- *Corresponding Author:
- J. Zhuo
Department of Pharmacology, School of Pharmaceutical Sciences, Hainan University, Haikou 570228, China
E-mail:zhuojinsheng@hainmc.edu.cn
| Date of Received | 10 November 2021 |
| Date of Revision | 18 January 2023 |
| Date of Acceptance | 18 May 2023 |
| Indian J Pharm Sci 2021;85(3):652-666 |
Abstract
The etiology and pathogenesis of Alzheimer's disease are extremely complicated and there is no effective cure available currently. Previous studies have proved the effectiveness of Shenwu capsule in treatment of Alzheimer's disease. However, its mechanism of action has not been systematically elucidated. Here, network pharmacology approach was used to revealing the underlying mechanism of Shenwu capsule in the treatment of Alzheimer's disease. The compound database of Shenwu capsule was constructed, and their potential targets were obtained. Alzheimer's disease gene set was acquired from databases. Subsequently, the potential therapeutic targets of Shenwu capsule on Alzheimer's disease by overlapping analysis, which validated by disease ontology and tissue enrichment analysis. Biological process and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis were performed to find the core biological processes and signaling pathways regulated by Shenwu capsule in treating Alzheimer's disease. By constructing and analyzing the protein-protein interaction network and compound-target network, the core targets and key components were identified. A total of 798 compounds and 1105 potential Shenwu capsule targets, 2566 Alzheimer's disease-related genes and 453 Shenwu capsule therapeutic targets were identified. 3102 biological processes were significantly enriched, such as membrane potential, oxidative stress, synaptic transmission, and neuronal death. 194 pathways of significance were identified, including neuroactive ligand-receptor interactions, lipid-atherosclerosis, Alzheimer's disease, cyclic adenosine 3′,5′-monophosphate signaling and calcium signaling. 35 core therapeutic targets and 55 key components were screened out. Network pharmacology provides an effective way to elucidate the mechanism of action and identify the core targets and key compounds of traditional Chinese medicine. Further studies are needed to validate the validity of this pr ediction.
Keywords
Traditional Chinese medicine, Shenwu capsule, Alzheimer's disease, network pharmacology, therapeutic mechanisms
Alzheimer's Disease (AD) is a highly complex and progressive neurodegenerative disease, which is characterized by memory loss and progressive neurocognitive dysfunction, accompanied by various neuropsychiatric symptoms and behavioral disorders[1,2]. It is one of the leading causes of dementia cases globally[3]. By 2050, it is estimated that a new case of AD will appear every 33 sec, resulting in nearly 1 million new cases each year, with an estimated prevalence between 11 and 16 million[2,3]. There are many hypotheses about the pathogenesis of AD, including cholinergic hypothesis, Amyloid β (Aβ) protein production and metabolism disorder hypothesis, τ-protein abnormal phosphorylation hypothesis, oxidative stress and free radical damage hypothesis, inflammation hypothesis, metal ion metabolism hypothesis and so on. However, at present, there is no hypothesis that can accurately and comprehensively explain the pathological characteristics of AD, which may be the result of multiple mechanisms[2-6]. Due to the lack of a full understanding of the pathophysiology of AD, especially the initial stage of the disease[7], there is no effective cure available currently[2]. Existing drugs usually target the symptoms, not the underlying pathology[8].
For the first time, our team put forward the biological basis of “Tonifying kidney and filling marrow” in Chinese medicine, including promoting energy metabolism and utilization, activating production of endogenous neurotrophic factor and promoting neuron survival and regeneration. Based on the Chinese medicine theory of "Tonifying kidney and filling marrow, supplemented by relieving phlegm and removing blood stasis", Shenwu Capsule (SWC) was developed and has obtained the national invention patent in China. It is a prescription for the treatment of AD and Mild Cognitive Impairment (MCI) with syndrome differentiation of kidney and spleen deficiency, phlegm turbidity and blood stasis. Like other Traditional Chinese Medicine (TCM) prescriptions, SWC is a complex system with multiple components, multiple targets, and multiple mechanisms. It is composed of six herbs, including Radix polygoni multiflori preparata (Chinese Pinyin: Zhi Shou Wu (ZSW)), Panax ginseng C. A. Mey (Chinese Pinyin: Ren Shen (RS)), Acorus tatarinowii (Chinese Pinyin: Shi Chang Pu (SCP)), Epimedium brevicornu Maxim. (Chinese Pinyin: Yin Yang Huo (YYH)), Pueraria lobate (Chinese Pinyin: Ge Gen (GG)), Rhizoma chuanxiong (Chinese Pinyin: Chuan Xiong (CX)). In the previous studies, SWC showed protective effect on peripheral blood lymphocyte DNA[9], retard aging process of the spinal cord through elevating the expression of neurotrophic factors in the lumbar spinal cord of aged rats[10]. In addition, SWC indicated effectiveness in AD-like mitochondrial deficiency model rats, suggesting its application in the treatment of AD[11]. However, the basic mechanism of action at the molecular level has not been systematically studied. The bioactive compounds, the potential targets and the related pathways of SWC remain unknown.
Network pharmacology, as a new emerging field that interprets the occurrence and development of diseases from the perspective of biological network balance[12,13], provides a new research strategy to explore TCM spanning multiple scales from the molecular and cellular level to the tissue and organism level. In recent years, Chinese scholars have made some important achievements and progress in the establishment of network pharmacology research methods and the application of them to study the scientific connotation of TCM, which has received great international response[14-19]. Therefore, we in this study employed network pharmacology methods to probe into mechanisms underlying curative effects of SWC on AD and mine the material basis, including the core targets and hub components.
Materials and Methods
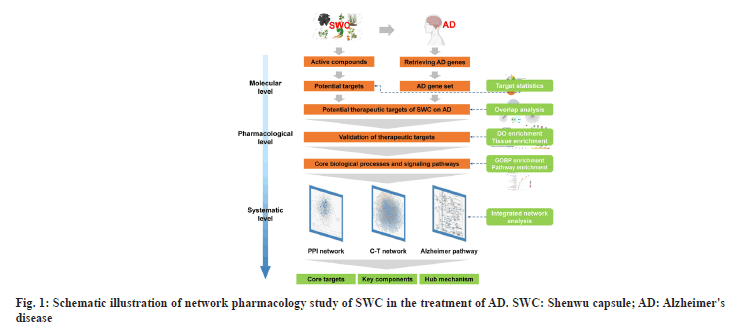
These have six main steps for the integrated system-based network pharmacology approach to disclose the curative effects of SWC as follows (fig. 1). Construction of molecular database for all six herbs in SWC from public databases, then the potential target data of the components were obtained, including known targets and putative targets. Acquisition of AD gene set from databases and then identification and validation of potential therapeutic targets of SWC on AD. Identification of core biological processes and signaling pathways regulated by SWC in treating AD based on enrichment analysis results, identification of core targets and key components through integrating network analysis. To better elucidate the holistic mechanisms of SWC using Alzheimer pathway as an example.
Identification of candidate targets of SWC in treating AD
Construction of a database of the SWC ingredients: In order to obtain the compound information as comprehensively as possible, we manually collected ingredients of SWC from TCM_ID[20], TCM_Mesh[21], TCMGeneDIT[22], TCMID[23,24], ETCM[25], TM_MC[26], TcmSP[27], BATMAN-TCM database[28]. Finally, the ingredients information was normalized through the compound identification number in PubChem database[29].
