- *Corresponding Author:
- Qingguo Li
Department of Cardiovascular Surgery, The Second Affiliated Hospital of Nanjing Medical University, Nanjing, Jiangsu Province 210011, China
E-mail: liqg@njmu.edu.cn
| This article was originally published in a special issue, “Recent Progression in Pharmacological and Health Sciences” |
| Indian J Pharm Sci 2024:86(2) Spl Issue “122-133” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Heart failure is a significant public health challenge, persisting as a prime contributor to mortality and morbidity. Autophagy, as a cellular process, plays a crucial role in maintaining an optimal intracellular environment and holds therapeutic potential in the context of heart failure, but the correlation between autophagy and heart failure has yet to be fully elucidated. Single cell ribonucleic acid sequencing is a powerful method for investigating cell-specific transcriptome changes in heart failure. The study aimed to find autophagy-related molecules, mechanisms, and pathways at the single-cell level. Additionally, we sought to assess the prognostic implications for individuals with heart failure. First, we downloaded the heart failure related datasets from the Gene Expression Omnibus database to identify autophagy-related differential genes. We identified 9 cell-specific transcriptional signatures associated with autophagy that might be involved in heart failure progression, and the expression of these genes was detected by single-cell strategy. Correlation analysis revealed the relationships between these genes and the expression patterns of various immune cell types. The results of functional enrichment analysis revealed a strong association of autophagy-related genes with cellular responses to external stimuli, as well as significant enrichment in lipid metabolism, atherosclerosis, and the phosphoinositide 3-kinase/protein kinase B signaling pathway. Potential targets were predicted by protein-protein interaction and molecular docking. This study has yielded novel insights into the role of autophagy-related cell-type-specific expression genes as biomarkers in the pathogenesis of heart failure, and established a valuable resource for further investigations into the underlying molecular mechanisms.
Keywords
Heart failure, cardiomyopathy, single cell ribonucleic acid sequencing, autophagy, myocarditis
Heart Failure (HF) represents a severe and terminal stage of diverse heart diseases, encompassing high morbidity and mortality rates. Patients with HF often experience progressive exercise intolerance and respiratory distress, whether at rest or during physical exertion, especially in cases of underlying coronary heart disease, hypertension, diabetes, or myocarditis, as well as those who have undergone intensive alcohol consumption, chemotherapy, and/ or radiation therapy. Additionally, the condition can induce fatigue, fluid overload, marked by occurrences like pulmonary edema or ankle edema, and lead to structural and functional abnormalities within the heart. Treatment strategies encompass a range from the implementation of diuretics to alleviate symptoms, to the utilization of an expanding array of diseasemodifying drugs and device therapies. Notably, the emergence of Sodium-Glucose Cotransporter 2 (SGLT2) inhibitors has ushered in a more recent enhancement of disease outcomes[1], but patients suffering from HF still have a poor prognosis and high mortality. It is necessary to explore and identify more effective prognostic biomarkers for therapeutic targets and clinical decisions[2].
Dilated Cardiomyopathy (DCM) is the main cause of HF. In DCM, a condition characterized by the enlargement and weakening of the heart’s chambers, the heart’s capacity to effectively pump blood throughout the body can be compromised. Consequently, these pathophysiological alterations can readily advance towards the development of HF. Therefore, it is critical to study the molecular mechanisms of DCM, enabling the identification of novel biomarkers for HF. Such endeavors hold the potential to unravel new treatment strategies. Autophagy, a process employed for the intracellular degradation of organelles and proteins, holds special significance for the enduring cardiomyocytes implicated in heart disease. Central to this phenomenon is the auto phagosome, a pivotal process that entails the encapsulation of cytoplasmic constituents within a specialized double-membraned vesicle known as an auto phagosome. The autophagy pathway can be delineated into distinct stages, including induction, vesicle nucleation, selective cargo recognition, auto phagosome formation, fusion between auto phagosomes and lysosomes, cargo degradation, and nutrient recycling. Autophagy, a conserved mechanism, profoundly contributes to sustaining intracellular equilibrium through the degradation of long-lived proteins and compromised organelles. Importantly, this mechanism is now widely recognized as a promising therapeutic target for several heart diseases[3]. Recent studies have shown that both HF and DCM have been extensively linked to the intricate pathogenesis of autophagy, with excessive activation of this process culminating in the degradation of essential molecules and organelles vital for cell survival. This dysregulation ultimately drives organ failure and precipitates patient mortality. Furthermore, autophagic phenomena have been observed in both HF patients and relevant animal models. Research has indicated the protective role played by autophagy in both HF and DCM[4,5]. Nonetheless, the intricate relationship between autophagy and HF remains elusive, particularly at the single-cell level. Little insight exists on other immune subsets involved in the cardio toxic response, and the pathway for autophagy and all its mediators also remain to be elucidated.
