- *Corresponding Author:
- S. K. Konidala
Department of Pharmaceutical Sciences, Vignan’s Foundation for Science Technology and Research, Guntur, Andhra Pradesh 522213, India
E-mail: sathishkonidala@gmail.com
| Date of Received | 01 June 2022 |
| Date of Revision | 07 June 2023 |
| Date of Acceptance | 02 November 2023 |
| Indian J Pharm Sci 2023;85(6):1551-1561 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The release of antimicrobials from the hospital, pharma industries, agriculture and animal husbandry waste into various environmental sources has considerably increased. This triggers the development of antimicrobial resistance and antimicrobial resistance genes in the ecosystem. Hence, high throughput simultaneous analytical methods are necessary to analyze multiple classes of antibiotics in aquaculture products, soil, meat, honey, milk, water resources and wastewater. This concise review describes various analytical methodologies and sample processing techniques frequently used to determine multiple classes of antibiotics in the diverse field of interest. This article also details the frequently detected antibiotics, various liquid-liquid and solid-phase extraction methods, their applications and the role of pKa values in the extraction efficiency of antibiotics.
Keywords
Antibiotics, liquid Chromatography with tandem mass spectrometry, extraction techniques, antimicrobial resistance, antimicrobial resistance genes, pKa values, solid-phase extraction cartridges
Worldwide, water sources are contaminated by pharmaceutically active compounds, especially antibiotics that enter the water sources via hospital waste, industrial waste, animal husbandry, and animal livestock. Based on the recent joint surveillance report on Antimicrobial Resistance (AMR) by World Health Organization (WHO) and European Center for Disease Prevention and Control (ECDC), more than 670 000 drug-resistant bacterial infections occurred in the Europe/European economic area alone and approximately 33 000 people died as a direct consequence of these infections, and the health burden of AMR is comparable to that of influenza, tuberculosis, and Human immunodeficiency virus (HIV) combined. Several countries have a rising concern about increasing resistance towards third-generation cephalosporins and carbapenems in Klebsiella pneumoniae and carbapenem-resistant Acinetobacter. The recommendation from WHO/ECDC includes disseminating resistant clones in healthcare and other potential areas and creating awareness of the severe limitations in treatment options for patients with infections caused by these pathogens[1].
Generally, pharmaceutical compounds, including antibiotics, Non-Steroidal Anti-Inflammatory Drugs (NSAIDs), steroids and analgesics are found at ng/l to µg/l in environmental resources such as sewage water, wastewater, surface water, groundwater and soil. This may be found in either parent form or a transformed state, i.e., metabolized and called ‘pseudo persistent’ as it contributes continuously to the environment system[2]. This is a concerning factor as the antibiotic load in the environment contributes to the proliferation of bacteria, which shows a resistant nature.
In most scenarios, wastewater treatment plants do not efficiently treat pharmaceutical contaminants, mainly antibiotics. Also, there are no strict regulatory guidelines to regulate wastewater treatments, especially in third-world countries. As per the Central Pollution Control Board, India, from the total of 53 998 million liters per day generated wastewater, only 19 827 million liters per day are treated in major metropolitan cities. It also raised concern that only 13.5% of sewage water is effectively treated out of 18.6 % of the total treatment capacity[3]. This shows the capability of water treatment plants in metropolitan cities. Apart from the above, the guidelines also recommend specific parameters like pH, biochemical oxygen demand, chemical oxygen demand, suspended solids, ammonical nitrogen, total nitrogen and fecal coliform in treated water. However, no parameters are found for the presence of active pharmaceutical ingredients.
Based on the report on ‘pharmaceuticals in drinking water by WHO, the human health risk has been assessed by chemical risk assessment methods such as acceptable daily intake or tolerable daily intake based on a variety of calculations which includes Point of Departure (PoD). According to the ‘United States Environmental Protection Agency’ policy, compounds with Margin of Exposure values >100 are generally considered a low level of concern. The PoD is the concentration at which no adverse effects are detected, the No-Observed-Adverse-Effect Level (NOAEL), or, less optimally, the lowest concentration at which adverse effects are seen Lowest-Observed-Adverse-Effect Level (LOAEL), in combination with an additional uncertainty factor[4]. The resistance in microorganisms is directly proportional to the consumption of potential antibiotics. This directly or indirectly led to an increase in the dosage or combination therapy or treatment with a new set of antibiotics. The existence of these potential components in water sources may be lethal, whether at low or high concentrations[5].
Antibiotics in the environment are sometimes extremely low. However, it is sufficient to create sublethal antibiotic pressure in the environment. Hence, a sensitive analytical method is necessary to detect them in the respective samples. Sample preparation is another critical factor in achieving sensitive methods. Hence, simultaneous analytical methods and sample processing techniques that can determine multi-class antibiotics with low sensitivity are critical analytical tools. This review will focus on estimating antibiotics by widely used analytical and processing technology and developing analytical methods to detect multiclass antibiotics.
