X. Huang, Z. Chen, G. Zhao, J. Shi, J. Chen, F. Chen, Y. Wei, J. Xia, G. Huang and S. Wang*
Department of Orthopaedic Surgery, Huashan Hospital, Fudan University, Shanghai, 200040, China
- *Corresponding Author:
- S. Wang
Department of Orthopaedic Surgery,
Huashan Hospital,
Fudan University,
Shanghai, 200040,
China
E-mail: wangsiqun_sh@126.com
| This article was originally published in a special issue: Special issue on “Trends in therapeutic Management of Various Conditions” |
| Indian J Pharm Sci 2020:82(3)Spl issue 6: 9-15 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
In order to study the expression of lysine-specific demethylase 6B, Kdm6b in differentiation of chondrocyte and chondrogenesis and the regulation effect of Kdm6b on chondrocyte anabolism, a mouse model of osteochondral defects was established in this study. The mice were subjected to antibiotic treatment and intestinal microbiota reconstruction by the method of specific knock out Kdm6b of embryonic cartilage. The mice were divided into Col2al-CreER Kdm6b experimental group and Kdm6b control group and then the destabilization of the medial meniscus operation was performed to model osteoarthritis in mice, which accelerated the abnormal metabolism of cartilage. In addition, transgenic mice with induced chondrocyte specific knockout of Kdm6b were constructed and embryonic samples were collected to study the effects of Kdm6b knockout on cartilage development. At the same time, the proliferation and apoptosis of chondrocytes after kdm6b gene knockout were also studied based on the obvious abnormalities in embryonic chondrogenesis. The results showed that 3 weeks after the mouse osteochondral defect modelling surgery, the mice in the Col2al-CreER; Kdm6b group still had an area without complete healing in the femoral trochle defect, while the defect areas in the Kdm6b group of mice were completely filled. After 12 weeks of mouse osteoarthritis modelling, mice in the Kdm6b group showed cartilage staining loss and cartilage defect. The mice in the Col2al-CreER Kdm6b group were more severe, and even a large area of wear and destruction of the cartilage layer appeared. However, after the growth plate thickness of mice in the Col2al-CreER Kdm6b group was observed, it was found that there was no statistical significance compared with the Kdm6b control group. qRT-PCR analysis showed that compared with the Kdm6b control group, the Col10al gene expression in the primary chondrocytes of the Col2a1-CreER Kdm6b group was significantly decreased. In addition, qRT-PCR and Western Blot sequencing verified that the expressions of Sox9, Col2al, Acan and other proteins related to anabolism were significantly reduced after Kdm6b knockout. Therefore, it can be concluded that Kdm6b directly regulated the expression of genes related to chondrocyte anabolism, and Kdm6b knockout could inhibit the anabolism of chondrocytes, impair the synthesis of cartilage matrix, and accelerate the progress of osteoarthritis.
Keywords
miR-128, athletic injury, myocyte membrane, CDKN1A, inflammatory cell
In recent years, as a degenerative disease, osteoarthritis has attracted greater attention [1]. Chondrocytes always maintain the stability of cartilage in cartilage tissue. Chondrocytes are distributed individually on the surface of cartilage tissue, while in the middle of cartilage tissue, these are distributed in groups [2]. Chondrocytes have the function of synthesizing and secreting substrates and fibers [3]. Studies published thus far have found that the synthesis and metabolism of chondrocytes are related to their environment. When chondrocytes are exposed to inflammatory environment, their synthesis and metabolism will be inhibited, which will accelerate the metabolic rate and accelerate the damage of arthritic cartilage [4]. Therefore, in the early pathological process of osteoarthritis, it is very important to find a way to improve the synthesis and metabolism of chondrocytes to prevent the progressive destruction of cartilage [5]. Studies on various proteins related to chondrocyte anabolism are helpful for people to understand the mechanism of growth and development of cartilage to provide new concepts for the diagnosis and treatment of clinical osteoarthritis [6]. Recent research showed that epigenetics can significantly regulate the formation process of cartilage and the pathological process of osteoarthritis. Epigenetics refers to the change of gene level expression through genomic imprinting, DNA methylation modification, maternal effect and RNA editing without changing the nucleotide sequence of genes, so as to produce a heritable phenotype [7]. Other studies have found that abnormal histone methylation in the SOX9 promoter region is closely related to the anabolism of chondrocytes in patients with hip osteoarthritis, suggesting that histone methylation may be involved in the pathologenisis of OA [8].