Acquisition of potential targets of SWC components: It is very important to obtain the interaction profiles between components in SWC and their potential targets to elucidate the mechanism of SWC[30]. In order to ensure the reliability of the target of the ingredients in SWC, we integrated the data from ten sources, which can be divided into four categories (Table 1). Marketed drug databases such as DrugBank[31] and TTD[32], activity assay databases such as ChEMBL[33] and PubChem[34], literature mining databases such as STITCH[35] and CTD[36], target prediction tools such as TargetNet[37], SwissTargetPrediction[38], ChEMBL_prediction tool[39] and BATMANTCM[ 28]. All the known targets obtained from DrugBank, TTD and ChEMBL, PubChem source was kept. Only the putative targets which can be predicted in at least two prediction models and validated by literature mining source at the same time were preserved as the potential targets of SWC. All the targets were normalized by UniProt database (http://www.uniprot.org)[40] and only the targets belonging to “Homo sapiens” were reserved for further analysis.
| # | Target_Source category | Target_Source | Interaction and target type | Selection criteria |
|---|---|---|---|---|
| 1 | Marketed drug database | DrugBank | Known | All |
| 2 | TTD database | Known | All | |
| 3 | Activity assay database | ChEMBL | Known | All |
| 4 | PubChem | Known | All | |
| 5 | Literature mining database | STITCH | Text-mining | Score≥0.9 |
| 6 | CTD | Text-mining | All | |
| 7 | Target prediction tool | TargetNet | Putative | Score=1 |
| 8 | SwissTargetPrediction | Putative | Score≥0.9 | |
| 9 | ChEMBL prediction tool | Putative | Confidence 90 % and active | |
| 10 | BATMAN-TCM | Putative | Score≥0.48 |
Table 1: 10 Target Sources Of The Ingredients In Swc.
Acquisition of the AD gene set. Based on current researches about the pathology of AD, the causal genes closely related to it were collected from MetaCore (https://portal.genego. com/), DisGeNET[41], Open Target Platform[42], MalaCards[43], OMIM[44], GeneCards[45], and CTD database[36]. The gene name from different source was standardized based on the Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.8 (https://david.abcc. ncifcrf.gov/)[46]. To ensure the credibility of AD gene set, we only retained genes that appeared in at least three data sources.
Identification of therapeutic targets of SWC in treating AD: Based on the potential targets of SWC components and AD gene set, we obtained their overlaps. This part can be regarded as the target set of SWC in the treatment of AD.
Enrichment analysis of SWC therapeutic targets in treating AD:
To verify whether the SWC target set in the treatment of AD is reliable, two kinds of enrichment analysis were performed. One is Disease Ontology (DO) enrichment, which checks whether the target set can be significantly enriched in AD or AD related diseases. clusterProfiler Version 4.0.3, an R Bioconductor package[47] was used to perform DO enrichment based on the disease annotation data in DO database (http://disease-ontology.org) [48]. The other is tissue enrichment, which examines whether currently identified SWC targets are over-represented by enriched expression in the disease-relevant tissue. Tissue Specific Expression Analysis (TSEA)[49] was taken to perform the tissue enrichment analysis by using Specificity Index thresholds (pSI) R package function to calculate pSI of varying stringency.
For the sake of interpreting the mechanisms of SWC for AD from a systematic perspective, clusterProfiler mentioned above was used to perform functional annotation and pathway enrichment analysis of the potential targets of SWC in the treatment of AD. The Gene Ontology (GO) (http://geneontology.org/), including terms of biological processes, cellular components and molecular functions, which could identify the possible biological mechanism of SWC targets[50]. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database (https://www.kegg. jp/), providing the molecular interaction/reaction network diagram, is a comprehensive knowledge base for functional interpretation from a systematic view[51].
Genes or proteins shared by different pathways may play an important role in cross-talk between different signaling pathways[52]. Therefore, in this study, we conducted frequency statistics on targets enriched into these differential pathways as one of the factors for core therapeutic target screening.
Construction and analysis of network:
In order to identify the hub targets and to explore its pharmacological mechanism, two kinds of networks were established.
Protein-Protein Interaction (PPI) network: The potential SWC targets in treatment of AD were submitted to STRING database (https://string-db.org/) to construct PPI network[53] with the highest confidence (0.9)[54]. To verify that the interactions between them were statistically valid, PPI interaction enrichment tests were conducted. Whole genome was assumed as the statistical background.
Compound-Target (C-T) network: The relationships between the compounds and targets were used to build C-T network. If a compound potentially acts on a target, they are connected by an edge. Cytoscape v3.8.1 was used to visualize the above two networks and compute the topological parameters[55]. In addition, NetworkAnalyzer plug-in of Cytoscape v3.8.1 was used to compute topological parameters. Hub nodes in the network were evaluated by the degree parameter, that is, the number of its adjacent nodes[56,57].
In order to identify the core target of SWC in the treatment of AD based on comprehensive consideration of multiple factors, a Combination Score (CS) formula for evaluating the importance of targets is defined as follow:
CS=D_1*0.2+D_2*0.4+N*0.3+T*0.1 (1)
In this formula, D1 and D2 indicates the degree value computed in PPI network and C-T network, respectively. N represents the number of significantly enriched pathways in which a target participates. T is the target type score. Here, the target type scores of known targets (T Known target) and putative targets (T Putative target) are set to 15 and 5 respectively. CS greater than 2.5 times the average CS was used as the threshold to screen the core therapeutic targets. We screen the key components based on the C-T network and take the degree greater than 2.5 times the mean as the threshold
Evaluation of Drug-Likeness (DL) of compounds in SWC:
To screen out the potential active compounds with favorable physicochemical properties and pharmacokinetics properties, SwissADME (http://www.swissadme.ch) was used to evaluate pharmacokinetics, DL and medicinal chemistry friendliness of compounds in SWC[58]. Six physicochemical properties were considered for a rapid appraisal of DL and the optimal range for each property was given below.
Lipophilicity: XLOGP3 between −0.7 and +5.0; size: MW between 150 and 500 g/mol; polarity: TPSA between 20 and 130 Å; solubility: log S no higher than 6; flexibility: no more than 9 rotatable bonds and saturation: fraction of carbons in the sp3 hybridization no less than 0.25[59,60]. Compounds fully meet the above screening criteria to be considered showing DL. This means that these compounds have potentially excellent Absorption, Distribution, Metabolism, Excretion (ADME) properties with a high development success rate and can be prioritized in subsequent studies. In this study, the predicted ADME parameters were only considered as one of the factors for screening potential active compounds.
Results and Discussion
After standardization and de duplication, a total of 798 compounds from six herbs of SWC were collected by comprehensive retrieval and integration of compound information from eight public databases; 75 in ZSW, 379 in RS, 123 in SCP, 78 in YYH, 74 in GG, and 206 in CX.