Classification of Antibiotics
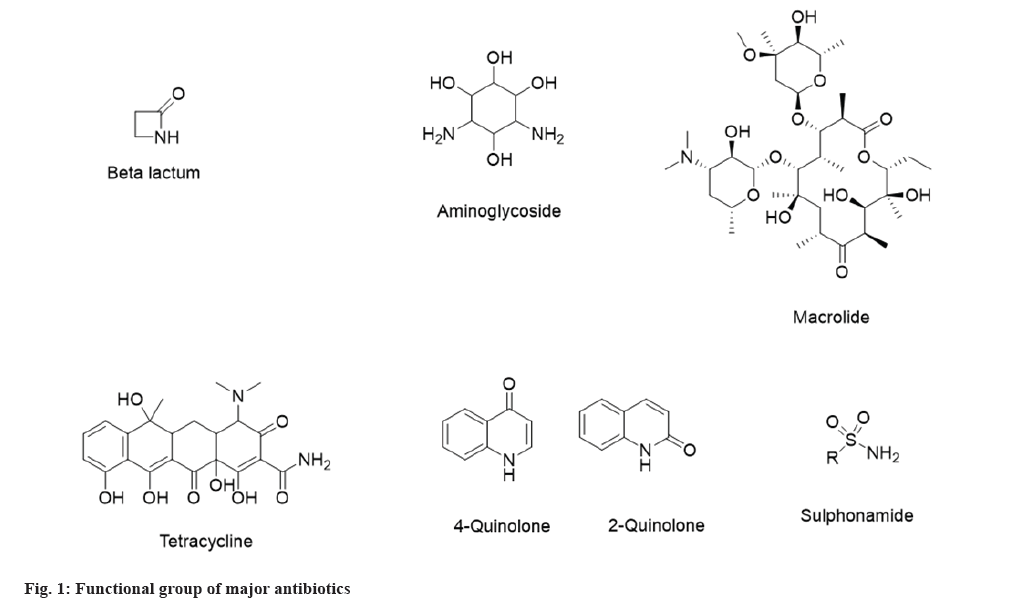
In general, antimicrobials are active against specific microorganisms in humans and animals by suppressing their proliferation or killing their ability to survive. According to the European Medicines Agency, animal antimicrobials were categorized into four categories. Avoid (category-A), Restrict (category-B), Caution (category-C), and Prudence (category-D) which include all major classifications[6]. Similarly, the World Organization for Animal Health has listed the specific classifications of antibiotics used in the veterinary field. The common antimicrobials used in veterinary are aminoglycosides, macrolides, cephalosporins, bicyclomycin, lincosamides, types of penicillin, phenicol, quinolones and tetracyclines[7]. Types of antibiotics are the broad and narrow spectrum that includes aminoglycoside, beta-lactam, tetracycline, various generations of cephalosporins, macrolides, sulphonamides, quinolones and specific large molecules are also acting as antibiotics such as peptide and oligonucleotides. An oligonucleotide is an efficient therapy against bacterial resistance to antibiotic treatment. Lipid Oligonucleotides (LONs) were proved to be efficient in delivering the oligonucleotide sequences in the prokaryotic cells and decreasing the minimum inhibitory concentration of resistant bacteria to a third-generation cephalosporin, the ceftriaxone[8]. Apart from a broad and narrow spectrum, antibiotics are also classified as bactericidal and bacteriostatic based on their activity against microorganisms. Table 1 describes the classification of antibiotics[9-11] in the simplest form. Also, the antiviral classification includes anti-herpes, anti-retrovirus, anti-influenza, etc., whereas the antifungal group consists of azole derivatives, polyene, echinocandins and allylamine.
| Classification | Spectrum | Nature | Synthetic/Natural |
|---|---|---|---|
| Beta-lactam* | Broad/narrow | Bactericidal | Natural/synthetic |
| Macrolides | Broad/narrow | Bacteriostatic | Natural/semi-synthetic |
| Aminoglycoside | Broad | Bactericidal | Natural/semi-synthetic |
| Tetracycline | Broad | Bacteriostatic | Natural/semi-synthetic |
| Quinolones | Broad | Bactericidal | Natural/synthetic |
| Sulphonamides | Broad | Bacteriostatic | Synthetic |
| Oxazolidinones | Broad | Bacteriostatic | Synthetic |
| Nitroimidazole | Broad | Bactericidal | Synthetic |
| Peptides | Broad | Bacteriostatic | Natural/synthetic |
Note: *includes penicillin, carbapenem, cephalosporin, beta-lactamase inhibitors
Table 1: Classification of antibiotics.
Based on their nature, antimicrobials act on either bacterial cell wall, nucleic acid synthesis, protein synthesis, bacterial folate synthesis, and cell membrane[12]. These key factors will decide whether the antibiotic is bactericidal or bacteriostatic against the specific microorganisms. Fig. 1 shows the functional groups of major antibiotics which dictate the mechanism of action in bacteria.
In 2019, the AWaRe[13] Classification Database was developed by WHO based on the recommendation of an expert committee on the ‘Selection and Use of Essential Medicines’.
AWaRe stands for Access, Watch and Reserve to emphasize the importance of their optimal uses and potential for antimicrobial resistance. It includes details of 180 antibiotics and is intended to be an interactive tool for countries to support antibiotic monitoring and optimal use better. The database also says that the WHO does not recommend antibiotics due to a lack of evidence on their reliability. The ‘Access’ antibiotics group includes antibiotics that have activity against a wide range of commonly encountered susceptible pathogens while showing lower resistance potential than antibiotics in the other groups. The ‘Watch’ antibiotics group includes antibiotics with higher resistance potential and most of the highest priority agents among the Critically Important Antimicrobials for Human Medicine and antibiotics at relatively high risk of selection of bacterial resistance. In contrast, the ‘Reserve’ group of antibiotics includes antibiotics and antibiotic classes that should be reserved to treat confirmed or suspected infections due to multi-drug-resistant organisms.
