- *Corresponding Author:
- Chunyan Fan
Xigao, Taiyuan University of Technology
53 West Mining Street, Wanbailin District
Taiyuan City, 030024
Shanxi Province, China
E-mail: fanchunyan9255@163.com
| This article was originally published in Special issue on “Trends in therapeutic Management of Various Conditions” |
| Indian J Pharm Sci 2020:82(3)spl issue 6;58-64 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In order to detect the changes in expression of miR-128 during skeletal muscle repair in a rat model of muscle injury and to provide a basis for treating skeletal muscle injury, a rat skeletal muscle injury model was established. Rats were randomly divided into the quiet control group, 0 h after exercise group, 6 h after exercise group, 12 h after exercise group, 24 h after exercise group, 48 h after exercise group, 72 h after exercise group, 1 w after exercise group and 2 w after exercise groups to explore the expression pattern of miR-128, Sp1, CDKN1A during skeletal muscle contusion repair. Results showed that muscle fibers in the control group were polygonal, closely arranged and regular in shape. The myocyte membrane was intact and the nucleus was located under the myometrium. At 0 h after exercise, cells were swollen and round, cell volume increased and the cell gap increased. At 6, 12, 24 and 48 h after exercise, the cells continued to swell and the intercellular space became smaller than that in group C. In the 72 h after exercise group, cell swelling was very high and there were a large number of infiltrated inflammatory cells and exudation of blood cells. In 1 w after exercise group, inflammatory cell infiltration decreased since exercise. When it reaches to 2w, the morphology and structure of the cells return to normal, and the repair process was completed. Successful establishment of the rat skeletal muscle repair model, it is found that the expression of muscle-specific miR-128 did not change significantly during the process of muscle repair. miR-128 might be involved in the process of regeneration after skeletal muscle injury.
Keywords
miR-128; sports injury; myocyte membrane; CDKN1A; inflammatory cells
Repair of skeletal muscle injury has been a hot research topic. Studies have shown that skeletal muscle satellite cells are promising muscle stem cells[1]. Satellite cells and growth factors play an important role in the growth and development of skeletal muscle, training adaptation, injury repair and transplantation[2]. Normally, resting satellite cells can be activated rapidly when skeletal muscle is damaged. However, some diseases or aginginduced satellite cell activation disorders will lead to skeletal muscle regeneration cannot be completed normally[3,4]. Recent studies demonstrated that endogenous satellite cells cannot complete the repair process alone, and some growth factors and appropriate stimulation are necessary conditions for skeletal muscle to complete the repair and regeneration process[5]. At present, the mechanism of skeletal muscle injury has so many proposed theories that include energy exhaustion theory, free radical theory, mechanical injury theory, inflammatory cell infiltration theory and so on while there is no consensus among academic circles. According to the theory of energy metabolic disorder of muscle fibers, the stored energy in skeletal muscle fibers is quickly exhausted while the intracellular energy generation rate is relatively slow, which cannot supply the energy needed by muscle in time[6,7]. The energy supply in muscle fibers cannot meet the energy required for muscle contraction. In addition, a large amount of waste generate during exercise accumulates in muscle cells, which has a great impact on the normal function of skeletal muscle, and may be an important cause of skeletal muscle injury. In skeletal muscle cells, calcium ion is also an important initiating factor for nerve impulse to transform into muscle contraction[8].
MicroRNAs (miRNA) are a large family of small noncoding single-stranded RNA with a length of about 22 amino acids. At present, there are about 38,000 kinds of miRNA discovered, among which there are about 2,800 kinds in human body[9]. MiRNAs widely exist in many eukaryotic organisms. MiRNAs play a wide range of regulatory roles in biological processes such as cell proliferation, differentiation, apoptosis, and have a very important impact on the growth and development of organisms. The biosynthesis of miRNAs in cells mainly involves 5 steps, transcription to form pri-miRNAs, processing of pri-miRNAs to form pre-miRNAs in the nucleus, pre-miRNA transport to the nucleus, truncation of pre-miRNAs in cytoplasm and formation of mature miRNAs[10]. MiR-128 is divided into 2 types, 1/2 (also called a/b), which are located on the 2q21.3 and 3p22.3 chromosomes of humans, respectively. Studies have shown that miR-128 is closely related to the formation of gliomas. When gliomas occur, their expression often shows abnormalities. MiR-128 may act by binding to its target gene and inhibiting the proliferation of glioma cells[11].