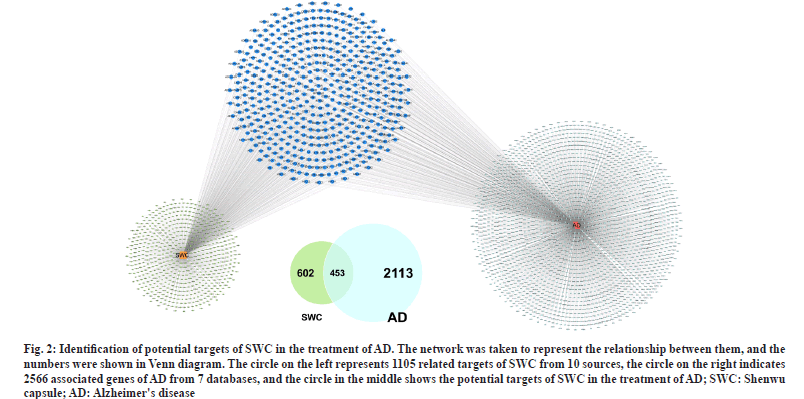
To ensure the accuracy and reliability of SWC targets, ten sources in four categories was used to obtain the corresponding target data for 798 compounds. According to the strict screening criteria, a total of 1105 SWC potential targets were obtained. Through standardization, deduplication, screening, and integration of AD gene information from seven databases, the AD gene set was constructed, which contained 2566 genes in total. The common gene shared between SWC potential targets and AD genes was considered as therapeutic targets of SWC on AD, and a total of 453 targets were obtained for subsequent analysis (fig. 2).
Fig. 2: Identification of potential targets of SWC in the treatment of AD. The network was taken to represent the relationship between them, and the numbers were shown in Venn diagram. The circle on the left represents 1105 related targets of SWC from 10 sources, the circle on the right indicates 2566 associated genes of AD from 7 databases, and the circle in the middle shows the potential targets of SWC in the treatment of AD; SWC: Shenwu capsule; AD: Alzheimer's disease.
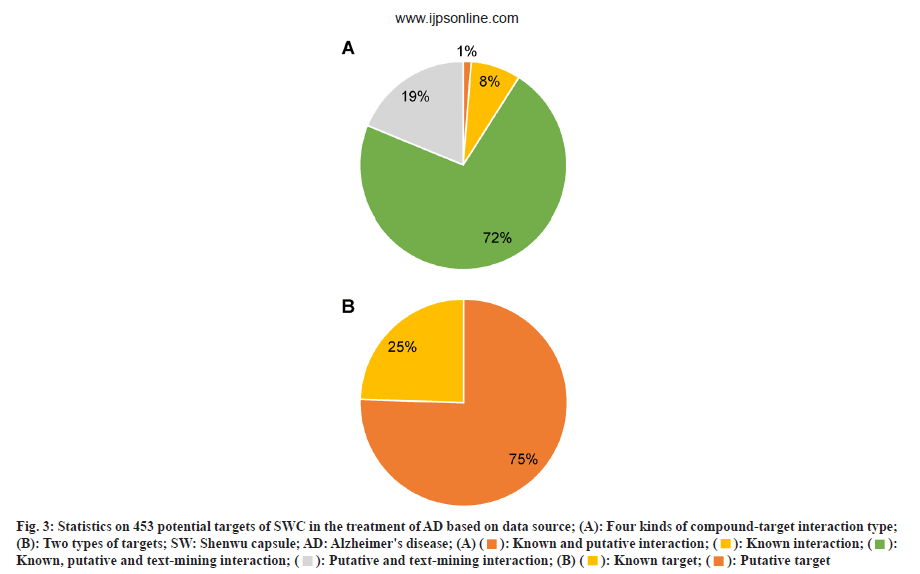
For the 453 therapeutic targets of SWC on AD, there are 12 184 compound-target interaction relationships, including 9893 known interactions (~81 %) and 2291 putative interactions (~19 %) (fig. 3A). There are 342 known targets and 111 (~75 %) putative targets (~25 %) respectively (fig. 3B). This target set can be divided into two types; 342 known targets and 111 (~75 %) putative targets (~25 %) (fig. 3B).
Fig. 3: Statistics on 453 potential targets of SWC in the treatment of AD based on data source; (A): Four kinds of compound-target interaction type; (B): Two types of targets; SW: Shenwu capsule; AD: Alzheimer's disease; (A) (  ): Known and putative interaction; (
): Known and putative interaction; ( ): Known interaction; (
): Known interaction; ( ): Known, putative and text-mining interaction; (
): Known, putative and text-mining interaction; ( ): Putative and text-mining interaction; (B) (
): Putative and text-mining interaction; (B) ( ): Known target; (
): Known target; ( ): Putative target
): Putative target
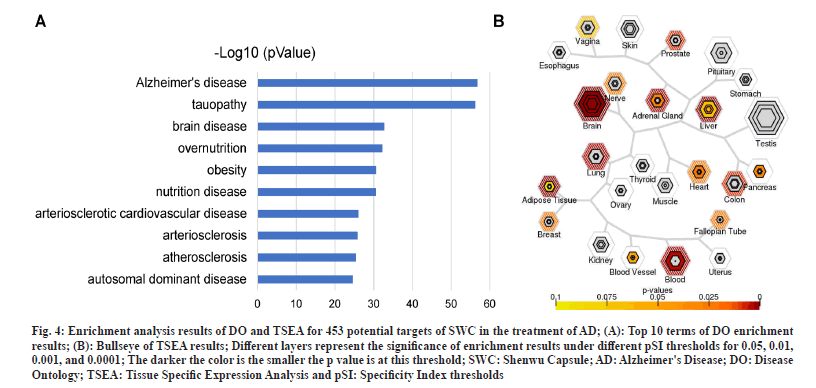
In order to verify that the above 453 therapeutic targets are indeed highly reliable and can be used for subsequent analysis, DO enrichment and tissue enrichment were performed (fig. 4A). The DO enrichment result showed that AD (P=1.88E-57) was significantly enriched. In addition, Tauopathy (P=5.92E-57) and brain disease (P=1.91E-33) ranked second and third, respectively, were both associated with AD (fig. 4A). Tissue enrichment results displayed that SWC targets in the treatment of AD could be enriched into brain tissues at different thresholds (fig. 4B). The analysis results from both angles show that 453 potential targets of SWC for the treatment of AD are highly reliable and can be used for subsequent analysis.
Fig. 4: Enrichment analysis results of DO and TSEA for 453 potential targets of SWC in the treatment of AD; (A): Top 10 terms of DO enrichment results; (B): Bullseye of TSEA results; Different layers represent the significance of enrichment results under different pSI thresholds for 0.05, 0.01, 0.001, and 0.0001; The darker the color is the smaller the p value is at this threshold; SWC: Shenwu Capsule; AD: Alzheimer's Disease; DO: Disease Ontology; TSEA: Tissue Specific Expression Analysis and pSI: Specificity Index thresholds
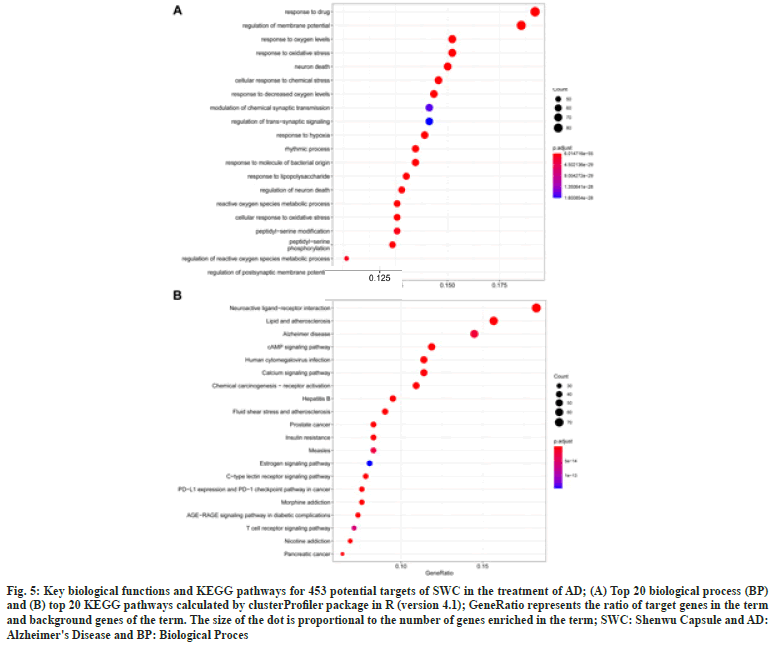
Through biological process and KEGG pathway enrichment analysis of these 453 common targets, 3102 biological processes and 194 pathways of significance were identified. The representative Gene Ontology Biological Process (GOBP) and KEGG pathway were visualized in fig. 5. The GOBP results indicated that most of these targets were associated with a variety of biological processes, such as membrane potential, oxidative stress, synaptic transmission, neuron death, hypoxia, etc. (fig. 5A). The top 5 significantly enriched KEGG pathway were neuroactive ligandreceptor interaction (P=7.37E-30), lipid and atherosclerosis (P=3.12E-34), AD (P=1.30E-15), cyclic Adenosine 3′,5′-Monophosphate (cAMP) signaling pathway (P=1.09E-19), human cytomegalovirus infection (P=1.22E-17), and calcium signaling pathway (P=1.92E-16) (fig. 5B).