Analytical Methodologies
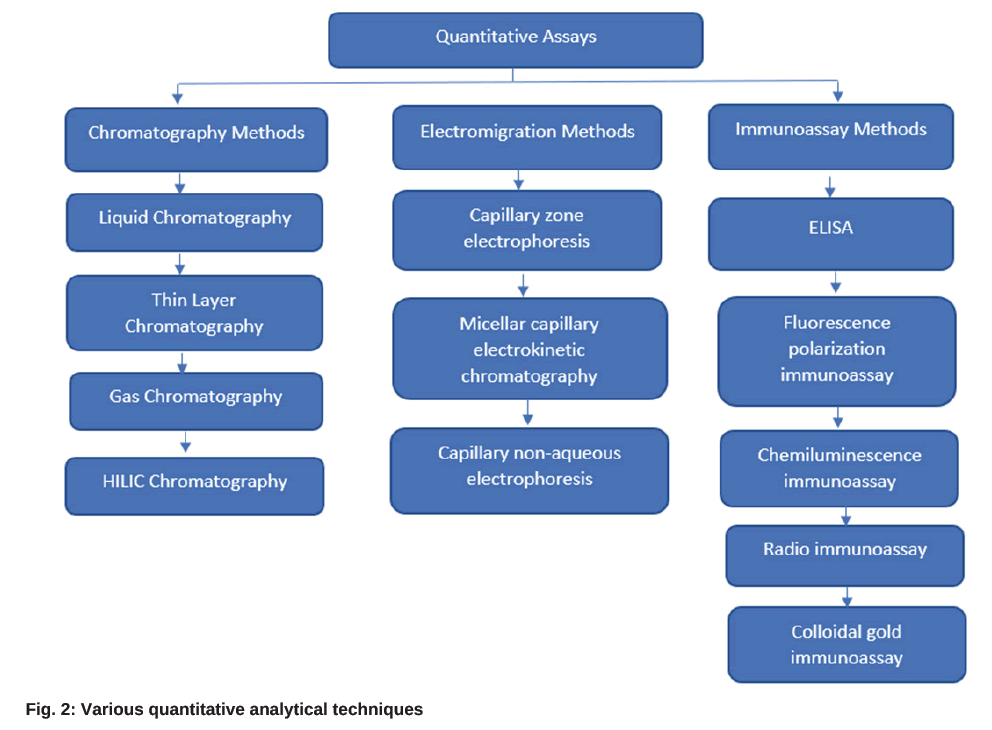
Residual antibiotics must be determined in various matrices such as food, agriculture products, animal husbandry products and portable water. Currently, several detection methods are available to identify the antibiotics from various environmental samples like wastewater, surface water, soil and other samples like milk, honey, etc., However, the right instrument must be chosen for data generation based on its application and detection limitations. The qualitative or rapid screening process and quantitative or confirmatory assays are the two types of detection methods used to determine antibiotics. Infrared detection, oscillographic polarography, and in vitro microbiological methods are generally used to determine the antibiotics qualitatively. Whereas spectrophotometry methods, including mass and Ultraviolet (UV) spectrometry, colorimetry, fluorimetry, conductometry, amperometry titration method, and chromatography methods, are applied for quantitative detection[14], and sometime Enzyme-Linked Immunosorbent Assay (ELISA) methods with fluorescent, chemiluminescent, or radiolabels are used for detection of residual antibiotics[15]. However, ELISA-based methods suffer from poor specificity issues. Fig. 2 represents the most followed quantitative analytical techniques in the determination of antibiotics in various fields of interest.
The separation technique, such as liquid chromatography, is mostly coupled with several analyzers, such as mass spectrometry, UV detections, Nuclear Magnetic Resonance (NMR) and Fourier Transform Infrared (FTIR). Along with this coupled technology, promising methods such as capillary electrophoresis-mass spectrometry, gas chromatography-mass spectrometery, and matrix-assisted laser desorption ionization-time of flight mass spectrometry are used for quantitative analysis[16]. Every technology has advantages and disadvantages; however, the applications are used based on the limitations concerning sample preparation.
A ‘nanomaterial-based’ Aptasensors (aptamer sensor) is used to detect ampicillin. It is a visible-light-driven self-powered photoelectrochemical sensor with carbon dots[17]. Aptamers are Deoxyribonucleic Acid (DNA) or Ribonucleic Acid (RNA) oligonucleotides that can bind with high affinity to specific targets. Aptasensors are developed for the detection of specific antibiotics that can be categorized into three types, i.e., electrical, fluorescent, and colorimetric[18]. However, due to its limitations on the experimental procedure, the technique remains an average methodology from a practical utility point of view.
Another approach in the determination of certain antibiotics is Hydrophilic Interaction Liquid Chromatography (HILIC). This is an emerging yet alternative approach to separate polar compounds through the polar stationary phase[19]. Though this technology is widely used to determine amino acids, peptides, biomarkers and oligonucleotides, it has limitations such as a high matrix effect, not being completely compatible with Liquid Chromatography-Atmospheric Pressure Chemical Ionization technology (LC-APCI) and being specific to the detection of only polar compounds. Hence the technology remains still under exploration.
Since Liquid Chromatography with Tandem Mass Spectrometry (LC-MS/MS) has a wide range of field applications, such as food, pesticides, pharmaceuticals, and biological, it plays an essential role in confirmatory assays. Tandem mass spectrometry is the most used detection technique with Selected Reaction Monitoring (SRM). However, based on the requirement, the liquid chromatography part can also be attached to other hybrid detection analyzers, such as quadrupole and time of flight and quadropule ion trap[20], for different applications.