The single cell Ribonucleic Acid-sequencing (scRNA-seq) is a novel method for high-throughput sequencing in the genome on the basis of a single cell[6], which can investigate the specific cell types and gene expression state of a single cell, revealing the heterogeneity between cells[7]. Currently, scRNA-seq has emerged as a pivotal tool, providing profound and novel insights into the intricate pathophysiological processes underlying cardiovascular diseases. A multitude of studies have harnessed the potential of scRNA-seq, uncovering crucial alterations in cellspecific gene expression that intricately contributing to the development of HF pathology. These alterations encompass a spectrum of conditions, including DCM, myocardial hypertrophy, and heart fibrosis. With its capacity to scrutinize gene expression patterns at the single-cell level, scRNA-seq technology holds great promise in identifying cell-specific molecular targets implicated in HF. However, it is essential to acknowledge that despite notable advancements in this field, the comprehensive elucidation of potential biomarkers within specific cell types, as discovered through scRNA-seq profiling of HF pathogenesis, necessitates further investigation and clarification.
In the present study, our aim was to identify the significant cell population and potential biomarker in HF. We downloaded the HF transcriptome dataset GSE5406 and GSE9128 as well as the scRNA-seq dataset GSE145154 related to HF from the Gene Expression Omnibus (GEO) database for bioinformatics analysis. The results provide insights into the cell-type-specific expression gene biomarker in HF development. Herein, we suggested a novel prognostic autophagy-related signature using Support Vector Machine (SVM) analysis followed by construction of Least Absolute Shrinkage and Selection Operator (LASSO) regression in HF. Significantly, we successfully identified a nine-gene signature associated with autophagy, and we utilized a single-cell strategy to detect the expression of these genes, such as Activating Transcription Factor 4 (ATF4) and Microtubule-Associated Protein 1 Light Chain (MAP1LC3B), in the context of HF. Studies have indicated that ATF4 plays a protective role against HF by counteracting oxidative stress. However, the relationship between MAP1LC3B and HF remains unclear. Investigating the mechanisms underlying the role of these genes in HF prognosis could offer novel insights into potential treatments for HF. Furthermore, these investigations could also provide a reference point for exploring the intricate relationship between autophagy and HF.
Materials and Methods
Data download and processing:
GSE5406[8] and GSE9128[9] microarray datasets, two data cohorts, and their corresponding sample clinical information were retrieved and downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/ geo/). The sequencing platform for two datasets was based on GPL96. Gene ID conversion was performed through the annotation file of the sequencing platform and the samples in the datasets were derived from Homo sapiens. 210 samples were included in the GSE5406 dataset, of which 16 samples were normal and 194 samples were HF; 11 samples were included in the GSE9128 dataset, of which 3 samples were normal and 8 samples were HF. We selected the normal control and HF samples from GSE5406 and GSE9128 datasets as the control group and disease group for analysis (n=221) and used the ComBat package to remove batch effects from the merged dataset and conduct subsequent analysis of the merged cohort[10].
The GSE145154 scRNA-seq dataset of HF was also downloaded from the GEO database[11], and the test platform was GPL20795 HiSeq X ten. It contains scRNA-seq data from 1 normal sample and 3 HF samples, all of which were derived from Homo sapiens. Only the DCM sample from the HF sample was used for analysis in this study.
