, where the values in the range of 0 to 3 shown to enhance the probability of good intestinal permeability.
Knowledge–based methods
Knowledge-based methods are based upon the concept of intrinsic binding energies and scoring of structural fragments. In this method mainly functional groups are used to classify drug and non-drug like molecules based on different scoring functional group fragment. Andrews et al., used a set of 200 drug molecules to derive a set of intrinsic binding energies for the 10 functional groups as shown in Table 1 [16,17]. The inherent binding of small molecules was then estimated by summing the intrinsic binding energies and subtracting an entropic factor; the method had been widely used previously for the reagent selection rather than drug likeness prediction. On similar lines, Muegge et al., in 2001 assigned a score to each molecule based on the presence of structural fragments typically found in drugs. The fragments used in this study were amines*, amides, alcohols, ketones, sulfones, sulfonamides, carboxylic acids*, carbamates, guanidine*, amidines*, urea, and esters. A molecule was given one point for each non-overlapping fragment. The molecules with a score between 2 and 7 were classified as drugs otherwise they were classified as non-drugs. Compounds containing a single pharmacophoric group would only be classified as drugs if they contained one of the groups marked with an asterisk in list of fragments [18].
| Functional groups | Score |
|---|---|
| Carboxylate | 8.2 |
| Phosphate | 10 |
| N+ | 11.5 |
| N | 1.2 |
| OH | 2.5 |
| O or S ether | 1.1 |
| Halogens | 1.3 |
| CO | 3.4 |
| C(Sp2) | 0.7 |
| C(Sp3) | 0.8 |
Table 1: Functional Groups Used In The Scoring Scheme Developed By Andrews
Functional group filters
A different approach is to identify functional groups that tend to be undesirable because of chemical reactivity and metabolic ability. Walter et al., briefly described an approach REOS (Rapid Elimination of Swill) to eliminate undesirable reagent in combinatorial libraries [1]. REOS is a hybrid method that combines some simple counting schemes similar to those in the RO5 with a set of functional groups filter to remove the reactive and otherwise under sizable moieties. The functional group filters implemented in REOS identify reactive, toxic, and otherwise undesirable moieties. Initial filtering is based on a set of seven property filters. Hydrogen bond donors, acceptors and charged groups are determined using a set of rules similar to those used in the PATTY program developed at Merck. Log P can be calculated based on a variety of schemes. A web-based interface makes it trivial to modify parameters to suit the needs of a particular drug discovery project. Examples of the functional group filters employed by REOS are listed in Table 2. In REOS, the functional groups filters are specified using the SMARTS [19] pattern matching language developed at Daylight Chemical Information Systems. SMARTS is extended version of the SMILES (Simplified Molecular Input Line Entry System) [20,21] notation developed specifically for sub-structure searching. Steps involved in REOS analysis are as follows: In the first step reagents are filtered; reactive and toxic reagents are removed in addition to the reagents that clearly will create a product that violates the molecular weight limits. In the next step, reagents checked for compatibility with chemistry- for example, when synthesizing amide one can simplify the chemistry by removing acids containing basic amines and amines containing acidic functionality. Finally the product is filtered considering the properties such as log P. This step is also incorporates a maximum count cutoff for the functional groups. The major advantage of SMARTS patterns is that they are simple ASCII text, which can be easily modified and used by a variety of applications. However, writing such patterns takes a bit of practice and the notation may not be immediately accessible to medicinal chemists.
| Functional groups | SMARTS notation |
|---|---|
| Sulfonyl halide | S(=O)(=O)[F, Cl, Br, I] |
| Acid halide | C(=O)[Cl, Br, I] |
| Peroxide | OO |
| Aldehyde | [HC]=O |
Table 2: Functional Group Filter Employed By Reos Program
Multi–property optimization
When designing a combinatorial library, drug– like character refers to only as a small number of properties, which must be optimized; it may also be necessary to optimize diversity, potency, selectivity or a number of other properties. Simultaneous optimization of multiple properties of as combinatorial library involves selection of random subset of reagents, construction of a virtual library of compounds from these reagents, calculation of the properties of combinatorial products, making modifications to the reagent subset and accepting the changes if they improve the like character of the library. This process is repeated until a predetermined stopping condition has been reached.
Gillet et al., [22] used a genetic algorithm to optimize both diversity and drug–like character of a combinatorial library. Libraries were scored by calculating frequency distribution for each of the five properties (log P, MWT, HBD and HBA) and comparing this distribution with that calculated from the CMC. The library whose frequency distribution most closely matched that of the CMC received the highest score. Zheng et al., used simulated annealing to optimize a set of four characteristics of a combinatorial library i.e. diversity, developing ease, focusing and practicality [23,24].