The antibiotic residuals in the unknown samples may be observed in pg/l. So, to determine such low concentrations in all types of matrices such as water, wastewater, aqua products and soil most sophisticated and sensitive equipment like LC-MS/MS is necessary to achieve the desired output. Many ionization techniques are available. However, electrospray ionization is a unique ionization technique where the ions are generated directly from within a solution that is sprayed from a fine needle at atmospheric pressure[21]. A simultaneous analytical method was developed using an LC-MS/MS to successfully estimate 9 different antibiotics such as cefaclor, cefazolin, cefpodoxime, ceftazidime, cefuroxime, ciprofloxacin, ertapenem, sulfamethoxazole, and trimethoprim. The proposed method was applied for therapeutic drug monitoring in plasma[22]. The method was sensitive to 0.05 mg/l to 1.5 mg/l for various antibiotics in biological samples. Similarly, another simultaneous method for nine compounds from four different classes of antibiotics, such as beta-lactam, fluoroquinolone, macrolide and nitro-imidazole, with a limit of detection set to 1-10, 0.8 to 10, and 0.8 to 25 ng/l in the treated ground and river water respectively, was developed using LC-MS/MS. This method was applied to determine various antibiotics in water samples[23]. Interestingly, a simultaneous method was developed to estimate 23 types of antibiotics using LC-MS/MS to determine antibiotics in bovine milk samples[24]. This method successfully achieved sensitivity as low as 0.17 ng/ml. At least 37 antibiotics from 6 different groups were determined from honey samples using LC-MS/MS with a 5 µg/kg sensitivity as the lower limit[25]. Using LC-MS/MS as a platform, simultaneous methods are being developed to estimate antibiotics in various samples and applied in diverse fields of interest. This shows that among different analytical technologies that are approached to determine antibiotics, LC-MS/MS is an exceptional and most sophisticated technology for estimating multi-class antibiotics with low sensitivity. Table 2 shows the list of antibiotics with various details in the respective matrices.
| Antibiotica | Processing method | Matrix | LOQb | % Recoveryc | Analytical system | Reference |
|---|---|---|---|---|---|---|
| Cephalosporins | PPT/SPE | Plasma/water | 0.1 to 1 mg/l | >75 | LC-MS/MS | [20,21] |
| Carbapenem | PPT | Plasma | 0.5 mg/l | 100 | LC-MS/MS | [20] |
| Quinolones | PPT/SPE | Plasma/water/honey | 0.05 mg/l | >72 | LC-MS/MS | [20,21] |
| Trimethoprim | PPT | Plasma | 0.125 mg/l | >80 | LC-MS/MS | [20] |
| Sulfamethoxazole | PPT | Plasma | 1.5 mg/l | >70 | LC-MS/MS | [20] |
| Macrolide | SPE | Water/honey | 2 ng/l | >50 | LC-MS/MS | [21,23] |
| nitro-imidazole | SPE | Water | 10 ng/l | >45 | LC-MS/MS | [21] |
| Tetracycline | SPE | Bovine milk/honey | 13.60 ng/ml | >80 | LC-MS/MS | [22,23] |
| Note: a,b,c: The list of antibiotics and their LOQ ranges in respective matrices with the percentage recovery | ||||||
Table 2: Antibiotics in various matrices.
Sample Processing Techniques
The scope of sample preparation is to separate the pharmaceutical residues from the sources like water, food and biological matrices. Usually, antibiotic residues are detected at lower levels for various reasons, such as the matrix factor and impurities. To achieve a sensitive method, the sample extraction process must be selective. An appropriate extraction process will lead to a better recovery. Hence, sample processing is the critical step in multi-class analysis. Determining multi-class antibiotics in a single run is a unique strategy, as the extraction procedure has a lot of limitations due to the various physicochemical properties of the compounds[26]. The sample processing involves three standard extraction techniques described below. However, many other hybrid technologies are also emerging to achieve better sensitivity. Protein Precipitation (PPT) is to separate protein that binds with other substances. Liquid-Liquid Extraction (LLE) is the technique makes a solute immiscible from one solvent to another. This is also known as partitioning. Solid-Phase Extraction (SPE) is the extraction technique adsorbs the analyte onto its solid stationary bed. The adsorption is based on the chemical nature of the compound towards the fixed bed.
PPT:
Though PPT is an inexpensive and cost-effective technology, the extraction method is rapid. However, achieving the low detection limit is difficult as the sample clean-up procedure is not a part of the precipitation technique. Due to this disadvantage, a sensitive method cannot be demonstrated using PPT. However, a combination of PPT followed by liquid-liquid or SPE can be followed to achieve the desired sensitivity. This was proven in the estimation of an New Chemical Entity molecule following a combinational processing technique[27]. Initially, the drug was precipitated using a mixture of methanol and acetonitrile along with formic acid. Post vortex and centrifugation, the supernatant was directly loaded into an Oasis Mixed-mode Cation Exchange (MCX) cartridge. Following this combination precipitation technique, the lower limit of quantitation was achieved as low as 0.5 ng/ml with 100 % recovery. Following the precipitation technique alone, paromomycin was analyzed in human plasma using LC-MS/MS with a limit of quantitation of 5 ng/ml[28]. The plasma samples were precipitated using 20 % trichloroacetic acid, and the supernatant was then diluted with water in a ratio of 1:1 for analysis. Using perchloric acid as a precipitating agent, beta-lactam and cephalosporins were determined in human plasma[29]. Initially, when adding 60 % perchloric acid, it was observed that the compounds were degraded. However, it was overcome by reducing to 40 % perchloric acid along with acetate buffer at pH 4.5 as a medium. To analyze samples from multi-drug resistant tuberculosis patients, a method was designed to analyze at least 20 antitubercular drugs using precipitating solvent as methanol and acetonitrile. With this precipitation method, the inter and intra-day precision was observed at 14.3 %, whereas the accuracy was observed between 84.8 %-113.0 %[30].
LLE:
LLE is the second most prioritized technique in sample processing. Due to its better extraction recovery and improved matrix effect than PPT, LLE is considered for multi-class antibiotics analysis. However, extraction recovery cannot be achieved in some cases due to the variable physicochemical nature of the analytes towards the extracting organic solvent. Also, there are several other obstacles such as loss of analytes, low sensitivity, need for a larger volume of extraction solvent, and environmental un-friendly[31]. Some powerful organic solvents used for this processing technology are methyl-tertiary butyl ether, diethyl ether, dichloromethane, n-hexane, toluene, and ethyl acetate. The excessive usage of organic solvents is also against the principles of Green Analytical Chemistry (GAC)[32], which is considered the major drawback of this technique. Other variations in the LLE technique are Liquid-Phase Microextraction (LPME), Salting-out LLE (SALLE), and Aqueous Two-Phase System (ATPS)[31]. These methods are followed to reduce the use of organic solvents and to reduce the processing time. However, due to limitations, these methods are less widely used.