Fig. 5: Key biological functions and KEGG pathways for 453 potential targets of SWC in the treatment of AD; (A) Top 20 biological process (BP) and (B) top 20 KEGG pathways calculated by clusterProfiler package in R (version 4.1); GeneRatio represents the ratio of target genes in the term and background genes of the term. The size of the dot is proportional to the number of genes enriched in the term; SWC: Shenwu Capsule and AD: Alzheimer's Disease and BP: Biological Proces.
The PPI network of SWC targets in the treatment of AD was developed to quantify the function of specific proteins and identify the hub targets at the systematic level. The PPI network consisted of 453 nodes and 1982 edges. The average node degree and the average local clustering coefficient were 8.75 and 0.397, respectively. PPI interaction enrichment result indicated that the PPI network has significantly more interactions than what would be expected for a random set of proteins of similar size, drawn from the genome (p<1.0e-16). This means that these proteins are closely connected biologically as a group. The degree of nodes in the network was calculated.
The relationships between 453 therapeutic targets of SWC on AD and the compounds in SWC were used to construct C-T network. There are 1224 nodes (including 453 targets and 771 compounds) and 12 184 edges. The degree of nodes in C-T network was calculated. In this network, the average number of targets per compound is 15.8, indicating the multi-target property of SWC ingredients.
Based on the definition of Combination Score (CS), we ranked and identified the core target of SWC in the treatment of AD. The mean value of CS is 16.7. According to the threshold, 35 core therapeutic targets were screened (Table 2), including 32 known targets and 3 putative targets. The average of degree of compounds in C-T network is 15.8. According to the threshold, we screened 55 key components and the top 30 compounds were displayed in Table 3.
Interpretation of the mechanism of SWC intervention in AD pathway:
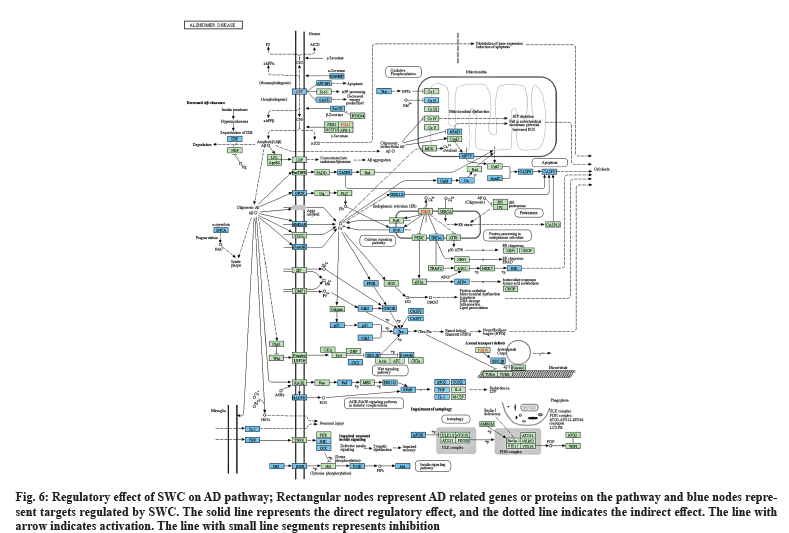
Based on the results of pathway enrichment, 61 therapeutic targets enriched in the AD pathway (hsa05010) were displayed (fig. 6). We found that targets were widely distributed in this pathway and were involved in regulating multiple biological processes, such as neuronal injury, apoptosis, energy production, synaptic dysfunction, reactive oxygen species, mitochondrial dysfunction, neurofibrillary tangles, long-term potentiation, autophagy and axonal transport defects, ultimately affecting the survival of neurons. In addition, we also found the cross-talk effect between this pathway and other pathways, such as calcium signaling pathway (p=1.92E-16), apoptosis (p=1.92E-16), Wnt signaling pathway (p=0.00097), AGE-RAGE signaling pathway in diabetic complications (p=1.31E-16), insulin signaling pathway (p=5.06E-07), autophagy (p=1.13E-05).
Fig. 6: Regulatory effect of SWC on AD pathway; Rectangular nodes represent AD related genes or proteins on the pathway and blue nodes represent targets regulated by SWC. The solid line represents the direct regulatory effect, and the dotted line indicates the indirect effect. The line with arrow indicates activation. The line with small line segments represents inhibition.
Nine of the 35 core targets are distributed in this pathway, all of which are known targets, including RELA, MAPK1, CASP9, APP, AKT1, PIK3CA, TNF, PIK3R1 and CASP3 (Table 2). Moreover, all the 30 key components in SWC have potential regulatory effects on this pathway (Table 3).