Kdm6b is a demethylase of histone H3K27me3, which can specifically remove the dimethylation and trimethylation sites (H3K27me2/3) of H3K27me3 [9]. It has been reported that kdm6b is involved in a variety of cellular processes, including cell proliferation, differentiation, aging and apoptosis and plays an important role in developmental disorders, neurodegenerative diseases, tumours, aging and inflammatory diseases [10]. Studies have shown that kdm6b expression is upregulated in the activation process of immature CD4+ cell. Kdm6b knockout affects the differentiation and maturation of Th17 cells, which can reduce the severity of experimental allergic encephalomyelitis, suggesting its important role in the inflammatory process [11].
Therefore, in this study, osteoarthritis was modeled in mice with cartilage-specific knockout of Kdm6b and destabilization of the medial meniscus (DMM) surgery was performed to model osteoarthritis in mice. The involvement of Kdm6b in the mechanism of cartilage development, regulation of chondrocyte anabolism and the maintenance of cartilage homeostasis in osteoarthritis were studied, providing a new theoretical basis for the future clinical treatment of OA.
Materials and Methods
Cell culture reagents used in this study included cell culture medium DMEM series, fetal bovine serum, trypsin, 100 U/ml penicillin/streptomycin (dual resistance), dexamethasone (Sigma), β-glycerol- 2-phosphatedisodiumsalt (Sigma), ascorbicacid-2- phosphate salt (VC, Sigma), sodium pyruvate (life technologies), Insulin-transferrin-selenium additive (ITS, Life Technologies), TGF-β3 (PepreoTech). Mouse mesenchymal stem cells were used as test system. Western blot-related antibodies were SOX9, KDM6B and actin. Immunohistochemical and fluorescent antibodies were BrdU, KDM6B, COLX, BrdU, Ki67 and caspase-3.
Male mice (20-30 g, 8 w old) procured through formal channels were used in this study. Mice were housed in the animal house for a week and fed with standard feed. Indoor temperature was controlled at 20±2° and environmental relative humidity was controlled at 54-60 %. The daily light time was controlled at 8-12 h and the mice were given water and feed regularly every day to help them adapt to the environment as soon as possible.
Grouping:
The mice were divided into DMM group and cartilage defect group. Sixteen mice aged 8 w in the Col2al- CreER Kdm6b group and 16 mice in the Kdm6b control group were intraperitoneally injected continuously for 5 d for TM initiation gene knockout. On w 9, DMM surgery was performed on osteoarthritis mice (5 mice at 8 w and 5 mice at 12 w) or sham surgery (3 mice at 8 w and 3 mice at 12 w) to obtain the DMM surgery group. The 12-w old mice were divided into Col2al-CreER Kdm6b mice and Kdm6b control mice. Then, the 12-w old mice were subjected to TM intraperitoneal injection for 5 consecutive days and underwent osteochondral defect surgery. The osteochondral defect group was obtained 3 w after surgery.
Osteochondral defect model:
Male mice were anesthetized by injecting 12 % chloral hydrate (420 mg/kg) intraperitoneally. After complete anaesthesia, the skin of the knee joint was disinfected with 2.5 % iodine and a scalpel was used to cut an incision about 0.6 cm along the inside of the patellar ligament. The patellar ligament was dislocated to the outside and the skin was cut layer by layer to completely expose the femoral trochle to the field of view. S vertical needle puncture was made in the center of the femoral trochle with an anatomical microscope. The depth was controlled by gently pressing until the needle was removed and slowly oozing blood was visible at the defect. Osteochondral defects were made. The debris from the osteochondral defect was rinsed with saline, disinfected and the wound was sutured layer by layer to obtain the osteochondral defect model.