Identification of autophagy differential genes between the normal group and HF group:
222 autophagy genes were obtained from the Human Autophagy Database (HADb)[12] (http:// www.autophagy.lu/). We used the R package limma to perform a differential analysis of these genes in the merged cohort (logFC=0, p<0.05) to identify autophagy differential genes between the normal group and the HF group.
Search Tool for the Retrieval of Interacting Genes (STRING) (https://string-db.org/) is a database for searching interactions between known proteins and predictive proteins[13,14], which contains not only experimental data, results from text mining of PubMed abstracts and data from other databases, but also results predicted using bioinformatics. In this study, the STRING database was used to perform Protein- Protein Interaction (PPI) network construction against the obtained autophagy differential genes.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis:
GO analysis is a common method for large-scale functional enrichment studies[15], including Biological Process (BP), Molecular Function (MF), and Cellular Component (CC). The KEGG is a widely used database that stores information about genomes[16], biological pathways, diseases, and drugs. We used the clusterProfiler package to perform GO and KEGG enrichment analysis of autophagy differential genes (p<0.05)[17].
Screening of hub autophagy genes with HF by Lasso regression and SVM:
For the obtained autophagy differential genes, we used Lasso regression and SVM to further screen the disease feature genes of HF, and then we took the intersection of the genes obtained from the two methods and defined them as the hub autophagy genes with HF. The lasso regression was based on the glmnet package with a seed of 123, while the SVM was based on the caret package with a seed of 123[18].
For the identified hub autophagy genes, the diagnostic efficacy of each gene for HF was assessed in the merged cohort using a Receiver Operator Curve (ROC).
Unsupervised clustering analysis:
Unsupervised clustering is a resampling-based clustering algorithm for identifying each member and its number of subgroups and verifying the clustering rationality[19]. We used the ConsensusClusterPlus[20] package to identify different disease subtypes based on hub autophagy gene expression profile data for HF samples in a merged cohorts using unsupervised clustering.
Gene Set Enrichment Analysis (GSEA):
GSEA was used to assess the trend of distribution of genes in predefined gene sets in a table of genes ranked with phenotypic relevance to determine their contribution to the phenotype[21]. Enrichment analysis of all Differentially Expressed Genes (DEGs) in both high and low phenotypic relevance groups was performed using the R package clusterProfiler, and the parameters used in this GSEA were as follows; seeds of 2020, number of calculations of 10 000, the minimum number of genes contained in each gene set of 10 and maximum number of genes contained in each gene set of 500, and p-value correction method of Benjamini-Hochberg (BH). The c2.cp. v7.2.symbols.gmt gene set was obtained from the Molecular Signatures Database (MSigDB) for GSEA analysis[22,23], and the screening standards for significant enrichment were p<0.05 and False Discovery Rate (FDR) value (q<0.05).
Assessment of immune infiltration status:
We assessed the immune infiltration status of HF samples using single sample GSEA (ssGSEA)[24]. The abundance of 28 TIICs (activated B cells (Ba), activated Clusters of Differentiation (CD)-4+ T cells (CD4+ Ta), activated CD8+ T cells (CD8+ Ta), activated Dendritic Cells (DCa), CD56bright Natural Killer cells (CD56+ NK), CD56dim NK cells (CD56− NK), central memory CD4+ T cells (CD4+ Tcm), central memory CD8+ T cells (CD8+ Tcm), effector memory CD4+ T cells (CD4+ Tem), effector memory CD8+ T cells (CD8+ Tem), eosinophils, Gamma Delta T cells (γδT), immature B cells (Bi), immature DC (iDC), mast cells, Myeloid-Derived Suppressor Cells (MDSC), memory B cells (Bm), monocytes, NK cells, NK-T, neutrophils, plasmacytoid DC (pDC), macrophages, regulatory T cells (Treg), follicular helper T cells (fhT), type-1 T helper cells (Th1), type-17 T helper cells (Th17), and type-2 T helper cells (Th2)) in a single sample of the merged cohort was predicted using the Gene Set Variation Analysis (GSVA) (R package)[25].