Chemistry space methods
The basic assumption of these methods is that drugs will tend to possess distinct values for certain properties and as a result will be shown to be distinct from non-drugs when analyzed in multi-dimensional space [25]. A chemistry space is typically defined by calculating a number of descriptors for each molecule and using the descriptor values as points in a multi-dimensional space. For example, let us assume that we have calculated molecular weight, log P and number of H–bond donors for a set of molecules. These three descriptor values can then be used to define a point in a three dimensional spaces which represents each molecule. Molecules are then assigned a drug –likeness index between 0 and 100% through a comparison of the descriptor vector for a given molecule with the cluster center.
Examination of building blocks in known drugs
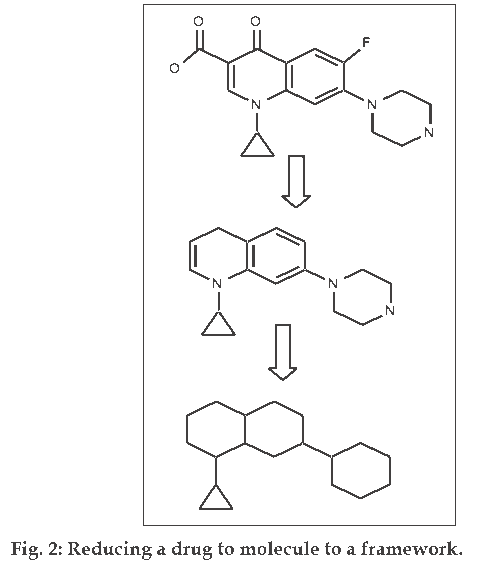
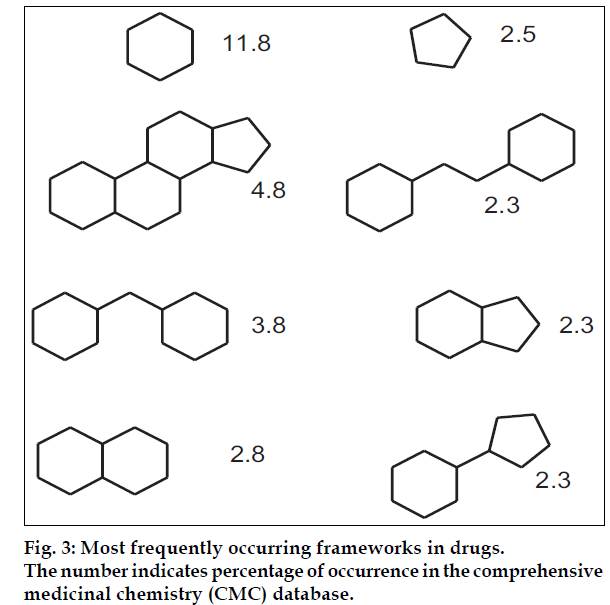
This approach does not directly distinguish drugs from non–drugs but it helps chemists to identify preferred moieties for library design. Bemis and Murcko [4,5] developed a method for organizing drugs by decomposing molecules into framework (fig. 2). A successful examination of 5120 compounds from the CMC yielded 1170 scaffolds. This suggests that drugs are rather diverse. However, when atom and bonds were considered equivalent, only 32 frameworks described the shapes of half of the drugs in the set. These frameworks are shown in (fig. 3). Then the frequency of occurrence of a particular framework in the entire database is compared to its frequency in a specific toxicity subset, this allows the discrimination between composition frameworks which occurs in a variety of molecules and toxicity conferring framework which occur primarily in molecules with a specific toxicity. An automated technique that uses a highly connected network to model the toxicity-conferring frameworks can then be used to screen a database and identify potentially toxic molecules. Toxicity is a major cause of failure for the drugs in clinical trials and this will undoubtedly continue to be an area of active research. A similar approach to assess the occurrence of structural motifs in drug molecules has been presented by Wang and Ramnarayan who have developed the concept of multilevel chemical compatibility (MLCC) between drug databases and a test molecule as a measure for drug-likeness. In MLCC, local atom environments are defined using up to tetra centered groups. The occurrence of these topological features is then tested for 11 704 compounds from the CMC and MDDR. A compound is recognized as drug-like if all of its topological motifs occur in the other known drugs.
Other Methods
The majority of the methods discussed above were developed by translating the collected knowledge of scientists involved in drug discovery into a computer programs. An alternate approach is to design a computer program for a set of drugs and non-drugs and allow the program to learn to distinguish these set of drugs and non-drugs.