Culture of primary chondrocytes in mice:
Mouse knee cartilage was removed and disinfected. Primary chondrocytes were obtained by enzymatic digestion method. The cartilage tissue cut into small pieces was digested with a 0.3 % type II collagenase (3 mg/ml, configured in a cartilage medium) for 3.5 h in a cell culture incubator. After digestion, it was centrifuged for 6 min in a low temperature centrifuge and the digestive juice was removed at a rotational speed of 1200 rpm. After the digestion solution was removed, cartilage medium was added, the ambient temperature in the incubator was controlled at about 35±2° and the carbon dioxide concentration was controlled at 6 %. After the digestive juices were removed, the solution added to the cartilage medium was placed in an incubator for culture. When the cell density reached the fusion degree of 75-85 %, chondrocytes were obtained by digestion with trypsin, and finally 1-3 generations of cells were adopted for relevant cell experiments.
Cell cryopreservation, cell resuscitation and preparation of cell protein samples:
When the cells were grown to a concentration of 75-85 %, the solution was frozen at -22° and thawed at 4°. Primary chondrocytes were obtained by enzyme digestion method as described in the above section. After the digestion solution was removed, cartilage medium was added, the ambient temperature in the incubator was controlled at about 35±2° and the carbon dioxide concentration was controlled at 6 %. Then, the solution added to the cartilage medium after the removal of digestive juice was placed in the incubator to warm up. After preheating, the cells were taken out from the liquid ammonia tank and quickly placed in a water bath at a temperature of 35±2° and gently shaken. When the cells in the water bath melted to half, the remaining cells were quickly transferred to the culture dish. The integrity of the cells of the culture dish was examined microscopically, and the culture dish was washed twice with the cartilage medium after 10 h. Finally, the culture dish was replaced with a new solution. This was the process of cell resuscitation. When the cell was resuscitated, it was necessary to prepare the protein sample of the cell. Firstly, cell proteins were extracted with lysate, cells were scraped off and the original medium was taken. If there was excess medium, the residual medium was washed away with pre-cooled PBS. The RIPA and protease inhibitor solution were then configured. 75 μl of the mixture was sequentially added to the 6 wells of the 6-well plate, and the cells were scraped off with the cell scraper and poured into an EP tube prepared in advance. The EP tube was weighed and the cells were shattered with an ultrasonic crusher. It was centrifuged for 29 min with a cryogenic centrifuge at 10 000 rpm. Three microliters of the supernatant was added to another new EP tube and stored at -20° for detection of total protein concentration. The remaining supernatant was added with 50 μl loading buffer containing bromophenol blue DTT and cooked for 20 min at 82° for denaturation.
Western Blot:
Separation gel with a concentration of 12 % was poured into a glass plate and capped with double distilled water. After all the polymerization, the double distilled water on the separation gel was poured off, and the concentrated glue was added to the separation rubber. It was then immediately inserted vertically into the glass plate and loaded after 0.5 h of complete polymerization. 20 μl of each sample of the experimental group and the control group was added to the sample well, and 5 μl of the low molecular weight protein standard was again added as a control. In the electrophoresis tank, 1X electrophoresis buffer was added, the power source was connected, the negative electrode was at the top, and the positive electrode was at the bottom. The gel concentration was 80 V, the gel separation voltage was 100 V, and electrophoresis was stopped until the bromophenol blue line reached the bottom of the electric pool after 2 h. A 0.2 μm polyvinylidene fluoride film and 12 sheets of filter paper were prepared, soaked in methanol for 1 minute, and then immersed in transfer buffer for 2 min. Then, according to the order of white paper, three-layer filter paper, nitrocellulose film, glue, three-layer filter paper, black paper, a sandwich was made and transferred. The fixture was placed in a transfer slot so that the black side of the fixture faced the negative pole and the white side faced the positive pole. The water tank was placed in an ice bath and transferred at 300 mA for 1.5 h. The membrane was transferred to a tray containing sealing solution and shaken on a decolorizing table at room temperature for 4 h. The antibody was diluted with TBST (the dilution rate was determined according to the antibody specification) and incubated at room temperature for 5° overnight. It was washed 3 times with TBST on a decolorizing shaker at room temperature for 15 min each time. A second antidiluting solution was prepared by the above method and contacted with a membrane, incubated for 1 h at room temperature, and washed 3 times with TBST. At normal temperature, it was washed three times on the colouring shaker, each time for 15 min. After ECL staining, the bands under the instrument were observed.