| Gene Symbol | Gene ID | D1 | D2 | N | T | CS | Target type | In AD pathway |
|---|---|---|---|---|---|---|---|---|
| HTR3A | 3359 | 0 | 280 | 2 | 15 | 114.1 | Known target | FALSE |
| TLR9 | 54106 | 1 | 231 | 7 | 15 | 96.2 | Known target | FALSE |
| MGLL | 11343 | 2 | 232 | 2 | 5 | 94.3 | Putative target | FALSE |
| MIF | 4282 | 2 | 226 | 2 | 15 | 92.9 | Known target | FALSE |
| RELA | 5970 | 51 | 142 | 73 | 15 | 90.4 | Known target | TRUE |
| PTGS1 | 5742 | 7 | 209 | 4 | 15 | 87.7 | Known target | FALSE |
| ALOX15 | 246 | 8 | 189 | 4 | 15 | 79.9 | Known target | FALSE |
| NOS3 | 4846 | 17 | 165 | 18 | 15 | 76.3 | Known target | FALSE |
| ESR1 | 2099 | 43 | 156 | 7 | 15 | 74.6 | Known target | FALSE |
| MAPK1 | 5594 | 59 | 45 | 116 | 15 | 66.1 | Known target | TRUE |
| ITGA4 | 3676 | 6 | 146 | 10 | 15 | 64.1 | Known target | FALSE |
| HDAC4 | 9759 | 8 | 147 | 5 | 15 | 63.4 | Known target | FALSE |
| ABCG2 | 9429 | 2 | 151 | 3 | 15 | 63.2 | Known target | FALSE |
| CASP9 | 842 | 8 | 123 | 32 | 15 | 61.9 | Known target | TRUE |
| APP | 351 | 20 | 138 | 3 | 15 | 61.6 | Known target | TRUE |
| AHR | 196 | 7 | 143 | 4 | 15 | 61.3 | Known target | FALSE |
| MAOA | 4128 | 6 | 134 | 11 | 15 | 59.6 | Known target | FALSE |
| AKT1 | 207 | 64 | 36 | 97 | 15 | 57.8 | Known target | TRUE |
| HSP90AA1 | 3320 | 67 | 97 | 13 | 15 | 57.6 | Known target | FALSE |
| CYP17A1 | 1586 | 4 | 137 | 4 | 5 | 57.3 | Putative target | FALSE |
| AR | 367 | 27 | 117 | 3 | 15 | 54.6 | Known target | FALSE |
| PIK3CA | 5290 | 51 | 31 | 101 | 15 | 54.4 | Known target | TRUE |
| CYP19A1 | 1588 | 4 | 126 | 1 | 15 | 53 | Known target | FALSE |
| ALPL | 249 | 0 | 128 | 0 | 15 | 52.7 | Known target | FALSE |
| MAOB | 4129 | 4 | 115 | 11 | 15 | 51.6 | Known target | FALSE |
| TNF | 7124 | 27 | 69 | 56 | 15 | 51.3 | Known target | TRUE |
| PIK3R1 | 5295 | 53 | 21 | 100 | 15 | 50.5 | Known target | TRUE |
| DNMT1 | 1786 | 10 | 116 | 1 | 15 | 50.2 | Known target | FALSE |
| SLC5A2 | 6524 | 1 | 118 | 0 | 15 | 48.9 | Known target | FALSE |
| TP53 | 7157 | 68 | 47 | 47 | 15 | 48 | Known target | FALSE |
| ESR2 | 2100 | 6 | 108 | 6 | 15 | 47.7 | Known target | FALSE |
| CAT | 847 | 6 | 108 | 7 | 5 | 47 | Putative target | FALSE |
| FAAH | 2166 | 3 | 109 | 1 | 15 | 46 | Known target | FALSE |
| PGR | 5241 | 8 | 98 | 5 | 15 | 43.8 | Known target | FALSE |
| CASP3 | 836 | 24 | 63 | 40 | 15 | 43.5 | Known target | TRUE |
Table 2: 35 Core Targets Of Swc In The Treatment Of Ad Based On Cs.
| CID | Name | D2 | Regulate AD pathway | Herb | Drug-like |
|---|---|---|---|---|---|
| 6623 | Bisphenol A | 279 | TRUE | RS | |
| 5280343 | Quercetin | 251 | TRUE | ZSW; YYH; GG; CX | |
| 445154 | Resveratrol | 228 | TRUE | ZSW | |
| 5957 | Adenosine-5'-triphosphate | 216 | TRUE | RS | |
| 5280961 | Genistein | 162 | TRUE | GG | |
| 985 | Palmitic acid | 122 | TRUE | RS; SCP; YYH; GG; CX | |
| 261 | Butyraldehyde | 111 | TRUE | CX | |
| 8343 | Bis(2-ethylhexyl) phthalate | 110 | TRUE | GG | |
| 446220 | Cocaine | 110 | TRUE | CX | TRUE |
| 611 | DL-Glutamic acid | 101 | TRUE | SCP | |
| 445639 | Oleic acid | 101 | TRUE | RS; CX | |
| 5280445 | Luteolin | 99 | TRUE | YYH | |
| 135398658 | Folic acid | 98 | TRUE | RS; CX | |
| 5280443 | Apigenin | 95 | TRUE | CX | |
| 119 | gamma-Aminobutyric acid | 87 | TRUE | SCP | |
| 5281672 | Myricetin | 83 | TRUE | ZSW | |
| 264 | Butyric acid | 82 | TRUE | CX | TRUE |
| 5280863 | Kaempferol | 82 | TRUE | ZSW; RS; YYH | |
| 60961 | Adenosine | 78 | TRUE | RS | |
| 5281707 | Coumestrol | 78 | TRUE | GG | |
| 6654 | alpha-Pinene | 72 | TRUE | RS; SCP; CX | |
| 3220 | Emodin | 67 | TRUE | ZSW; SCP | |
| 753 | Glycerol | 66 | TRUE | ZSW | |
| 1110 | Succinic acid | 65 | TRUE | RS; YYH | TRUE |
| 3893 | Lauric acid | 65 | TRUE | RS | |
| 5281708 | Daidzein | 65 | TRUE | YYH; GG | |
| 9064 | Cianidanol | 62 | TRUE | ZSW; YYH | |
| 370 | Gallic acid | 61 | TRUE | ZSW; YYH; CX | |
| 7127 | Methyleugenol | 61 | TRUE | SCP; CX | |
| 5280450 | Linoleic acid | 61 | TRUE | RS; SCP; YYH; CX |
Table 3: Top 30 Key Components In Swc With Therapeutic Effects On Ad
In this study, the key biological processes and KEGG signaling pathways, core target set and key active ingredients of SWC in the treatment of AD were identified based on the network pharmacology research strategy, and the regulatory mechanism of SWC was illustrated by the AD pathway. These results provide the possibility to understand the complex mechanism of TCM from the perspective of system and provide the direction for further research.
In the process of obtaining the therapeutic targets of SWC for AD, we used the data of 10 sources in 4 categories for integration, and adopted very strict screening criteria for the data of predicted sources before integration. Of the 453 targets, ~75 % were known targets, and the other ~25 % predicted targets were verified by data from the two text mining sources, which ensured the reliability of subsequent analysis results. In addition, we performed DO and tissue enrichment analysis to further verify and ensure the reliability of 453 therapeutic targets used for mechanism interpretation. It is worth mentioning that the definition of known targets in this study is those verified by relevant activity tests, among which activity tests at the biochemical level account for a large proportion. Therefore, verification can be carried out on corresponding cells or animal models according to actual needs in subsequent studies.
In order to elucidate the main regulatory pathways of SWC in the treatment of AD from a systematic perspective, biological processes and signaling pathways were enriched. The therapeutic targets are mainly enriched in membrane potential[61], oxidative stress[62], synaptic transmission[63], neuron death[64], hypoxia[65] are biological processes closely related to the physiological process of AD. SWC may exert pharmacological effects on AD by integrating and regulating these biological processes.
According to CS definition and screening threshold, a total of 35 core targets were obtained. The first two known targets are HTR3A and Toll-Like Receptors (TLR)9. As we know, 5-Hydroxytryptamine (5-HT) is primarily involved in the regulation of learning and memory. HTR3A (CS=114.1) was found to be the highest ranked, which has been found that relative mRNA expression of HTR2A, HTR3A (CS=114.1) and MAOA (CS=59.6) were significantly lower in PBMCs of patients with Late-Onset AD (LOAD) compared with controls[66]. At present, 5-HT receptor antagonists as a symptomatic treatment for cognitive and/or behavioral symptoms of AD have been studied[67,68]. TLR signaling pathway(s) may be involved in clearance of Aβ-deposits and direct or indirect activation of specific TLR such as TLR7, TLR8, and TLR9 (CS=96.2), can induce Aβ uptake or inflammatory response, which can be a therapeutic target for AD[69]. MGLL (CS=94.3), a putative target in this study, can regulate lipid metabolism and influence adult neurogenesis, thereby predisposing to AD during aging. It has been treated as a potential target for neurodegenerative diseases[70]. MIF (CS=92.9), a pro-inflammatory cytokine, was upregulated in the brain of AD patients. Its overexpression can significantly protect neuronal cells from Aβ-induced cytotoxicity[71]. RELA (CS=90.4) participates in the AD pathway, and is associated with Inflammatory process[72]. We will not discuss the relationship between these core targets and AD one by one. The above analysis proves that the core targets of SWC are highly reliable and representative, and that SWC treatment of AD is achieved through multiple targets and processes.