Expression of hub autophagy differential genes on scRNA-seq data:
We used the Seurat R package to import a singlecell dataset of HF, DCM samples and create the Seurat objects for this analysis[26]. We filtered for low-quality cells with gene counts <200 or >3000. The proportion of mitochondrial genes to all genetic material may indicate whether a cell is in homeostasis. We generally consider that when a cell has a higher proportion of mitochondrial genes than all genes, it may be in a state of stress. Therefore, we filtered cells with >10 % mitochondrial genes. After the above steps, we obtained 3555 cells.
The data were normalized using log normalize. After controlling the relationship between mean expression and dispersion, highly variable genes were identified in single cells. We then decentered all genes and used Principal Component Analysis (PCA) to cluster all cells, visualized the obtained cell subgroups using T-distributed Stochastic Neighbor Embedding (TSNE)[27], and used the human primary cell atlas dataset from single R to identify the cell type of each cluster[26,28]. The expression of hub autophagy differential genes was also assessed in each of the identified cell subgroups.
Statistical analysis:
All data calculations and statistical analyses were performed using R programming (https://www.rprojec t.org/, version 4.2.1). Comparisons between two groups were performed by the Wilcoxon rank sum test, while comparisons between three or more groups were performed by the Kruskal-Wallis test. The correlation was performed by the Spearman. The survival package of R was used for survival analysis, the Kaplan-Meier survival curve was used to show survival differences, and the log-rank test was used to assess the significance of the difference in survival time between the two groups of patients. Both univariate and multivariate Cox analysis were based on the survival R package, and all statistical p values were two-sided, with p<0.05 considered statistically significant.
Results and Discussion
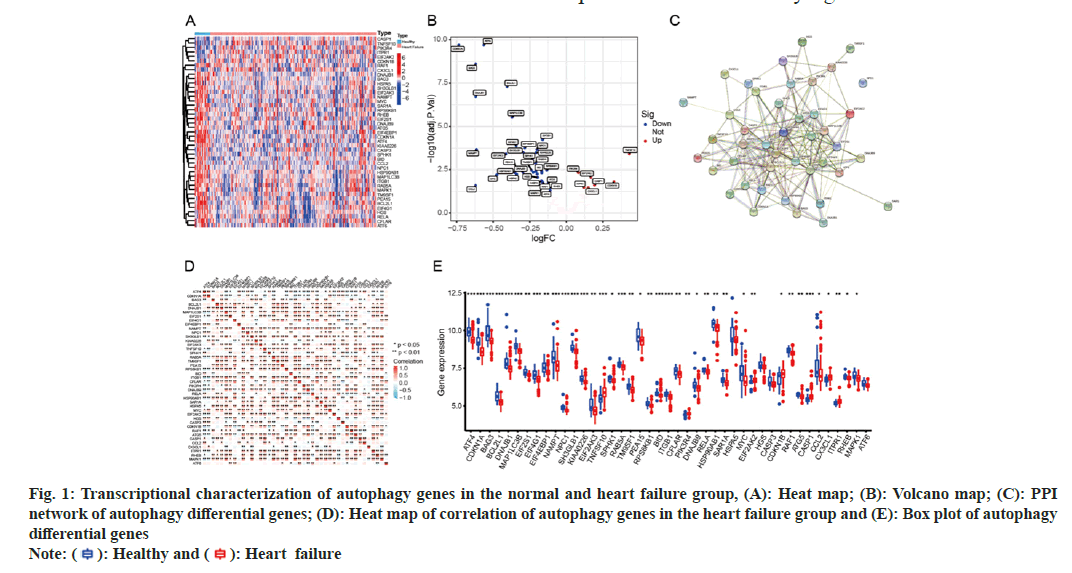
We observed autophagy-related transcriptional changes between healthy controls and DCM samples. We first extracted the expression of 222 autophagy genes in the merged cohort, and we found a total of 187 autophagy genes expressed in the merged cohort (fig. 1). The acquired autophagy genes were subjected to differential analysis and 43 DEGs were obtained, and their expression heat map (fig. 1A) and volcano map (fig. 1B) were plotted for genes with significant expression differences. The PPI showed how these 43 autophagy genes interact with each other and we found that the human kinase Ribosomal Protein S6 Kinase Beta-1 (RPS6KB1) is at the center of the network (fig. 1C). The correlation among differentially expressed autophagy genes within the HF group has been shown and it can be found that ATF4 and Cyclin-Dependent Kinase Inhibitor (CDKN1A) have a significant negative correlation with other genes (R<0, p<0.05) (fig. 1D). The box plot visualizes the differential expression of autophagy genes in the normal group and before HF, it was found that there were 43 differentially expressed autophagy genes (p<0.05), among which 7 were upregulated and 36 were downregulated (fig. 1E).