A hybrid technique of the Solid-Liquid Extraction (SLE) followed by the SPE method was used to extract certain antibiotics and their transformation products from anthropogenically altered solid environmental matrices[33]. While the SLE technique was followed to separate analytes from soil sources, the SPE method was followed for the clean-up procedure. However, this combined technique's recovery ranged from 45 %-85%. A unique extraction technique, SALLE was followed to estimate ciprofloxacin using high-performance liquid chromatography-diode array detector[34]. Magnesium sulphate was used as a salting-out agent for this processing technology. However, this method is applied only to determine one compound from the soil samples. The main advantage of using the SALLE technique is improved extraction efficiency of polar compounds using polar solvents. The combination of LLE and dispersive-LLME showed an efficient extraction using acetonitrile as an initial extracting solvent. Followed by this, a newly synthesized water-immiscible deep eutectic solvent was used for subsequent extraction steps. Using the Ion Mobility Spectrometry instrument, this method was applied to estimate oxytetracycline, penicillin G, and tilmicosin from sausage samples[35]. This instrumental technique characterizes molecules based on the gaseous phase mobility of their ions formed at ambient pressure and under an electric field. With this new combination technique, the limit of quantification was achieved at around 5.1 ng/g. An integrated PLE and SPE extraction technique was followed to extract antibiotics from soil samples[36]. In this method, the PLE was followed for extraction and diol SPE cartridges for the clean-up process. The method successfully achieved as low as 0.2 µg/kg with a recovery rate ranging from 38 %-94 % for macrolides, 76 %±24 % for ionophore salinomycin, and 118 %±21 % for pleuromutilin tiamulin using LC-APCI-MS/MS.
SPE:
SPE is the most widely used processing technology. Due to its reliability, reproducibility, a wide range of applications and use of compound-target stationary phase cartridges, the SPE method is the most familiar technique among scientists. SPE techniques include solid-phase microextraction, dispersive micro-SPE, magnetic SPE and matrix solid-phase dispersion[5,23,37,38].
Before selecting sorbents, it is important to check the pKa values of the selected antibiotics. pKa is a method used to identify the strength of the acid. It is a negative value of the acid dissociation constant. A lower pKa value indicates a stronger acid. pKa values for some critical antibiotics are provided in Table 3. Many antibiotics have acidic, and some have basic functional groups. Hence, pH plays a vital role in the extraction recovery of antibiotics. While the pH can affect the stability and interaction of analytes with sorbents, the same can also influence the speciation of antibiotics due to their acidic and basic functional groups in their structures[39]. Since antibiotics are acidic in general, these are in a ‘soluble’ form at a lower pH. Hence, the pH should be maintained lower than the pKa value to achieve a better extraction recovery[40].
| Classificationa | Compound | Molecular weight | Molecular formula | pKa valueb |
|---|---|---|---|---|
| Beta-lactam | Amoxicillin | 365.4 | C16H19N3O5S | 3.32 |
| Cephalosporin | Cephalexin | 347.39 | C16H17N3O4S | 4.5 |
| Fluoroquinolones | Ciprofloxacin | 331.35 | C17H18FN3O3 | 5.76 |
| Macrolides | Azithromycin | 748.98 | C38H72N2O12 | 8.74 |
| Nitroimidazole | Metronidazole | 171.15 | C6H9N3O3 | 15.44 |
| Aminoglycoside | Streptomycin | 581.58 | C21H39N7O12 | 11.09 |
| Oxazolidinone | Linezolid | 337.35 | C16H20FN3O4 | 1.8 |
| Carbapenem | Meropenem | 383.5 | C17H25N3O5S | 2.90 for Carboxylic acid and 7.4 for Pyrrolidinyl amino |
| Azole derivatives | Voriconazole | 349.31 | C16H14F3N5O | 1.76 |
| Sulphonamides | Trimethoprim | 290.32 | C14H18N4O3 | 7.12 |
| Tetracycline | Tetracycline | 444.4 | C22H24N2O8 | 3.3 |
Note: a,b: Commonly used antibiotics with their molecular weight and the respective pKa values
Table 3: pKa values for key antibiotics.
Various types of SPE sorbents are being used in the extraction technique. Also, each cartridge has a distinctive feature relating to the molecule. The various SPE sorbents are categorized into three prominent SPE techniques: polar, non-polar, and ion exchange SPE. The different types of cartridges and their applications are tabulated in Table 4[41]. Among every other cartridge C18, HLB, MCX, and Mixed-mode Anion exchange (MAX) are the most used cartridges in all applications. However, the most widely used cartridges are HLB specific to antibiotics determination. This is because the HLB cartridge shows both hydrophilic and lipophilic retention features. And hence, both polar and non-polar compounds can be retained in the sorbent.