Machine learning programs
Machine-learning approaches have been applied most successfully today to distinguish between drugs and non-drugs. Assuming that compounds structurally similar to known drug molecules are potential drug candidates themselves. Databases of drugs such as the CMC or MDDR and reagent-like databases such as the ACD can be statistically analyzed to identify criteria that distinguish drugs from non-drugs. Drug-classification models that are based on this idea include neural network approaches as well as recursive partitioning approaches.
Recursive partitioning approach
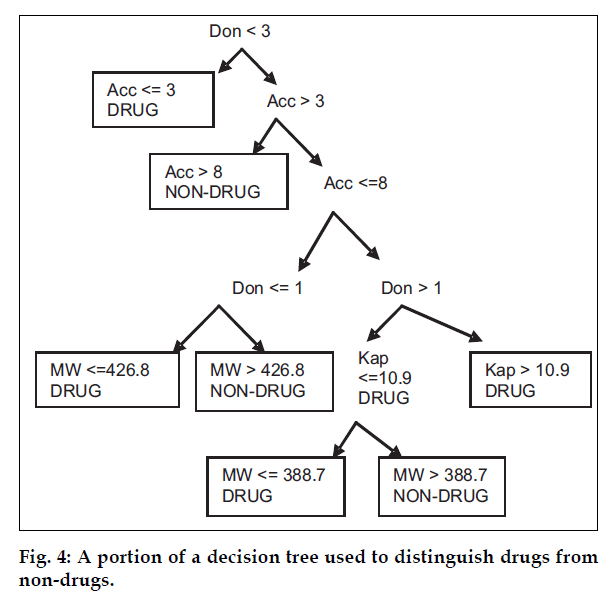
The machine-learning program (i.e. recursive partitioning approach) was used with a set of seven, one-dimensional descriptors to produce a decision tree which was able to correctly classify ~80% of CMC compounds and ~70% of ACD compounds. The rules for such trees can be identified by walking up the tree from bottom to top. An example of such a set of rules for a decision tree can be that if parameter like molecular weight (MW >388.7), kappa index (Kap <= 10.924), number of donor atoms (Don >1) and number of acceptor atoms (Acc >3) or number of acceptor atoms (Acc <=8) and number of donor atoms (Don <=3), then Class is called as Drug (fig. 4). The primary disadvantage of this method is its tendency to over train and produce rules based on chance correlation in the data.
Neural network approach
Neural network simulates the biological nervous system to create an output classification based on a set of input values. Simple neural networks use Ghose and Crippen atom types as topological descriptors. Ninety one statistically significant atom types correspond to 91 input neurons of the neural net. Typically, the hidden layer consists of five neurons which are used in the net design. The result from single neuron output layer can vary between 0.1 (non-drugs) or 0.9 (drugs). Trained on 5,000 drugs taken from the WDI and 5,000 compounds labeled as non-drugs taken from the ACD, the resulting neural net has been shown to correctly classify ~80% of other drugs/non-drugs. However the possible drawbacks of neural nets are that, discernible rules as to why a given compound is classified as drug or non-drug cannot be derived, also the neural net will strongly reflect its database heritage.
Applications
Compounds and screening set selection
One important application of these techniques is in the context of compounds and screening set selection. For example, techniques such as those already described can be used to filter a set of compounds from an external supplier prior to purchase. The genetic algorithm-based method applied to the selection of compounds from the corporate database for generation of a screening set. Profiling using the RO5 and PSA criteria can also provide a valuable indicator of the likely absorption characteristics of a combinatorial library or screening set.
Combinatorial library design
In addition to simply profiling libraries, this approach has been taken one step further and has been applied to the design of combinatorial libraries. In one example, a chemist had selected reagents for a combinatorial library (LIB1) in an oral drug discovery program to optimize parameters such as MW and ClogP in an approximate manner. A follow-up library (LIB2) was designed to optimize PSA and RO5 criteria much more rigorously with reagent selection being performed by a Monte Carlo search procedure. Both libraries were subsequently tested in a Caco-2 monolayer absorption system and both of the designed libraries shown much improved absorption. These results showed the added value of quantities such as PSA in compound (library) design in addition to more traditional computed descriptors such as ClogP and MW.
Virtual screening of chemical databases
Drug likeness is used as one of the filters in the virtual screening of chemical databases with the purpose to screen in those molecules which have the property similar to that of known drugs. The rule of five is used as primary filter for screening the chemical databases, which are modified in accordance with the potent molecule, to performed efficient searching procedure to yield only those molecules having drug like character.