Fig. 1: Transcriptional characterization of autophagy genes in the normal and heart failure group, (A): Heat map; (B): Volcano map; (C): PPI
network of autophagy differential genes; (D): Heat map of correlation of autophagy genes in the heart failure group and (E): Box plot of autophagy
differential genes
Note: ( ): Healthy and (
): Healthy and (  ): Heart failure
): Heart failure
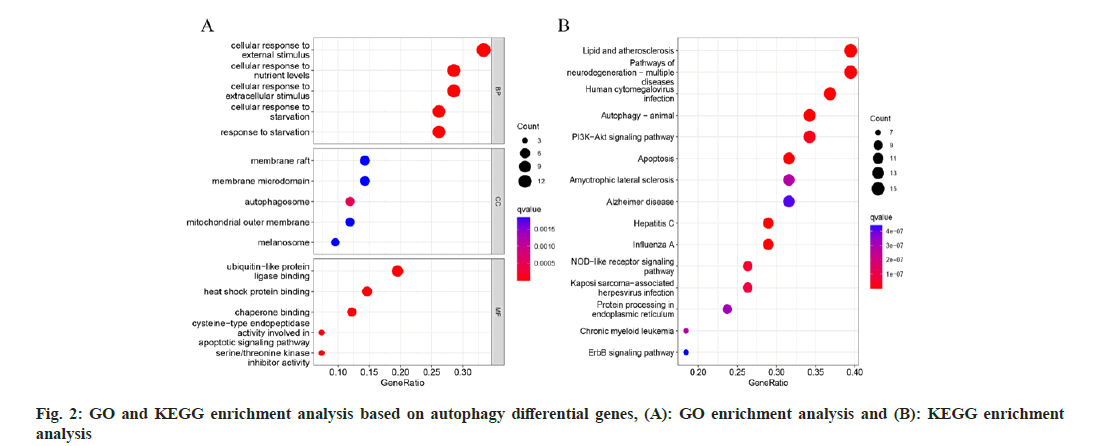
To identify distinct pathways connecting autophagy and HF. The autophagy differential genes in GO and KEGG enrichment analysis has been presented in the context (fig. 2). The autophagy differential genes were mainly enriched in cellular response to external stimulus, cellular response to nutrient levels, ubiquitin-like protein ligase binding and other pathways in GO analysis (fig. 2A); autophagy differential genes were mainly enriched in lipid and atherosclerosis, Phosphoinositide 3-Kinase (PI3K)/ Protein Kinase B (AKT) signaling pathway, apoptosis, and other pathways (fig. 2B). The exploration of the roles played by differential genes can provide valuable insights for subsequent studies related to HF. Genes identified in single-cell transcriptome analyses, such as ATF4, may also be implicated in these pathways.