| Types of Sorbenta | Structureb | Functional groupc | Mechanismd | Applications |
|---|---|---|---|---|
| Anionic exchange (mixed mode-SAX) |  |
C8 | Reverse phase or anion exchange selectivity | Suitable for an alkaline aqueous solution or neutral compounds in urine, and blood |
| Cationic exchange (mixed mode-SCX) |  |
C8 | Reverse phase or cation exchange selectivity | Suitable for an alkaline aqueous solution or neutral compounds in urine, and blood |
| Amino Cartridges | 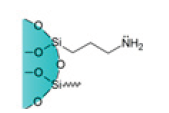 |
NH2 | Strong polar adsorption in non-polar organic solvents | Suitable to analyze peptides, drugs, mono and polysaccharides, steroids, and cholesterol. Widely used in food, environmental, pharmaceutical, and medical applications |
| Cyanopropyl Cartridges | 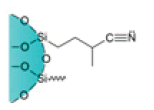 |
CN | Weak hydrophobic silica matrix used as normal or reverse phase adsorbent | Suitable to analyze drugs, pesticides, veterinary drug residues on food and dairy products, oil pollution, and pesticides in environmental samples |
| C18 Cartridges |  |
C18 | Retain non-polar compounds by strong hydrophobic interaction | Suitable to analyze drugs, poisons, pollutants, mycotoxins in food, preservatives in cosmetics |
| HLB Cartridges | 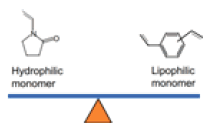 |
Monomers of N-vinylpyrollidone and divinylbenzene | Universal sorbent for acidic, basic, and neutral compounds. Enhanced reverse-phase capability with retention of polar analytes | Suitable to analyze drugs, environmental samples, agrochemical, and food samples |
Note: a,b,c,d: Widely used SPE cartridges and their specifications, along with structures, functional groups, etc
Table 4: Types of SPE cartridges.
Hybrid SPE-Phospholipid columns were used to determine 32 antibiotics in fish and shrimp muscles[42]. The recovery for these antibiotics ranges between 70 %-120 %, with the lower limit of quantitation at 0.062 µg/kg. This method was successfully applied to detect various antibiotics in aquaculture products. Antibiotics and some antivirals were determined in water using HLB, MCX, and MAX cartridges. With the LOQ set at 5 ng/l, the recovery was observed between 41 %-116 %. Following this method to detect antibiotics in wastewater, almost all target compounds were observed in the range between 10 and 570 ng/l[43]. However, a comparison study between Oasis HLB and Strata-X[44] for the determination of 18 different classes of antibiotics was performed with an optimized pH. It was observed that the best extraction efficiency was achieved in Oasis HLB in acidic pH. The LOQ for the antibiotics ranged between 0.8 ng/l-245.1 ng/l. This method determined all proposed compounds in wastewater samples up to 6.2 µg/l concentration. An optimized Solid-Phase Microextraction (SPME) technique was used to determine sulphonamides, macrolides, and trimethoprim with the LOQ set at 16 and 35 ng/l for influent and effluent samples. To compare the results, the SPE method's LOQ was set at 4.7 and 0.86 ng/l for the same samples[45]. The main advantages of the SPME method are decreased sample volume, less processing time, cost-effectiveness, reduced matrix effect and repeated usage. A similar comparison study between the SPME and SPE techniques to determine 10 sulphonamide antibiotics[46] in wastewater also confirms the advantages of the SPME method with the added advantage of good extraction efficiency. A study specifically compared the extraction efficiency of HLB and MCX cartridges used in the determination of certain antibiotics and their metabolites in water sources using linear-ion trap tandem mass spectrometry[47]. This study further proved that HLB cartridges showed better recovery and lesser matrix effect compared with MCX cartridges. This was succeeded as the HLB cartridges contain the composition of lipophilic divinylbenzene units and the hydrophilic N-vinylpyrrolidone units[48], allowing the efficient extraction of organic contaminants in a wide range of pH (from pH 1 to 14). Before extraction, a chelating agent such as sodium ethylenediaminetetraacetic acid must be used to complex the metals or multivalent cations (residual metal ions) soluble in water on SPE cartridges and glassware. The key advantages and disadvantages of the above-mentioned extraction techniques are provided in Table 5.
| Extraction techniques | Advantages | Disadvantages |
|---|---|---|
| PPT | 1. Crude processing technology | 1. High matrix effect |
| 2. Mostly used in drug discovery for faster processing | 2. The matrix effect may impact recovery | |
| 3. Cost-effective | 3. High chances of blockage due to precipitated particles in the supernatant | |
| 4. The extraction solvent is either methanol or acetonitrile | 4. Affects HPLC column performance | |
| LLE | 1. Higher recovery than the PPT technique | 1. Longer processing time |
| 2. Lesser matrix effect | 2. More chances of inhaling organic solvents | |
| 3. Better sensitivity due to concentrating the sample | 3. Not following green analytical chemistry | |
| SPE | 1. Alternative for PPT and LLE | 1. Expensive |
| 2. Sample clean-up | 2. Single-use cartridges | |
| 3. Lesser matrix effect and higher recovery | 3. Many steps in the extraction process | |
| 4. Analyte target sorbent can be used | 4. Complex methodology |
Table 5: Advantages and Disadvantages of various processing techniques.
Irrespective of the applications, the SPE technique is being followed widely for sample clean-up making the method free from the matrix effect and achieving the expected recovery. Researchers applied the developed analytical method for determining multi-class antibiotic residuals in various samples such as aquaculture products, meat, raw milk, wastewater and water resources, honey and soil.
Conclusion
This review concludes that various analytical methodologies and sample processing techniques are key factors in determining sub-lethal levels of antimicrobials in environmental sources. The methods must be sensitive enough to detect pharmaceutical ingredients in the respective samples. SPE techniques play a major role in determining antimicrobials in diverse samples compared to low-sensitive methods. The key challenges include simultaneous estimation, varied polarities of the compounds, acid dissociation constant (pKa) of varied classes of antimicrobials and other factors such as insoluble matters in the environmental samples.