The discovery of the role of miR-128 in the repair of muscle damage has brought new hope for better treatment of muscle damage. In order to understand the role of miR-128 in skeletal muscle repair, the research on miR-128 in the process of skeletal muscle satellite cell proliferation and differentiation and muscle damage repair process is reviewed.
Materials and Methods
Experimental animal grouping:
Sprague-Dawley (SD) rats used in the experiments were purchased through regular channels. Rats were adaptively fed in the animal room for 2 w after purchase. Rats were housed in cages, no more than 5 per cage and were given free access to standard rodent diet and water. The temperature was controlled at 21±2°. The illumination time was controlled at 8-12 h per day and the relative humidity was maintained at 50 to 55%. Rats were weighed regularly and when the body weight reached 260 g, the rats were rescreened to exclude rats with over or under body weight. Six days before the formal experiment, all rats were trained on a treadmill for 5 min at a speed of 10 m/min on a slope of 0°. After the training, the rats that could not complete the training were excluded. Finally, 72 rats were retained which were randomly divided into 9 groups of 8 rats each. According to the chosen time points after exercise, these were grouped as 0, 6, 12, 24, 48, and 72 h, and 1 w and 2 w after exercise, along with a control group (C).
The exercise program specifically referred to Armstrong’s downhill running training model. Except for the quiet control group rats, all the other 8 group rats were simultaneously subjected to a continuous acute treadmill training. The running platform slope was -16º, the speed was 16 m/min, and the time was 90 min. The purpose is to familiarize the rats with the environment of the animal running platform[12].
Animal model preparation and materials:
Each rat was made to complete a one-time downhill running motion to establish a downhill running damage model. In the 0, 6, and 12 h groups after exercise, 2 rats were added every 15 min to ensure that the rats were taken out at the exact time after exercise. All rats in the other groups were started at the same time.
Rats were anesthetized with 12 % chloral hydrate (420 mg/kg) given intraperitoneally. The rats were fixed in the supine position on the operating table and the abdominal cavity was opened to expose the inferior vena cava. About 5 ml of blood was collected into a blood collection tube without any additives. After 30 min, the blood was centrifuged for 18 min in a low temperature centrifuge at a speed of 3200 rpm. The serum separated was transferred to a centrifuge tube, and stored in an ultra-low temperature freezer. After collecting the blood, a scalpel was used to expose the rectus femoris muscle, which was carefully removed and placed in pre-cooled saline. The blood on the surface of the muscle was washed away, the fascia and fat were removed with a scalpel. Filter paper was used to absorb the moisture on the surface of muscle and the muscle was dipped in liquid nitrogen cooled isopentane, stirred slowly for about 8 s. Fully frozen muscle was quickly transferred from isopentane to dry ice. After about 20 min, the isopentane on the muscle surface was evaporated. The muscle was packed in tin foil, labelled and stored in a deep freezer at -80±2° for later use.
Hematoxylin and eosin (H and E) staining of muscle tissue:
The muscle tissue was fixed in 5 % paraformaldehyde, rinsed under running water for 6 min. The gastrocnemius muscle mass was reconstructed into 0.4×0.3×0.2 cm size. Gradient dehydration with alcohol was carried out, 50 % ethanol for 0.5 h, 75 % ethanol for 0.5 h, 85 % ethanol for 1 h, 95 % ethanol for 2 h and 100 % ethanol for 20, 30 and 40 min. Transparent treatment was carried out for 10, 15 and 10-20 min in xylene. Wax immersion was done in xylene+paraffin (1:1) for 20 min in a 60° oven, paraffin I for 30 min in a 60° oven and paraffin II for 1h. Tissue blocks were transversely embedded in a Leica EG1160 paraffin embedding machine. Continuous tissue slices of 5 μm thickness were cut on a Leica RM2135 paraffin slicing machine. Sections were spread with Leica HI1210 spreader. Slices are dried with Leica HI1220 dryer. The slides on the dryer were collected with a dyeing rack and stored in an oven at 37±1°.