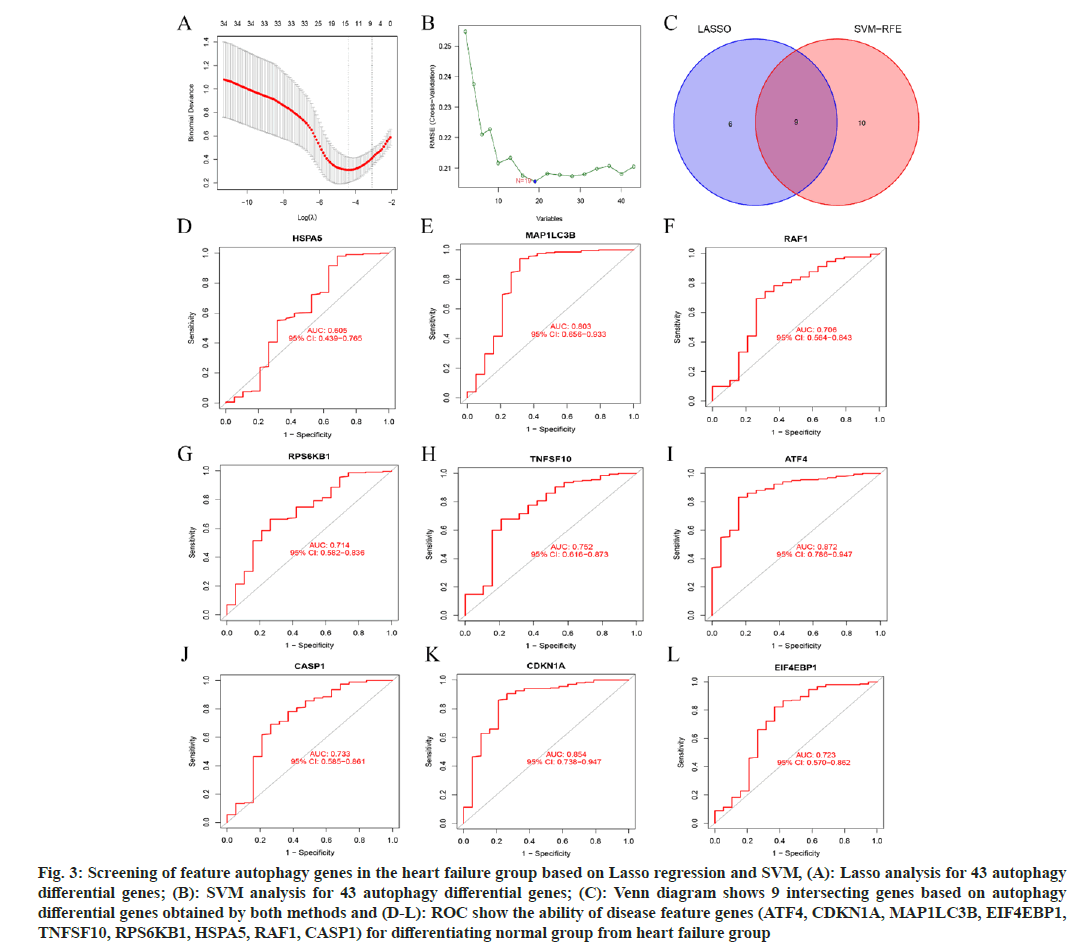
Lasso-based regression and SVM models have demonstrated greater accuracy and utility in predicting the relationship between autophagy- related genes and HF patients. 15 disease feature genes were identified by Lasso; ATF4, CDKN1A, MAP1LC3B, Eukaryotic Translation Initiation Factor 4E Binding Protein 1 (EIF4EBP1), SH3 domain containing GRB2 like, endophilin B1 (SH3GLB1), KIAA0226, TNF superfamily member 10 (TNFSF10), RAB5A, RPS6KB1, PIK3R4, Heat shock protein family (Hsp70) A member 5 (HspA5), RELA, RAF1, Caspase 1 (CASP1), C-X3-C Motif Chemokine Ligand 1 (CX3CL1) (fig. 3A); 19 disease feature genes were identified by SVM; BAG Cochaperone 3 (BAG3), Proliferation and Apoptosis Adaptor Protein 15 (PEA15), ATF4, CDKN1A, MAP1LC3B, BCL2L1, EIF2S1, Niemann-Pick Disease C1 (NPC1), Mitogen-Activated Protein Kinase 1 (MAPK1), EIF4G1, Myelocytomatosis Oncogene (MYC), ATF6, CASP8 and FADD Like Apoptosis Regulator (CFLAR), EIF4EBP1, TNFSF10, RPS6KB1, HSPA5, RAF1, CASP1 (fig. 3B); then we took the intersection of the two identified genes and screened 9 hub autophagy genes with HF; ATF4, CDKN1A, MAP1LC3B, EIF4EBP1, TNFSF10, RPS6KB1, HSPA5, RAF1, and CASP1 (fig. 3C). Finally, we used ROC to show the ability of these 9 genes for differentiating normal from HF patients (fig. 3D-fig. 3L). The expression of these nine autophagy-related genes within HF cell subsets will undergo verification through single-cell assays. Importantly, certain genes from this set may serve as potential therapeutic targets, thereby facilitating the exploration of the underlying mechanisms.