Since the concentration of residual compounds is usually very low in various environmental samples, achieving a sensitive method is necessary. LC-MS/MS, or any other high-resolution mass spectrometry, is the most preferred analytical instrument to achieve the required sensitivity. The developed and validated analytical method must provide the concentration of the antimicrobials present in the environmental samples. This will help detect and take necessary actions to eliminate trace contaminations from required sources such as wastewater, surface water, soil, honey, milk products, poultry products, and aquaculture products.
Authors' contributions:
My heartfelt thanks to the management and academic guide for supporting me in writing and publishing this review article that further supports my research work.
Conflict of interests:
The authors declared no conflict of interests.
References
- Report on AMR surveillance in Europe. World Health Organization and European Centre for Disease Prevention and Control, 2022.
- Seifrtova M, Novakova L, Lino C, Pena A, Solich P. An overview of analytical methodologies for the determination of antibiotics in environmental waters. Anal Chim Acta 2009;649(2):158-79.
[Crossref] [Google Scholar] [PubMed]
- Schellenberg T, Subramanian V, Ganeshan G, Tompkins D, Pradeep R. Wastewater discharge standards in the evolving context of urban sustainability: The case of India. Front Environ Sci 2020;8:30.
- Report on pharmaceuticals in drinking water. World Health Organization; 2011.
- Faleye AC, Adegoke AA, Ramluckan K, Bux F, Stenström TA. Identification of antibiotics in wastewater: current state of extraction protocol and future perspectives. J Water Health 2017;15(6):982-1003.
[Crossref] [Google Scholar] [PubMed]
- Categorization of antibiotics in the European Union, 2019; p. 6-7.
- OIE. OIE list of antimicrobials of veterinary importance. World Organ Anim Health 2021;2021:5-9.
- Kauss T, Arpin C, Bientz L, Vinh Nguyen P, Vialet B, et al. Lipid oligonucleotides as a new strategy for tackling the antibiotic resistance. Sci Rep 2020;10(1):1054.
- Calhoun C, Wermuth HR, Hall GA. Antibiotics. In: StatPearls. Treasure Island (FL): StatPearls Publishing; 2022.
- Pancu DF, Scurtu A, Macasoi IG, Marti D, Mioc M, Soica C, et al. Antibiotics: conventional therapy and natural compounds with antibacterial activity-a pharmaco-toxicological screening. Antibiotics 2021;10(4):401.
[Crossref] [Google Scholar] [PubMed]
- Ullah H, Ali S. Classification of anti-bacterial agents and their functions. Antibacterial agents 2017;31;10:1-6.
- Samanidou V, Nisyriou S. Multi‐residue methods for confirmatory determination of antibiotics in milk. J Sep Sci 2008;31(11):2068-90.
[Crossref] [Google Scholar] [PubMed]
- AWaRe antibiotic classification. World Health Organization; 2021.
- Abramova AA, Grakhova EV, Isakov VG. Migration of antibiotics in natural aquatic environment. In: IOP Conference Series: Mater Sci Eng 2020;962(4):042076.
- Ahmed S, Ning J, Peng D, Chen T, Ahmad I, Ali A, et al. Current advances in immunoassays for the detection of antibiotics residues: A review. Food Agri Immunol 2020;31(1):268-90.
- Pauter K, Szultka-Młyńska M, Buszewski B. Determination and identification of antibiotic drugs and bacterial strains in biological samples. Molecules 2020;25(11):2556.
[Crossref] [Google Scholar] [PubMed]
- Yang Z, Ding X, Guo Q, Wang Y, Lu Z, Ou H, et al. Second generation of signaling-probe displacement electrochemical aptasensor for detection of picomolar ampicillin and sulfadimethoxine. Sens Actuators B Chem 2017;253:1129-36. [Crossref] [Google Scholar] [PubMed]
- Parthasarathy R, Monette CE, Bracero S, S. Saha M. Methods for field measurement of antibiotic concentrations: Limitations and outlook. FEMS Microbiol Eco 2018;94(8):fiy105.
[Crossref] [Google Scholar] [PubMed]
- Buszewski B, Noga S. Hydrophilic interaction liquid chromatography (HILIC)—a powerful separation technique. Anal Bioanal Chem 2012;402:231-47.
[Crossref] [Google Scholar] [PubMed]
- Chen GL, Fang YY. The LC-MS/MS methods for the determination of specific antibiotics residues in food matrices. In: Mass spectrometry food safety: methods and protocols 2011:309-55.
- Bush KL. Mass Spectrometry, Instrumentation and launch points for the next fifty years. Kennesaw State Univ 2001; pg 1-14.
- Rehm S, Rentsch KM. LC-MS/MS method for nine different antibiotics. Clin Chim Acta 2020;511:360-7.
[Crossref] [Google Scholar] [PubMed]
- Mirzaei R, Yunesian M, Nasseri S, Gholami M, Jalilzadeh E, Shoeibi S, et al. An optimized SPE-LC-MS/MS method for antibiotics residue analysis in ground, surface and treated water samples by response surface methodology-central composite design. J Environ Health Sci Eng 2017;15:1-6.
[Crossref] [Google Scholar] [PubMed]
- Alija G, Hajrulai-Musliu Z, Uzunov R. Development and validation of confirmatory LC–MS/MS method for multi-residue analysis of antibiotic drugs in bovine milk. SN Appl Sci 2020;2:1-3.
- Bohm DA, Stachel CS, Gowik P. Validation of a multi-residue method for the determination of several antibiotic groups in honey by LC-MS/MS. Anal Bioanal Chem 2012;403:2943-53.
[Crossref] [Google Scholar] [PubMed]
- Moreno-Bondi MC, Marazuela MD, Herranz S, Rodriguez E. An overview of sample preparation procedures for LC-MS multiclass antibiotic determination in environmental and food samples. Anal Bioanal Chem 2009;395:921-46.