Tissue sections were removed from the oven and treated with 100 % alcohol for 3 min (twice), 95 % alcohol for 3 min (twice) and 85 % alcohol for 3 min (twice). Tissue sections were washed with running water for 6 min. Tissue sections were stained with hematoxylin for 3 min, differentiated with 1 % hydrochloric acid alcohol for 2 se and washed with running water for 3 min. Tissue sections were treated with Eosin for 3 min, 85 % alcohol for 3 min, 95 % alcohol for 3 min (twice), 100 % alcohol for 3 min (twice) and xylene for 3 min. Finally, neutral resin glue was used to seal the slices and then the slices were flattened until they were aired. The slices were observed under BX53F optical microscope and the images were collected by computer software.
Total RNA extraction and reverse transcription:
Muscle tissue was taken in a 2 ml centrifugal tube and 1 ml TRIzol reagent was added immediately. Two 5 mm steel beads were added, Qiagen tissue homogenizer was used to fully homogenize the tissue. The homogenate was kept at room temperature for 3 to 5 min, 200 μl chloroform was added to the initial amount of 1 ml TRIzol, centrifuged for 15 min at 4°, 12 000 rpm. The upper water phase was carefully aspirated and added to another centrifugal tube. The initial amount of 500 μl isopropanol per ml TRIzol was added, placed at -20° for 30 min to complete precipitation. Centrifuged at 4°, 12 000 rpm for 10 min, the supernatant was carefully removed. To the pellet 1 ml of 75 % ethanol was added, centrifuged again at 4°, 12 000 rpm for 5 min, 75 % ethanol was used to wash the RNA pellet, supernatant was removed, pellet was dried partially, and when the RNA was slightly transparent appropriate volume of RNase-free water was added to completely dissolve. The yield of RNA was measured on an UV spectrophotometer at 260 nm, OD=40 μg/ml. The concentrated RNA was diluted in an appropriate ratio to a final concentration of 200 ng/μl.
A PCR tube was taken and a solution containing 2 μg of RNA was added and 1 μl oligo (dT)18 was added. It was made up to 12 μl with deionized water without ribonuclease. The mixture was incubated at 60° for 8 min on a PCR machine and rapidly cooled on ice. Buffer 4 μl, 2 μl 10 mM triphosphate deoxynucleotides (dNTPs), 1 μl RNA inhibitor and 1 μ1 reverse transcriptase were sequentially added and mixed, incubated at 40-42° for 60 min on a PCR instrument and the reverse transcriptase was inactivated at 80±2° for 6 min.
Tissue protein extraction and content detection:
To the minced tissue, appropriate amount of a 20:1 mixture of cytoplasmic protein extraction reagents A and B was added. A homogenate was prepared by adding phenylmethylsulfonyl fluoride (PMSF) to a final concentration of 1 mM. The tissue and tissue homogenates are mixed in a ratio of 200 microliters of tissue homogenate per 60 mg of tissue and thoroughly homogenized in a glass homogenizer on an ice bath at 4°. The homogenate was transferred to a plastic centrifuge tube and placed in an ice bath for 15 min, centrifuged at 4°, 1500 g for 5 min. The supernatant was transferred to a pre-cooled plastic tube. The protein concentration was measured using the BCA method.
The tissue was placed in a cell pulverizer and 100 μl of protein extract was added. After lysing on ice for 10 min, the lysate was collected in a 1.5 mL microcentrifuge tube (EP) and centrifuged at 4°, 14 000 g for 15 min. The supernatant was stored at -70±2°. Bovine serum albumin (BSA, 1 mg/ml) was taken from -20° and thawed at room temperature. centrifuge tubes (1.5 ml, 54) were taken, with 6 in a group, were labeled as 0, 2.5, 5.0, 10.0, 20.0 and 40.0 μg. Various reagents were added to each tube, mixed and kept at room temperature for 2 min. Colorimetric analysis was performed on an enzyme-linked immunosorbent assay (ELISA) instrument. At the time of sample measurement, firstly, 1.5 mL centrifuge tubes were taken, 1 ml of Coomassie Brilliant Blue solution was added to each tube, the protein was measured for 30 min at room temperature. Then, a tube of Coomassie Brilliant Blue was taken, 100 μ1 of 0.15 mM NaCl solution was added, mixed and placed for 2 min as a blank sample. Finally, a tube of Coomassie Brilliant Blue was taken, 95 μl of 0.15 mM NaCl solution and 5 μl of the sample were added, mixed and placed for 2 min, and analyzed by colorimetry on an ELISA instrument.