As for the key components obtained by the degree parameter, bisphenol A (D2=279) from Panax ginseng C.A.Mey (RS) ranked highest. It can mediate AD-like neurotoxicity by affecting brain insulin resistance[73]. Quercetin (D2=251) from four herbs possesses anti-Alzheimer, antidiabetic and anti-obesity effects with clinical evidence and has been suggested as a promising therapeutic agent[74]. Several studies have shown that resveratrol (D2=228) from Radix polygoni multiflori preparata (ZSW) regulates many aspects of AD, such as neuro-inflammation, adaptive immunity, autophagy, antioxidants and estrogen and has excellent therapeutic potential[75,76]. Genistein (D2=162), a representative compound from Pueraria lobate (GG), has been found to target directly the Aβ and tau to control the intracellular signaling pathways responsible for neurons death in the AD brain[77]. Part of Chinese herbal medicine prescription toxic side effects, and under the guidance of the theoretical system of TCM compatibility application can reduce the toxic side effects, at the same time to achieve synergistic effect[78]. In this study, we can explain the theory of synergism and toxicity reduction at the molecular level. As for the synergy mechanism, quercetin, resveratrol, apigenin (D2=95), and kaempferol (D2=82) are flavonoids have shown the potential activity against AD. They possess anti-amyloidogenic and fibril-destabilization activity, as well as being able to act as metal chelators and to suppressing oxidative stress[79]. For mechanism of toxicity reduction, quercetin can abrogate bisphenol A induced altered neurobehavioral response and oxidative stress in zebrafish by modulating brain antioxidant defense system[80]. In addition, resveratrol can prevent bisphenol A-induced autism, type 2 diabetes mellitus, and metabolic syndrome by augmenting brain-derived neurotrophic factor synthesis and action[81]. Through the above analysis, we can identify the active ingredients in SWC, both toxic and beneficial, in a relatively reliable manner. This not only provides the material basis for explaining the mechanism of TCM action, but also provides the possibility for understanding the compatibility theory of TCM from the molecular level, such as the theory of synergism and toxicity reduction.
Data analysis and mining in this study provide many verifiable directions for subsequent research. Although there are few studies on SWC and AD at present, a series of studies can be carried out based on the highly reliable findings of this study in the future, such as the relationship between some core targets and key components, the synergistic mechanism of key components, and the regulatory effects of key targets in a specific signaling pathway.
The purpose of this study is to elucidate the mechanism of SWC in the treatment of AD from a systematic perspective and explore the key material basis through network pharmacology technology. The results shows that SWC exerted its pharmacological effects on AD by regulating a variety of biological processes, such as membrane potential, oxidative stress, synaptic transmission, neuron death, hypoxia, etc., and multiple pathways, including neuroactive ligand-receptor interaction, lipid and atherosclerosis, AD, cAMP signaling pathway, human cytomegalovirus infection, and calcium signaling pathway, etc. HTR3A, TLR9, MGLL, MIF, RELA, PTGS1, ALOX15, NOS3, ESR1, MAPK1, ITGA4, HDAC4, ABCG2, CASP9, APP, AHR and so on are the main therapeutic targets of SWC. The key regulatory and pharmacological components of SWC include quercetin, resveratrol, genistein, palmitic acid, butyraldehyde, bis (2-ethylhexyl) phthalate, cocaine, DL-Glutamic acid, oleic acid, luteolin, folic acid, apigenin, gamma-aminobutyric acid, etc. This study provides a theoretical basis and research paradigm for elucidating the synergistic effects of multiple components, multiple targets, and multiple pathways in TCM treatment of diseases from a systematic perspective, which will undoubtedly promote the modernization of TCM. Although the data used in this study is highly reliable, it is only based on data mining and data analysis. Therefore, further clinical verification studies on the role of SWC in AD should be carried out, and multi-level verification should be carried out in combination with actual research needs, such as cell models and animal models.
Acknowledgments:
We gratefully acknowledge financial support from the Hainan Province Science and Technology Special Fund (No. ZDYF2022SHFZ304 and No. ZDKJ2021034). We also wish to extend our thanks to any individual for their assistance in the conduction of this study.
Author Contributions:
Yitao Xing and Nianhong Chen conducted and completed the data analysis and manuscript writing and contributed equally to this work. Dingguo Wang, Xueying Lin and Tiandong Lin contributed to the data of predictive target for components. Yanting Song contributed to the systematic search and study selection. Jinsheng Zhuo provided some good suggestions and supervision. All authors contributed to the article and approved the submitted version.
Funding:
This research was funded by the Hainan Province Science and Technology Special Fund (No. ZDYF2022SHFZ304 and No. ZDKJ2021034).
Conflict of interests:
The authors declare no competing interests.
References
- Scheltens P, Blennow K, Breteler MM, De Strooper B, Frisoni GB, Salloway S, et al. Alzheimer's disease. Lancet 2016;388(10043):505-17.
- Tiwari S, Atluri V, Kaushik A, Yndart A, Nair M. Alzheimer’s disease: Pathogenesis, diagnostics, and therapeutics. Int J Nanomed 2019:5541-54.
[Crossref] [Google Scholar] [PubMed]
- Alzheimer's Association. 2015 Alzheimer's disease facts and figures. Alzheimer's Dement 2015;11(3):332-84.
- Lane CA, Hardy J, Schott JM. Alzheimer's disease. Eur J Neurol 2018;25(1):59-70.
- Jack Jr CR, Bennett DA, Blennow K, Carrillo MC, Dunn B, Haeberlein SB, et al. NIA-AA research framework: Toward a biological definition of Alzheimer's disease. Alzheimer's Dement 2018;14(4):535-62.
[Crossref] [Google Scholar] [PubMed]
- Hane FT, Lee BY, Leonenko Z. Recent progress in Alzheimer’s disease research, part 1: pathology. J Alzheimer's Dis 2017;57(1):1-28.
[Crossref] [Google Scholar] [PubMed]
- Gao LB, Yu XF, Chen Q, Zhou D. Alzheimer's disease therapeutics: Current and future therapies. Minerva Med 2016;107(2):108-13.
[Google Scholar] [PubMed]
- Imbimbo BP, Ottonello S, Frisardi V, Solfrizzi V, Greco A, Seripa D, et al. Solanezumab for the treatment of mild-to-moderate Alzheimer’s disease. Expert Rev Clin Immunol 2012;8(2):135-49.
[Crossref] [Google Scholar] [PubMed]
- Wang H, Li L, Ye CF, Zhang L. Effect of shenwu capsule on the peripheral blood lymphocyte DNA of rat model induced by beta-amyloid injection. Zhongguo Zhong Yao Za Zhi 2004;29(2):176-9.