- Xue YJ, Akinsanya JB, Liu J, Unger SE. A simplified protein precipitation/mixed‐mode cation‐exchange solid‐phase extraction, followed by high‐speed liquid chromatography/mass spectrometry, for the determination of a basic drug in human plasma. Rapid Commun Mass Spectrom 2006;20(18):2660-8.
[Crossref] [Google Scholar] [PubMed]
- Roseboom IC, Thijssen B, Rosing H, Mbui J, Beijnen JH, Dorlo TP. Highly sensitive UPLC-MS/MS method for the quantification of paromomycin in human plasma. J Pharm Biomed Anal 2020;185:113245.
- do Nascimento TG, Soares Aragão CF, Dantas de Medeiros F, de Jesus Oliveira E, Oliveira Macêdo R. Validation of a method for determination of ampicillin in human plasma using LC-DAD. J Chromatogr Sci 2009;47(9):749-55.
[Crossref] [Google Scholar] [PubMed]
- Kim HJ, Seo KA, Kim HM, Jeong ES, Ghim JL, Lee SH, et al. Simple and accurate quantitative analysis of 20 anti-tuberculosis drugs in human plasma using liquid chromatography–electrospray ionization–tandem mass spectrometry. J Pharm Biomed Anal 2015;102:9-16.
[Crossref] [Google Scholar] [PubMed]
- Khatibi SA, Hamidi S, Siahi-Shadbad MR. Application of liquid-liquid extraction for the determination of antibiotics in the foodstuff: Recent trends and developments. Critic Rev Anal Chem 2022;52(2):327-42.
[Crossref] [Google Scholar] [PubMed]
- de la Guardia M, Garrigues S. Past, present and future of green analytical chemistry. 2020;2:p.1-18.
- Bajkacz S, Felis E, Kycia-Słocka E, Harnisz M, Korzeniewska E. Development of a new SLE-SPE-HPLC-MS/MS method for the determination of selected antibiotics and their transformation products in anthropogenically altered solid environmental matrices. Sci Total Environ 2020;726:138071-7.
[Crossref] [Google Scholar] [PubMed]
- Gezahegn T, Tegegne B, Zewge F, Chandravanshi BS. Salting-out assisted liquid–liquid extraction for the determination of ciprofloxacin residues in water samples by high performance liquid chromatography–diode array detector. BMC Chem 2019;13(1):1-28.
[Crossref] [Google Scholar] [PubMed]
- Saei A, Javadi A, Mogaddam MR, Mirzaei H, Nemati M. Development of homogeneous liquid–liquid extraction combined with dispersive liquid–liquid microextraction based on solidification of floating droplets of a ternary component deep eutectic solvent for the analysis of antibiotic residues in sausage samples prior to ion mobility spectrometry. Anal Method 2020;12(34):4220-8.
- Schlüsener MP, Spiteller M, Bester K. Determination of antibiotics from soil by pressurized liquid extraction and liquid chromatography–tandem mass spectrometry. J Chromatogr A 2003;1003(1-2):21-8.
[Crossref] [Google Scholar] [PubMed]
- Li KM, Rivory LP, Clarke SJ. Solid-phase extraction (SPE) techniques for sample preparation in clinical and pharmaceutical analysis: A brief overview. Curr Pharm Anal 2006;2(2):95-102.
- Khatibi SA, Hamidi S, Siahi-Shadbad MR. Current trends in sample preparation by solid-phase extraction techniques for the determination of antibiotic residues in foodstuffs: A review. Critic Rev Food Sci Nutr 2021;61(20):3361-82.
- Heidari M, Kazemipour M, Bina B, Ebrahimi A, Ansari M, Ghasemian M, et al. A qualitative survey of five antibiotics in a water treatment plant in central plateau of Iran. J Environ Public Health 2013;2013:1-9.
- Aydin S, Aydin ME, Ulvi A, Kilic H. Determination of antibiotics by SPE-LC-MS/MS in wastewater and risk assessment. Adv Environ Res 2018;7(3):201-12.
- Types of SPE cartridges, Hawach; 2023.
- Fedorova G, Nebesky V, Randak T, Grabic R. Simultaneous determination of 32 antibiotics in aquaculture products using LC-MS/MS. Chem Papers 2014;68:29-36.
- Ngumba E, Kosunen P, Gachanja A, Tuhkanen T. A multiresidue analytical method for trace level determination of antibiotics and antiretroviral drugs in wastewater and surface water using SPE-LC-MS/MS and matrix-matched standards. Anal Method 2016;8(37):6720-9.
- Rossmann J, Schubert S, Gurke R, Oertel R, Kirch W. Simultaneous determination of most prescribed antibiotics in multiple urban wastewater by SPE-LC–MS/MS. J Chromatogr B 2014;969:162-70.
[Crossref] [Google Scholar] [PubMed]
- McClure EL, Wong CS. Solid phase microextraction of macrolide, trimethoprim, and sulfonamide antibiotics in wastewaters. J Chromatogr A 2007;1169(1-2):53-62.
[Crossref] [Google Scholar] [PubMed]
- Balakrishnan VK, Terry KA, Toito J. Determination of sulfonamide antibiotics in wastewater: A comparison of solid phase microextraction and solid phase extraction methods. J Chromatogr A 2006;1131(1-2):1-0.
[Crossref] [Google Scholar] [PubMed]
- Gros M, Rodríguez-Mozaz S, Barceló D. Rapid analysis of multiclass antibiotic residues and some of their metabolites in hospital, urban wastewater and river water by ultra-high-performance liquid chromatography coupled to quadrupole-linear ion trap tandem mass spectrometry. J Chromatogr A 2013;1292:173-88.
[Crossref] [Google Scholar] [PubMed]
- Solid phase extraction with Oasis HLB sorbent: Simple procedures with Superior sample preparation, Waters Inc. 1998