DNA extraction:
The 220 ml NaOH solution was added to the EP tube containing mouse tissue samples to soak the samples. The tube was then placed in a metal bath at 95° for 0.5 h, and the solution was shaken and mixed uniformly. Twenty millilitres of Tris-HC1 solution was added to the test tube and shaken evenly. Finally, the cells were centrifuged for 2 min at 2200 rpm and the supernatant was taken as a tissue DNA sample.
Statistical analysis:
All data were expressed in the form of mean±standard deviation (SD). SPSS22.0 statistical software was used for processing and one-way analysis of variance was used to test the significance of differences among groups. When p value was less than 0.05, the data difference was statistically significant. And statistical differences were expressed as *p<0.05.
Results and Discussion
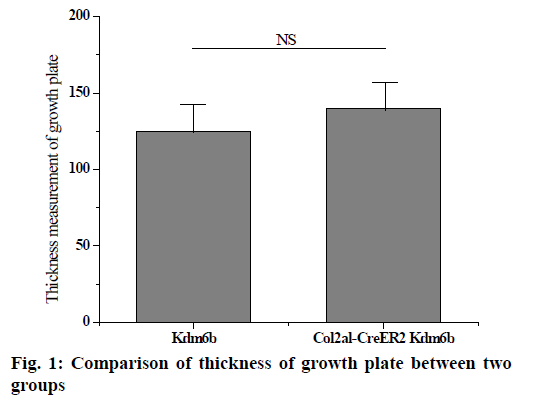
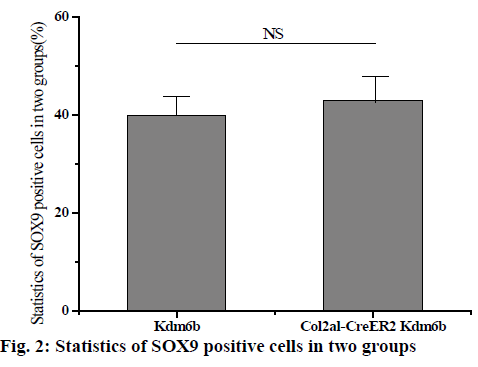
The knee joints of mice aged 1 mo and 8 w were stained and analysed and the knee joints of mice aged 2 and 12 mo were removed by tm intraperitoneal injection. Safranin-o staining results of knee cartilage in 2-mo old Col2al-CreER Kdm6b mice showed no significant matrix degradation or cartilage surface damage and there were no significant differences in the Osteoarthritis Research Society International (OARSI) scores between the 2 groups. As shown in fig. 1 below, considering that Kdm6b knockout results in abnormal growth during cartilage development stage, the thickness of growth plate of Col2al-CreER Kdm6b mice was also observed, and the results showed no statistical difference compared with the control group (p>0.05). At the same time, cartilage was also observed in aged mice in the Col2al-CreER Kdm6b group. Safranin O staining results showed slight damage to the cartilage surface of the knee joint in mice aged 12 mo in the Col2al-CreER Kdm6b group. In contrast, chondrocyte specific knockout of Kdm6b mice showed fewer hypertrophic chondrocytes in the hyaline cartilage layer than in the control group. As shown in fig. 2 below, immunohistochemical staining results of COLII and SOX9 showed no abnormalities in the cartilage matrix of the 2-mo old mice in Col2al-CreER Kdm6b group. Immunohistochemical examination of common proteins in articular cartilage revealed that the expression of proteins related to cartilage matrix synthesis decreased in mice in the col2al-creer Kdm6b group, the staining of COLII became lighter, and the number of AGGRECAN positive cells decreased. Staining results of proteins related to cartilage catabolism showed no significant changes in the expression of MMP13 or ADAMTS5 in mice in the Col2al-CreER Kdm6b group compared with those in the Kdm6b control group. COLX, the cartilage hypertrophy index of Col2al-CreER Kdm6b mice, was found to be significantly lower than that of the Kdm6b control group, which was consistent with the finding that Kdm6b knockout inhibited the maturation of osteocytes during development. These results suggested that knocking out kdm6b after cartilage maturation could reduce the ability of chondrocytes to synthesize matrix, leading to spontaneous cartilage destruction in elderly mice. However, OARSI scores of knee joints of 12-mo old mice showed significant cartilage damage and increased OARSI scores in mice in the Col2al-CreER Kdm6b group, but there was no statistical difference.