SDS-PAGE protein electrophoresis:
The glass plate and the comb were washed, rinsed with double distilled water, dried and fixed on the glue maker. A certain concentration of separation glue is prepared and poured in to the two glass plates fixed in advance. Double distilled water was slowly added to flatten the surface of the gel, which is placed at room temperature for about 25-30 min. Separating gel (10 %) was configured with tetramethylethylene diamine (TEMED). The gel was shaken and poured immediately till the glue surface is raised to 2/3 of the whole glass plate height and then a layer of water was added to the glue. After 30 min at room temperature when the separating gel solidified sufficiently, the upper layer of water was removed and dried with filter paper. The concentrate (5%) is configured and TEMED is shaken immediately after it is added. The gel is shaken and poured immediately. After 30 minutes at room temperature the concentrated gel solidified sufficiently, the comb was pulled out vertically and steadily. The prepared gel was put into the electrophoresis tank, fresh electrophoresis buffer was added into the inner tank to ensure that at least the inside of the small glass plate is immersed. According to the protein concentration of the extract, a sample of 100 μg protein solution was used. Before sampling, the protein was denatured by placing the sample in boiling water for 3-5 min. The sample was taken in an EP tube, 2× loading buffer was added until the final concentration is 1×. Sampling began after sufficient electrophoretic buffer is added to the outer trough of the electrophoresis tank. Ten microliters adder was used to insert the sample into the hole and the sample was slowly added. Marker was added to the last side. At a constant voltage of 60V, the voltage is increased to 100V after separating the gel and the electrophoresis is terminated as soon as bromophenol blue run out of the gel.
Statistical analysis of experimental data:
All data are expressed in the form of mean±standard deviation (SD). SPSS22.0 and Excel statistical software are used to process the data. One-way ANOVA was used to test the difference between groups. The level of significance was set at P<0.05.
Results and Discussion
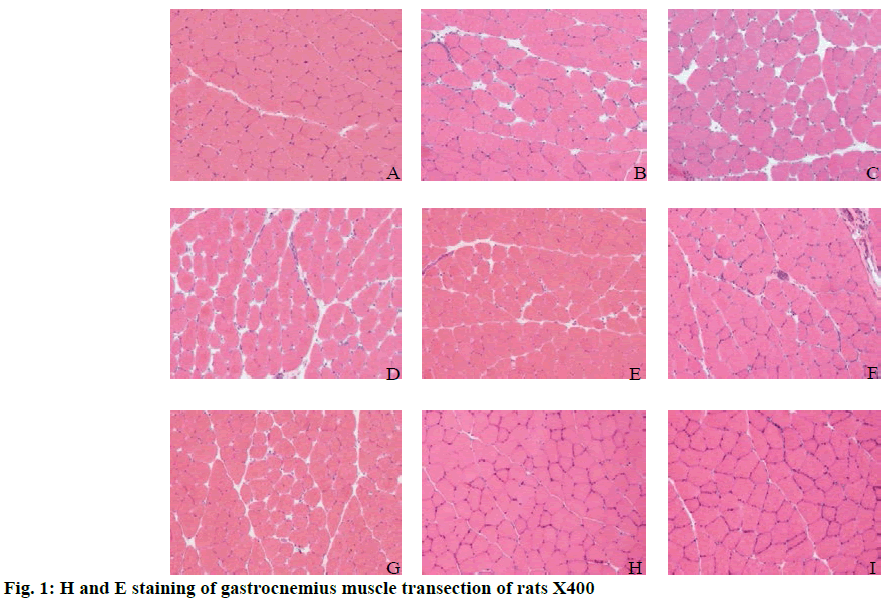
In fig. 1A, the muscle fibers in the control group were uniform in size and polygonal in shape. The muscle fibers were arranged closely, the cell boundaries were clear and the nuclei were uniformly around the cells. In fig. 1B, the muscle fibers in the 0 h group were mostly swollen and round and the cell sizes were quite different. The cell gap was large and the arrangement was loose. In fig. 1C, the swelling of muscle fibers was similar to that of round muscle fibers at 6 h after exercise and the cell size was different. The cell gap was also large and loosely arranged, which was similar to that of the 0 h group after exercise. In