Fig. 3: Screening of feature autophagy genes in the heart failure group based on Lasso regression and SVM, (A): Lasso analysis for 43 autophagy differential genes; (B): SVM analysis for 43 autophagy differential genes; (C): Venn diagram shows 9 intersecting genes based on autophagy differential genes obtained by both methods and (D-L): ROC show the ability of disease feature genes (ATF4, CDKN1A, MAP1LC3B, EIF4EBP1, TNFSF10, RPS6KB1, HSPA5, RAF1, CASP1) for differentiating normal group from heart failure group
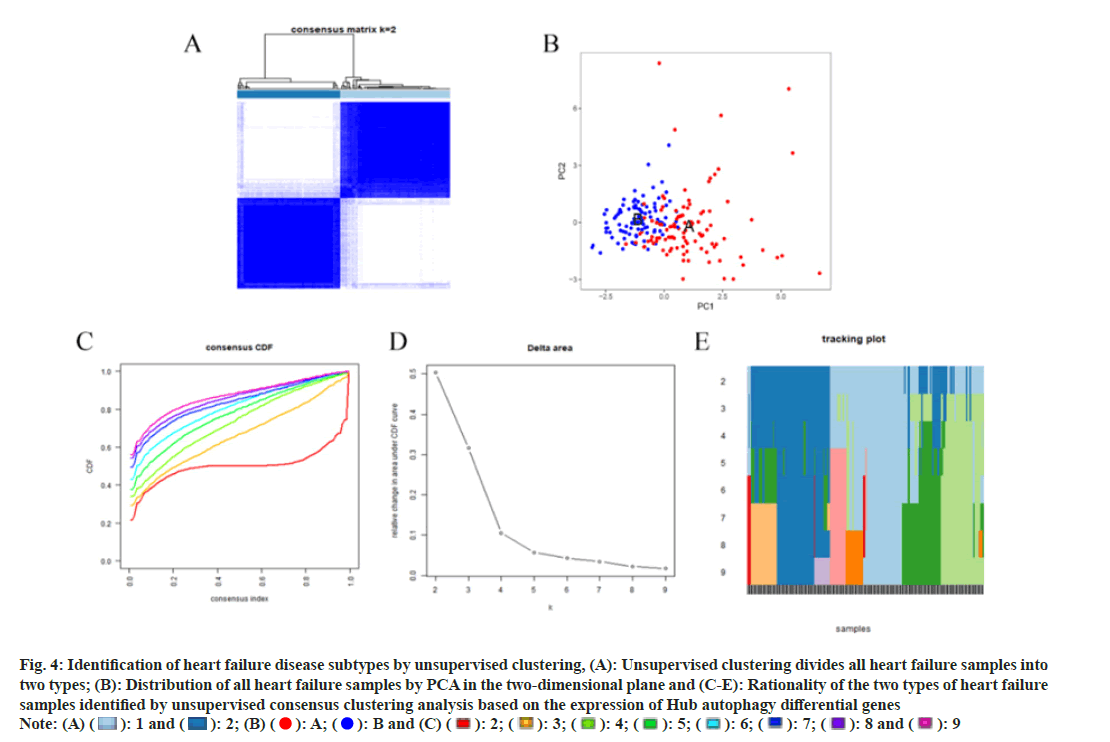
We performed unsupervised clustering analysis on HF samples based on the Hub autophagy gene, and the distribution of samples (fig. 4A). PCA analysis shows the distribution of each sample in the twodimensional plane when they are divided into two types, and it can be found that the two types of patient samples are clearly distinguished in the PCA (fig. 4B). All HF samples can be well distinguished when they are divided into two types, with two types being the ideal and reasonable classification (fig. 4C-fig. 4E). By amalgamating data from diverse sources, we achieved more reasonable disease classification outcomes pertinent to HF and autophagy.