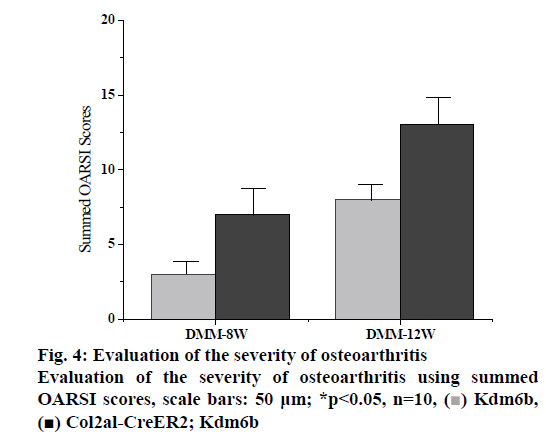
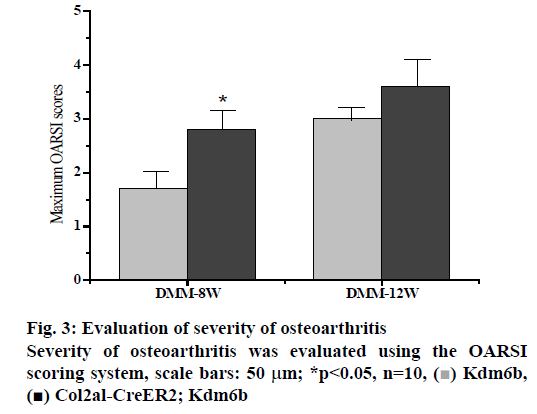
DMM osteoarthritis model was established in the col2al-creer kdm6b group and the kdm6b control group, respectively and knee joint specimens were collected for analysis at 8 and 12 w after surgery. Immunofluorescence detection of the cartilage of the knee joint showed that Kdm6b was expressed in a large amount in the cartilage of mice, while the expression of Kdm6b in the mice of the Col2al-CreER Kdm6b groupwas significantly decreased, demonstrating the success of chondrocyte-specific knockout of Kdm6b. Safranin O staining results showed that 8 w after DMM, the hyaline cartilaginous layer in the Col2al-CreER Kdm6b group of mice became significantly lighter stained and had significant cartilage damage. Twelve weeks after DMM surgery, there was obvious cartilage staining loss and cartilage defect in Kdm6b mice. Cartilage damage in the Col2al-CreER Kdm6b group was significantly worse than that in the control group, with wear and large area damage of hyaline cartilage layer. As shown in the figs. 3 and 4, OARSI scoring of all Safranin O staining samples showed that the Col2al-CreER Kdm6b group had the most severe injuries, which were higher than the control group 8 w after the operation and there was a statistical difference (p<0.05). After scoring the cartilage injury in different parts of the knee joint, it was found that the severity of OA in the Col2al-CreER Kdm6b group was significantly higher than that in the Kdm6b group at 8 and 12 w after DMM. According to Safranin O staining, immunohistochemical staining results of cartilage matrix synthesis related proteins showed COLII staining loss in the hyaline cartilage layer of tibia of mice in the Col2al-CreER Kdm6 group, and more positive cells in Kdm6b mice than those in AGGRECAN and SOX9. These results suggested that the specific knockout of Kdm6b of chondrocytes may impair the synthesis of cartilage matrix during the pathological process of osteoarthritis.