fig. 1D, the muscle fibers in the 12 h group were still swollen, the difference of cell size was recovered compared with that in the 6 h group and the cell gap was still much larger than that in the control group. In fig. 1E, the muscle fibers of 24 h group recover slightly compared to the 12 h group, but were slightly swollen compared to the control group. The cell gap was smaller than that of 12 h group or larger than that of the control group. The cell arrangement was tighter and neater than that of the 12 h group. In fig. 1E, the muscle fibers in the 24 h group recovered slightly compared to the 12 h group, but the muscle fibers in the control group were slightly swollen. The cell gap was smaller than that in the 12 h group, or larger than that in the control group. The cell arrangement was tighter and neater than that in the 12 h group. In fig. 1F, the swelling of some muscle fibers in 48 h after exercise is similar to that of the control group. The gap between cells was restored, but larger than that of the control group. The arrangement of muscle fibers was also restored, which was more orderly, but worse than that of the control group. The number of nuclei around muscle fibers was more than that of the control group. In fig. 1G, at 72 h after exercise, some cells of muscle fibers were slightly swollen. The intercellular space recovered, but the intercellular space was larger than that of the control group. The arrangement of muscle fibers was also better than that of 48 h after exercise. In fig. 1H, the muscle fibers in group 1W after exercise were uniform in size and polygonal. The intercellular space was small and arranged tightly. The nuclei were uniformly arranged around the cells. In fig. 1I, the muscle fibers in the 2W group were uniform in size and polygonal. The cell gap was small and arranged closely. The nuclei were evenly distributed around the cells and the muscle fibers returned to the control group.
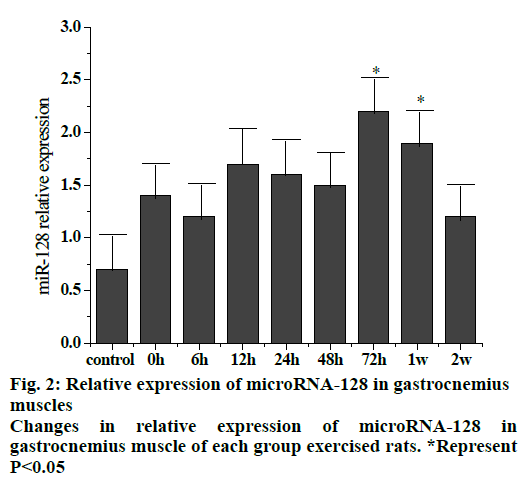
Fig. 2 shows the trend of relative expression of miR- 128 at different stages of gastrocnemius muscle injury in rats. The results showed that compared to the control group, the overall expression of miR-128 increased after exercise and returned to a slightly higher level after 2 w. Compared to the control group, the expression at 72 h and 1 w after exercise were significantly higher (p < 0.05).
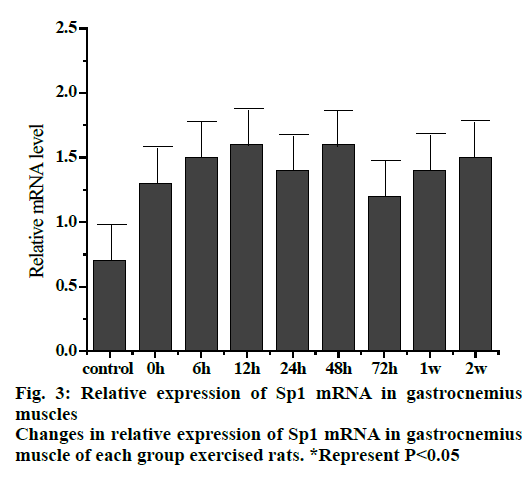
Fig. 3 shows the trend of relative expression of Sp1 mRNA at different stages of gastrocnemius muscle injury in rats. The results showed that compared to the control group, the level of Spl mRNA expression was higher. The expression of Sp1 mRNA was continuously increased but not significantly at 0, 6 and 12 h groups, which reached a maximum in 12 h group. The relative expression of Sp1 gene remained more or less slightly but not significantly elevated or decreased in 24, 48 and 72 h, 1 and 2 w groups.