[Google Scholar] [PubMed]
- Zhang L, Zhao N, Wang R, Li L. Effects of Shenwu capsule and its component tetrahydroxystilbene glucoside on expression of neurotrophic factors in lumbar spinal cord of aged rats. Zhongguo Zhong Yao Za Zhi 2009;34(12):1557-61.
[Google Scholar] [PubMed]
- Zhang L, Zhang RY, Li YL, Ye CF, Li L. Effects of Shenwu capsule on learning-memory ability and cholinergic function of brain in AD-like rat model induced by chronic infusion of sodium azide by minipump. Zhongguo Zhong Yao Za Zhi 2013;38(9):1300-5.
[Google Scholar] [PubMed]
- Hopkins AL. Network pharmacology. Nat Biotechnol 2007;25(10):1110-1.
- Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol 2008;4(11):682-90.
- Shao LI, Zhang B. Traditional Chinese medicine network pharmacology: Theory, methodology and application. Chin J Nat Med 2013;11(2):110-20.
[Crossref] [Google Scholar] [PubMed]
- Zhao S, Li S. A co-module approach for elucidating drug–disease associations and revealing their molecular basis. Bioinformatics 2012;28(7):955-61.
[Crossref] [Google Scholar] [PubMed]
- Li S, Zhang B, Zhang N. Network target for screening synergistic drug combinations with application to traditional Chinese medicine. BMC Syst Biol 2011;5(1):S10.
[Crossref] [Google Scholar] [PubMed]
- Zhao S, Li S. Network-based relating pharmacological and genomic spaces for drug target identification. PloS One 2010;5(7):e11764.
[Crossref] [Google Scholar] [PubMed]
- Liang X, Li H, Li S. A novel network pharmacology approach to analyse traditional herbal formulae: The Liu-Wei-Di-Huang pill as a case study. Mol Biosyst 2014;10(5):1014-22.
[Crossref] [Google Scholar] [PubMed]
- Zheng J, Wu M, Wang H, Li S, Wang X, Li Y, et al. Network pharmacology to unveil the biological basis of health-strengthening herbal medicine in cancer treatment. Cancers 2018;10(11):461.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Zhou H, Liu YB, Wang JF, Li H, Ung CY, et al. Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br J Pharmacol 2006;149(8):1092-103.
[Crossref] [Google Scholar] [PubMed]
- Zhang RZ, Yu SJ, Bai H, Ning K. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Sci Rep 2017;7(1):2821.
- Fang YC, Huang HC, Chen HH, Juan HF. TCMGeneDIT: A database for associated traditional Chinese medicine, gene and disease information using text mining. BMC Complement Altern Med 2008;8(1):58.
[Crossref] [Google Scholar] [PubMed]
- Xue R, Fang Z, Zhang M, Yi Z, Wen C, Shi T. TCMID: Traditional Chinese medicine integrative database for herb molecular mechanism analysis. Nucl Acids Res 2012;41(D1):D1089-95.
[Crossref] [Google Scholar] [PubMed]
- Huang L, Xie D, Yu Y, Liu H, Shi Y, Shi T, Wen C. TCMID 2.0: A comprehensive resource for TCM. Nucl Acids Res 2018;46(D1):D1117-20.
[Crossref] [Google Scholar] [PubMed]
- Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucl Acids Res 2019;47(D1):D976-82.
- Kim SK, Nam S, Jang H, Kim A, Lee JJ. TM-MC: A database of medicinal materials and chemical compounds in Northeast Asian traditional medicine. BMC Complement Altern Med 2015;15(1):218.
[Crossref] [Google Scholar] [PubMed]
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:134.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H, et al. BATMAN-TCM: A bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci Rep 2016;6(1):21146.
[Crossref] [Google Scholar] [PubMed]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2019 update: Improved access to chemical data. Nucl Acids Res 2019;47(D1):D1102-9.
[Crossref] [Google Scholar] [PubMed]
- Rix U, Superti-Furga G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol 2009;5(9):616-24.
[Crossref] [Google Scholar] [PubMed]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucl Acids Res 2018;46(D1):D1074-82.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Zhang S, Li F, Zhou Y, Zhang Y, Wang Z, et al. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res 2020;48(D1):D1031-41.
- Gaulton A, Hersey A, Nowotka M, Bento AP, Chambers J, Mendez D, et al. The ChEMBL database in 2017. Nucl Acids Res 2017;45(D1):D945-54.
[Crossref] [Google Scholar] [PubMed]
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, et al. PubChem substance and compound databases. Nucl Acids Res 2016;44(D1):D1202-13.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Santos A, Von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucl Acids Res 2016;44(D1):D380-4.
[Crossref] [Google Scholar] [PubMed]
- Davis AP, Grondin CJ, Johnson RJ, Sciaky D, McMorran R, Wiegers J, et al. The comparative toxicogenomics database: Update 2019. Nucleic Acids Res 2019;47(D1):D948-54.
[Crossref] [Google Scholar] [PubMed]
- Yao ZJ, Dong J, Che YJ, Zhu MF, Wen M, Wang NN, et al. TargetNet: A web service for predicting potential drug–target interaction profiling via multi-target SAR models. J Computer Aided Mol Des 2016;30(5):413-24.
[Crossref] [Google Scholar] [PubMed]
- Gfeller D, Grosdidier A, Wirth M, Daina A, Michielin O, Zoete V. SwissTargetPrediction: A web server for target prediction of bioactive small molecules. Nucl Acids Res 2014;42(W1):W32-8.
[Crossref] [Google Scholar] [PubMed]
- Bosc N, Atkinson F, Felix E, Gaulton A, Hersey A, Leach AR. Large scale comparison of QSAR and conformal prediction methods and their applications in drug discovery. J Cheminform 2019;11:1-6.
- UniProt C. UniProt: A worldwide hub of protein knowledge. Nucl Acids Res 2019;47(D1):D506-15.
[Crossref] [Google Scholar] [PubMed]
- Piñero J, Ramírez-Anguita JM, Saüch-Pitarch J, Ronzano F, Centeno E, Sanz F, et al. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucl Acids Res 2020;48(D1):D845-55.
[Crossref] [Google Scholar] [PubMed]
- Carvalho-Silva D, Pierleoni A, Pignatelli M, Ong C, Fumis L, Karamanis N, et al. Open Targets Platform: New developments and updates 2 y on. Nucl Acids Res 2019;47(D1):D1056-65.
- Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J, et al. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucl Acids Res 2017;45(D1):D877-87.
[Crossref] [Google Scholar] [PubMed]
- Amberger JS, Bocchini CA, Scott AF, Hamosh A. OMIM. org: Leveraging knowledge across phenotype–gene relationships. Nucl acids Res 2019;47(D1):D1038-43.
[Crossref] [Google Scholar] [PubMed]
- Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, et al. GeneCards Version 3: The human gene integrator. Database 2010;2010:baq020.
[Crossref] [Google Scholar] [PubMed]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol 2003;4(9):P3.
[Crossref] [Google Scholar] [PubMed]
- Yu G, Wang LG, Han Y, He QY. Cluster profiler: An R package for comparing biological themes among gene clusters. Omics 2012;16(5):284-7.
[Crossref] [Google Scholar] [PubMed]
- Schriml LM, Arze C, Nadendla S, Chang YW, Mazaitis M, Felix V, et al. Disease ontology: A backbone for disease semantic integration. Nucl Acids Res 2012;40(D1):D940-6.