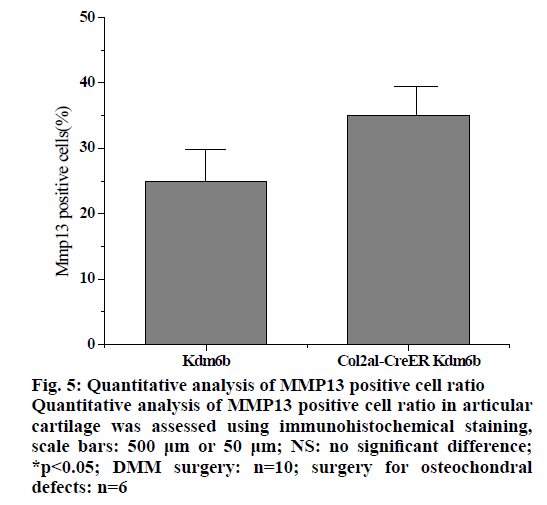
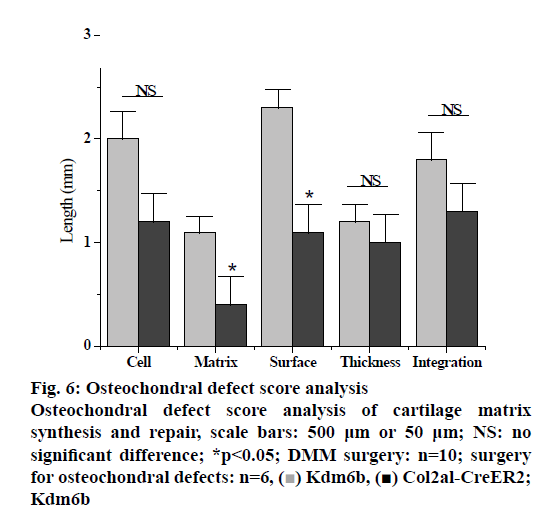
In the pathologenesis of osteoarthritis, the maintenance of intra-cartilage balance is not only related to cartilage metabolism, but also plays an important role in the process of cartilage destruction. Therefore, the effect of kdm6b knockdown on the catabolism of chondrocytes was investigated, as shown in fig. 5. Immunohistochemical staining results of tibial cartilage of mice in the Col2a1-CreER Kdm6b group after DMM surgery showed decreased COLX expression and increased MMP13 expression. However, there was no significant difference in ADAMTS5 expression compared with the control groups. Comparing adjacent sections with Safranin O staining and COLX immunohistochemical staining, it was found that cartilage destruction was more severe in Col2a1-CreER Kdm6b group. Mice in the Col2al-CreER Kdm6b group were compared with those in the Kdm6b control group, and it was found that the chondrocyte hypertrophy of mice in the Col2al-CreER Kdm6b group was less obvious than that in the control group, and the number of COLX positive cells was much less than that in the control group. Consistent with in vivo staining results, qRT-PCR analysis also demonstrated that the gene expression of Col10al in primary chondrocytes of Col2a1-CreER Kdm6b mice was decreased compared with the control group. Considering that the anabolism and catabolism of chondrocyte both change and interact with each other during the pathological process of osteoarthritis, the DMM model can’t fully prove that Kdm6b knockout accelerates the progress of osteoarthritis by inhibiting chondrocyte anabolism. Therefore, a model of osteochondral defect was added to illustrate the effect of Kdm6b knockout on cartilage matrix synthesis. Three weeks after the modelling of the defect, gross observation was made on the femoral trochle defect and it was found that the defect in Kdm6b mice was filled in the appearance, while the unhealed area remained in the defect of the mice in the Col2al-CreER Kdm6b group. H and E staining results of the sections at the center of the defect showed that the control group had flat articular surfaces and dense deposition of matrix components. However, the defects in the Col2al-CreER Kdm6b group were uneven and almost filled by fibroblasts, lacking matrix components. Similarly, the Safranin O staining and COLII immunohistochemical staining results showed that the defects in the Kdm6b group had more cartilage matrix, while the defects in the col2al-creer Kdm6b group were incomplete and the cartilage matrix was also significantly less than those in the control group. As shown in fig. 6, the osteochondral defect score also showed that compared with the control group, the repair ability of mice in the Col2al-CreER Kdm6b group and Kdm6b group was decreased, especially in terms of matrix deposition and surface integrity of cartilage defect repair. According to the results of DMM osteoarthritis model and osteochondral defect repair model, it was believed that kdm6b gene knockout can accelerate the progress of osteoarthritis by inhibiting the synthesis and metabolism of chondrocytes.