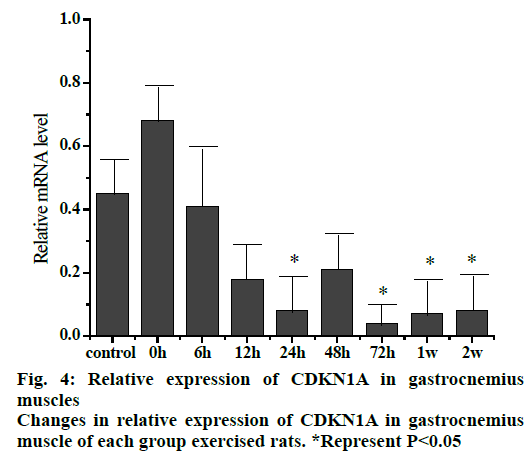
The relative expression of CDKN1A mRNA (fig. 4) in gastrocnemius muscle of rats at different stages after exercise injury indicated that the expression of CDKN1A mRNA in gastrocnemius muscle decreased as a whole after exercise compared to the control group. The expression of CDKN1A mRNA in the 0 hour group increased initially, then decreased gradually in the rest of the groups with the decrease in 24 and 72 h, 1 and 2 w groups was significant compared to the control group (p<0.05).
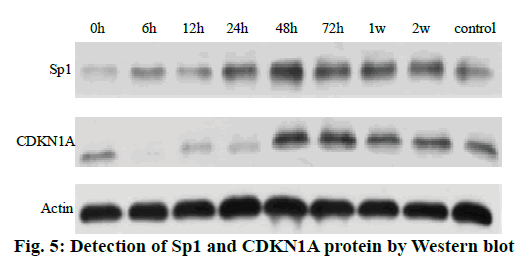
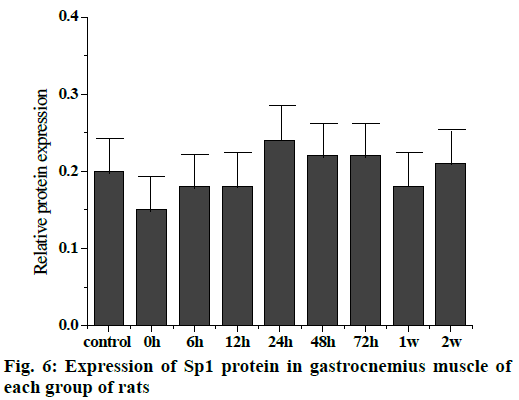
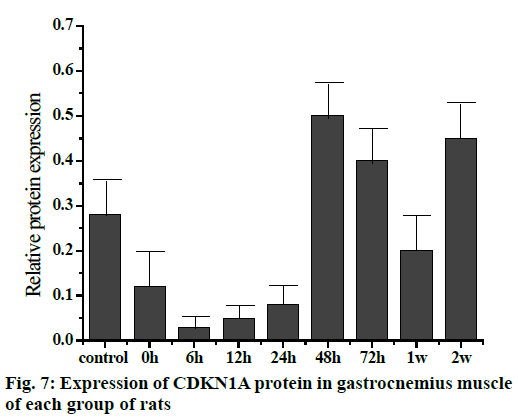
The Western blot results of the Sp1 and CDKN1A protein levels are shown in fig. 5. Compared to the control group, the Sp1 protein level in the gastrocnemius muscle decreased gradually but not significantly at 0, 6 and 12 h, The Sp1 protein levels increased and reached a maximum at 24 h and then returned to the normal level. There was no significant difference between the two groups as shown in fig. 6. Compared to the control group, the expression of CDKN1A protein decreased in the 0, 6, 12 and 24 h groups during the repair of skeletal muscle injury, which reaches the maximum in the 48 h group. There was no significant difference between the two groups. After that, the expression of CDKN1A protein decreased further in the 72 h and 1 w group but increased in the 2 w group but none of these changes were significant (fig. 7).