[Crossref] [Google Scholar] [PubMed]
- Dougherty JD, Schmidt EF, Nakajima M, Heintz N. Analytical approaches to RNA profiling data for the identification of genes enriched in specific cells. Nucleic acids Res 2010;38(13):4218-30.
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. Nat Genet 2000;25(1):25-9.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucl Acids Res 2017;45(D1):D353-61.
[Crossref] [Google Scholar] [PubMed]
- Guo X, Wang XF. Signaling cross-talk between TGF-β/BMP and other pathways. Cell Res 2009;19(1):71-88.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucl Acids Res 2016, 2017. 45(D1):D362-8.
[Crossref] [Google Scholar] [PubMed]
- Talwar P, Gupta R, Kushwaha S, Agarwal R, Saso L, Kukreti S, et al. Viral induced oxidative and inflammatory response in Alzheimer's disease pathogenesis with identification of potential drug candidates: A systematic review using systems biology approach. Curr Neuropharmacol 2019;17(4):352-65.
[Crossref] [Google Scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google Scholar] [PubMed]
- Assenov Y, Ramírez F, Schelhorn SE, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics 2008;24(2):282-4.
[Crossref] [Google Scholar] [PubMed]
- Karbalaei R, Allahyari M, Rezaei-Tavirani M, Asadzadeh-Aghdaei H, Zali MR. Protein-protein interaction analysis of Alzheimers disease and NAFLD based on systems biology methods unhide common ancestor pathways. Gastroenterol Hepatol Bed Bench 2018;11(1):27-33.
[Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7(1):42717.
[Crossref] [Google Scholar] [PubMed]
- Lovering F, Bikker J, Humblet C. Escape from flatland: Increasing saturation as an approach to improving clinical success. J Med Chem 2009;52(21):6752-6.
[Crossref] [Google Scholar] [PubMed]
- Ritchie TJ, Ertl P, Lewis R. The graphical representation of ADME-related molecule properties for medicinal chemists. Drug Discov Today 2011;16(1-2):65-72.
[Crossref] [Google Scholar] [PubMed]
- Reiss AB, Arain HA, Stecker MM, Siegart NM, Kasselman LJ. Amyloid toxicity in Alzheimer’s disease. Rev Neurosci 2018;29(6):613-27.
[Crossref] [Google Scholar] [PubMed]
- Jiang T, Sun Q, Chen S. Oxidative stress: A major pathogenesis and potential therapeutic target of antioxidative agents in Parkinson’s disease and Alzheimer’s disease. Prog Neurobiol 2016;147:1-9.
[Crossref] [Google Scholar] [PubMed]
- Selkoe DJ. Alzheimer's disease is a synaptic failure. Science 2002;298(5594):789-91.
[Crossref] [Google Scholar] [PubMed]
- Butterfield DA, Halliwell B. Oxidative stress, dysfunctional glucose metabolism and Alzheimer disease. Nat Rev Neurosci 2019;20(3):148-60.
[Crossref] [Google Scholar] [PubMed]
- Peers C, Pearson HA, Boyle JP. Hypoxia and Alzheimer’s disease. Essays Biochem 2007;43:153-64.
[Crossref] [Google Scholar] [PubMed]
- Neshan M, Campbell A, Malakouti SK, Zareii M, Ahangari G. Gene expression of serotonergic markers in peripheral blood mononuclear cells of patients with late-onset Alzheimer's disease. Heliyon 2020;6(8):e04716.
- Ferrero H, Solas M, Francis PT, Ramirez MJ. Serotonin 5-HT 6 receptor antagonists in Alzheimer’s disease: Therapeutic rationale and current development status. CNS Drugs 2017;31(1):19-32.
[Crossref] [Google Scholar] [PubMed]
- Lalut J, Karila D, Dallemagne P, Rochais C. Modulating 5-HT4 and 5-HT6 receptors in Alzheimer’s disease treatment. Future Med Chem 2017;9(8):781-95.
[Crossref] [Google Scholar] [PubMed]
- Tahara K, Kim HD, Jin JJ, Maxwell JA, Li L, Fukuchi KI. Role of toll-like receptor signalling in Aβ uptake and clearance. Brain 2006;129(11):3006-19.
[Crossref] [Google Scholar] [PubMed]
- Syal C, Kosaraju J, Hamilton L, Aumont A, Chu A, Sarma SN, et al. Dysregulated expression of monoacylglycerol lipase is a marker for anti-diabetic drug metformin-targeted therapy to correct impaired neurogenesis and spatial memory in Alzheimer's disease. Theranostics 2020;10(14):6337.
[Crossref] [Google Scholar] [PubMed]
- Zhang S, Zhao J, Zhang Y, Zhang Y, Cai F, Wang L, et al. Up regulation of MIF as a defense mechanism and a biomarker of Alzheimer’s disease. Alzheimer's Res Ther 2019;11(1):54.
[Crossref] [Google Scholar] [PubMed]
- Xie L, Zhang N, Zhang Q, Li C, Sandhu AF, Williams III G, et al. Inflammatory factors and amyloid β-induced microglial polarization promote inflammatory crosstalk with astrocytes. Aging 2020;12(22):22538.
[Crossref] [Google Scholar] [PubMed]
- Wang T, Xie C, Yu P, Fang F, Zhu J, Cheng J, et al. Involvement of insulin signaling disturbances in bisphenol a-induced alzheimer’s disease-like neurotoxicity. Sci Rep 2017;7(1):7497.
[Crossref] [Google Scholar] [PubMed]
- Ebrahimpour S, Zakeri M, and Esmaeili A. Crosstalk between obesity, diabetes, and Alzheimer ’s disease: Introducing quercetin as an effective triple herbal medicine. Ageing Res Rev 2020;62:101095.
[Crossref] [Google Scholar] [PubMed]
- Turner RS, Thomas RG, Craft S, Van Dyck CH, Mintzer J, Reynolds BA, et al. A randomized, double-blind, placebo-controlled trial of resveratrol for Alzheimer disease. Neurology 2015;85(16):1383-91.
[Crossref] [Google Scholar] [PubMed]
- Yan Y, Yang H, Xie Y, Ding Y, Kong D, Yu H. Research progress on Alzheimer's disease and resveratrol. Neurochem Res 2020;45(5):989-1006.
[Crossref] [Google Scholar] [PubMed]
- Uddin MS, Kabir MT. Emerging signal regulating potential of genistein against Alzheimer’s disease: A promising molecule of interest. Front Cell Dev Biol 2019;7:197.
[Crossref] [Google Scholar] [PubMed]
- Wang KX, Gao Y, Gong WX, Ye XF, Fan LY, Wang C, et al. A novel strategy for decoding and validating the combination principles of Huanglian Jiedu decoction from multi-scale perspective. Front Pharmacol 2020;11:567088.
[Crossref] [Google Scholar] [PubMed]
- Simunkova M, Alwasel SH, Alhazza IM, Jomova K, Kollar V, Rusko M, et al. Management of oxidative stress and other pathologies in Alzheimer’s disease. Arch Toxicol 2019;93(9):2491-513.
[Crossref] [Google Scholar] [PubMed]
- Sahoo PK, Pradhan LK, Aparna S, Agarwal K, Banerjee A, Das SK. Quercetin abrogates bisphenol A induced altered neurobehavioral response and oxidative stress in zebrafish by modulating brain antioxidant defence system. Environ Toxicol Pharmacol 2020;80:103483.
[Crossref] [Google Scholar] [PubMed]