Figure 5: Quantitative analysis of MMP13 positive cell ratio Quantitative analysis of MMP13 positive cell ratio in articular cartilage was assessed using immunohistochemical staining, scale bars: 500 μm or 50 μm; NS: no significant difference; *p<0.05; DMM surgery: n=10; surgery for osteochondral defects: n=6
In this study, the regulatory effect of Kdm6b on the anabolism of chondrocytes in a mouse cartilage injury repair model was taken as a foothold. Firstly, the effects of chondrocyte-specific Kdm6b knockout on the expression of various proteins were studied, and the mechanism of spontaneous knee cartilage injury in elderly mice with chondrocyte-specific Kdm6b knockout was investigated. Then, the effect of kdm6b knockout on the catabolism of chondrocytes, as well as the expression and effect of kdm6b knockout on the differentiation of chondrocytes were detected, revealing the function and mechanism of kdm6b in regulating cartilage anabolism. Finally, DMM surgery was used to model osteoarthritis in mice, and it was found that Kdm6b knockout accelerated the destruction of cartilage matrix in osteoarthritis by inhibiting the anabolism of chondrocytes, providing a new theoretical basis for the clinical diagnosis and treatment of OA patients in the future. It was found that kdm6b can regulate chondrocyte matrix synthesis and maintain joint cartilage homeostasis, which may be involved in chondrocyte metabolism by sequencing screening. Some studies found that kdm6b was involved in the regulation of chondrocyte metabolism and cartilage matrix synthesis. Other studies showed that kdm6b was combined to the promoter region of genes related to chondrocyte anabolism and directly regulated gene expression. In summary, the following conclusions could be drawn.
Firstly, the expression of kdm6b increased during chondrogenesis induction in vitro in mesenchymal cell lines and kdm6b was widely expressed in chondrocytes during embryonic cartilage development. Kdm6b was involved in the differentiation of chondrocytes and the development and maturation of normal cartilage. After kdm6b was knocked out during development, chondrocyte malformation, decreased proliferation ability, reduced matrix synthesis, and abnormal limb development were found, suggesting that kdm6b was essential in maintaining normal cartilage development. Secondly, in the process of chondrocyte anabolism, a series of expressed genes were related to Kdm6b, which can regulate the expression of related proteins synthesized by cartilage matrix.
References
- Dai J, Yu D, Wang Y. Kdm6b regulates cartilage development and homeostasis through anabolic metabolism. Ann Rheum Dis 2017;76(7):1295-303.
- Wang X, Tang D, Shen P. Analysis of DNA methylation in chondrocytes in rats with knee osteoarthritis. BMC Musculoskelet Disord 2017;18(1):377.
- Li C, Xu MM, Wang K. Macrophage polarization and meta-inflammation. Transl Res 2018;191:29-44.
- Bayarsaihan D. Epigenetic mechanisms involved in modulation of inflammatory diseases. Curr Opin Clin Nutr Metab Care 2016;19(4):263-69.
- Van Meurs J B J, Boer C G, Lopez?Delgado L. Role of Epigenomics in Bone and Cartilage Disease. J Bone Miner Res 2019;34(2):215-30.
- Fu G, Ren A, Qiu Y. Epigenetic regulation of osteogenic differentiation of mesenchymal stem cells. Curr Stem Cell Res Ther 2016;11(3):235-46.
- Krstic J, Trivanovic D, Obradovic H. Regulation of Mesenchymal Stem Cell Differentiation by Transforming Growth Factor Beta Superfamily. Curr Protein Pept Sci 2018;19(12):1138-54.
- Marini F, Cianferotti L, Brandi M. Epigenetic mechanisms in bone biology and osteoporosis: Can they drive therapeutic choices. Int J Mol Sci 2016;17(8):1329.
- Zhuang H, Zhang X, Zhu C. Molecular mechanisms of PPAR-γ governing MSC osteogenic and adipogenic differentiation. Curr Stem Cell Res Ther 2016;11(3):255-64.
- Zhu S, Dai J, Liu H. Down?regulation of Rac GTPase?activating protein OCRL1 causes aberrant activation of Rac1 in osteoarthritis development. Arthritis Rheum 2015;67(8):2154-63.
- Im GI. Gene transfer strategies to promote chondrogenesis and cartilage regeneration. Tissue Eng Part B Rev 2015;22(2):136-148.
 Kdm6b, [
Kdm6b, [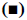 Col2al-CreER2; Kdm6b
Col2al-CreER2; Kdm6b