In this study, continuous downhill running training was used to cause skeletal muscle injury in rats. H and E staining showed that the muscle cells in gastrocnemius muscle showed slight swelling and inflammatory cell infiltration after exercise compared to the control group and the degree of injury was most serious at 3 d. The ultrastructure of muscle cells and the abnormalities of sarcomere rupture, disappearance and filament degradation were also found after exercise. Morphological evidence directly suggests that exercise (downhill running) does cause skeletal muscle damage in rats. However, compared to the degree of injury, the morphological results showed that the sports injury of skeletal muscle is far less serious than that caused by drug injection. In conclusion, H and E staining and morphological evidence from previous studies on ultrastructure intuitively confirmed that the established model of motor muscle microinjury and repair in rats is successful, but it is different from the model of muscle injury caused by drug injection. In fact, the microinjury model built in this study was closer to the changes of muscle injury caused by sports activities and this model is more representative in the field of sports science.
It is speculated that miR-128 might participate in the regeneration process of exercise-induced skeletal muscle injury and promote the repair of skeletal muscle injury by regulating the expression of CDKN1A. The specific regulation mechanism of Spl and CDKN1A could not be fully explained only by the changes in expression of miR-128, Spl and CDKN1A, but the actual phenomena on the surface could be seen. Moreover, the degree of muscle injury in this experimental model is slight, and the nerve injury is slighter. In combination with histological results, no myotubes with nuclei located in the center of the cell were found. Therefore, to further understand the specific mechanisms of miR-128, Spl and CDKN1A in this experimental model, more indepth research is needed.
In summary, it could be concluded that the expression of miR-128 was not significantly changed in exercise-induced skeletal muscle injury and the expression of miR-128 might be involved in the regeneration of exercise-induced skeletal muscle injury. The expression of CDKN1A might be affected by the regulation of Sp1 expression and the expression of CDKN1A promoted repair of injured skeletal muscle.
References
- Sun JH, Zhang L, Wang XW. Relation between Injury Time and the Expression of COX6C mRNA in Skeletal Muscle of Rats after Contusion. Fa Yi Xue Za Zhi 2015;31(3):177.
- Zhang H, Yongli XU, Wang S. The early histomorphological changes and expressions of atrogin-1 and MuRF-1 mRNA in skeletal muscle of paralytic limb of cerebral ischemia/reperfusion injury rats. Chinese J Rehabil Med 2014;29(7):610-14.
- Trajano LASN, Trajano ETL, Thomé AMC. Pulsed low-level infrared laser alters mRNA levels from muscle repair genes dependent on power output in Wistar rats. Laser Phys Lett 2017;14(10):105603.
- Wright CR, Brown EL, Della Gatta PA. Regulation of Granulocyte Colony-Stimulating Factor and Its Receptor in Skeletal Muscle Is Dependent Upon the Type of Inflammatory Stimulus. J Interferon Cytokine Res 2015;35(9):710.
- Alves AN, Ribeiro BG, Fernandes KP. Comparative effects of low-level laser therapy pre- and post-injury on mRNA expression of MyoD, myogenin, and IL-6 during the skeletal muscle repair. Lasers Med Sci 2016;31(4):679-685.
- Xu TP, Liu XX, Xia R. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene 2015;34(45):5648-61.
- Sun KT. Overexpression of Mechano-Growth Factor Modulates Inflammatory Cytokine Expression and Macrophage Resolution in Skeletal Muscle Injury. Front Physiol 2018;9:999.
- Liu X, Liu Y, Zhao L. Macrophage depletion impairs skeletal muscle regeneration: The roles of regulatory factors for muscle regeneration. Cell Biol Int 2017;41(3):228-38.
- Mounier R, Chazaud B. PPARγ transcription factor controls in anti-inflammatory macrophages the expression of GDF3 that stimulates myogenic cell fusion during skeletal muscle regeneration. Med Sci 2017;33(5):466.
- Dey BK, Pfeifer K, Dutta A. The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 2014;28(5):491-501.
- Yu T, Chang Y, Gao XL. Dynamic Expression and the Role of BDNF in Exercise-induced Skeletal Muscle Regeneration. Int J Sports Med 2017;38(13):959-66.
- Ferraro E, Giammarioli AM, Chiandotto S. Exercise-Induced Skeletal Muscle Remodeling and Metabolic Adaptation: Redox Signaling and Role of Autophagy. Antioxid Redox Signal 2014;21(1):154-76.