Conclusion
It is clear that there are many research groups currently engaged in identifying drug-like and non-drug-like molecules. A common theme is to learn from history i.e. to examine databases of known compound with biological activity and draw conclusions from the data for the properties, such as toxicity and oral bioavailability etc. However neither are available databases extensive; nor have the data been experimentally determined in a consistent manner. Thus there is a need for the generation of larger data sets of diverse compounds for such properties in area of the toxicity prediction. Future advances in the field shall involve the combination of general drug-likeness with specific properties of small molecules to hit specific gene families (such as GPCRs or kinases).
References
- Walters WP, Stah MT, and Murcko MA. Virtual screening: An overview. Drug Discov Today 1998;3:160-78.
- Walters WP, Murcko A, Murcko MA. Recognizing molecules with drug-like properties. Curr Opin Chem Biol 1999;3:384-7.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 1997;23:3-25.
- Bemis GW, Murcko MA. The properties of known drugs, 1: Molecular frameworks. J Med Chem 1996;39:2887-93.
- Bemis GW Murcko MA. Properties of known drugs, 2: Side chains. J Med Chem 1999;42:5095-9.
- Lutz MW, Menius JA, Choi TD, Laskody RG, Domanico PL, Goetz AS, et al. Experimental design for high troughput screening. Drug Discov Today 1996;1:277-86.
- Gallop MA, Barrett RW, Dower WJ, Foder SP, Gorden AM. Applications of combinatorial technologies to drug discovery, 1: Background and peptide combinatorial libraries. J Med Chem 1994;37:1233-51.
- Gorden EM, Barrett RW, Dower WJ, Foder SP, Gallop MA. Applications of combinatorial technologies to drug discovery, 2: Combinatorial organic synthesis, library screening strategies and future directions. J Med Chem 1994;37:1385-401.
- Martin EJ, Blaney JM, Siani MA, Splellmeyer DC, Wong AK, Moos WH. Measuring diversity: Experimental design of combinatorial libraries for drug discovery. J Med Chem 1995;38:1431-6.
- Warr WA. Combinatorial chemistry and molecular diversity: An overview. J Chem Inf Comput Sci 1997;37:134-40.
- Brown RD, Martin YC. Designing combinatorial library mixtures using a genetic algorithm. J Med Chem 1997;40:2304-13.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 2001;46:3-26.
- Ghose AK, Viswanadhan VN, Wendolowski JJ. Prediction of hydrophobic (lipophilic) propertoies of small moleculesusing fragmental methods: An analysis of ALOGP and CLOGP methods. J Phys Chem 1998;102:3762-72.
- Ghose AK, Viswanadhan VN, Wendolowski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery, 1: A qualitative characterization of known drug databases. J Comb Chem 1999;1:55-67.
- Oprea TI. Property distribution of drug related chemical databases. J Comput Aided Mol Des 2000;14:251-64.
- Andrews PR, Craik DJ, Martin JL. Functional group contributions to drug-receptor interactions. J Med Chem 1984;27:1648-57.
- Leach AR, Bradshaw J, Green DV, Hann MM, Delany JJ 3rd Implementation of a system for reagent selection and library enumeration, profiling and design. J Chem Inf Comput Sci 1999;39:1161-72.
- Muegge I, Heald SL, Brittelli D. Simple selection criteria for drug-like chemical matter. J Med Chem 2001;44:1841-6.
- James CA, Weininger D, Delany J. Daylight theory Manual, 4.71. Available from: http://www.daylight.com/dayhtml/doc/theory/theory. smarts.html.
- Weininger D. SMILES, a chemical language and information system, 1: Introduction to methodology and encoding rules. J Chem Inf Comput Sci.1988;28:31-6.
- Wieninger D, Wieninger A, Wieninger JL. SMILES 2: Algorithm for generation of unique SMILES notation. J Chem Inf Comput Sci 1989;29:97-101.
- Gillet VJ, Willett P, Bradshaw J, Green DV. Selecting combinatorial libraries to optimize diversity and physical properties. J Chem Inf Comp Sci 1999;39:169-77.
- Zheng W, Hung ST, Saunders JT, Seibel GL. PICCOLO: A tool for combinatorial library design via multicriterion optimization. Pac Symp Biocomput 2000;5:585-96.
- Press WH, Teukolssky SA, Vetterling WT, Flannery BP. Neumerical Recipes in C. 2nd ed. Cambridge University Press: 1992. 444-51.
- Agrafiotis DK, Myslik JC, Salemme FR. Advances in diversity proÞling and combinatorial series design. Mol Divers 1998;4:1-22.