Fig. 4: Identification of heart failure disease subtypes by unsupervised clustering, (A): Unsupervised clustering divides all heart failure samples into
two types; (B): Distribution of all heart failure samples by PCA in the two-dimensional plane and (C-E): Rationality of the two types of heart failure
samples identified by unsupervised consensus clustering analysis based on the expression of Hub autophagy differential genes
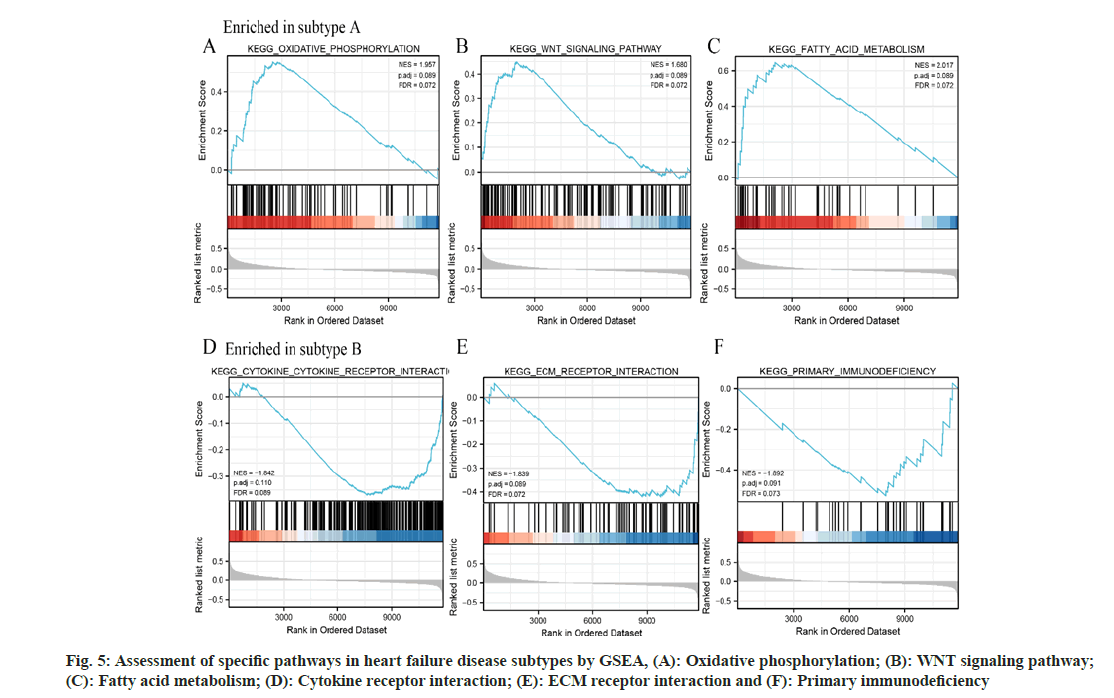
We used the GSEA to assess the feature pathways in the identified HF disease subtype A and subtype B in HF samples from the merged cohort based on the c2.cp.v7.2.symbols.gmt gene set from the MSigDB. The results showed that the oxidative phosphorylation (fig. 5A), WNT signaling pathway (fig. 5B), and fatty acid metabolism (fig. 5C) pathway were significantly upregulated in subtype A, while the cytokine receptor interaction (fig. 5D), Extracellular Matrix (ECM) receptor interaction (fig. 5E), and primary immunodeficiency (fig. 5F) pathways were significantly upregulated in subtype B. Metabolismrelated signaling pathways predominantly exhibit associations with subtype A HF, whereas subtype B HF is primarily linked to cell-cell interaction signaling pathways. This observation posits that distinct pathogenic mechanisms may underlie the two divergent HF subtypes.
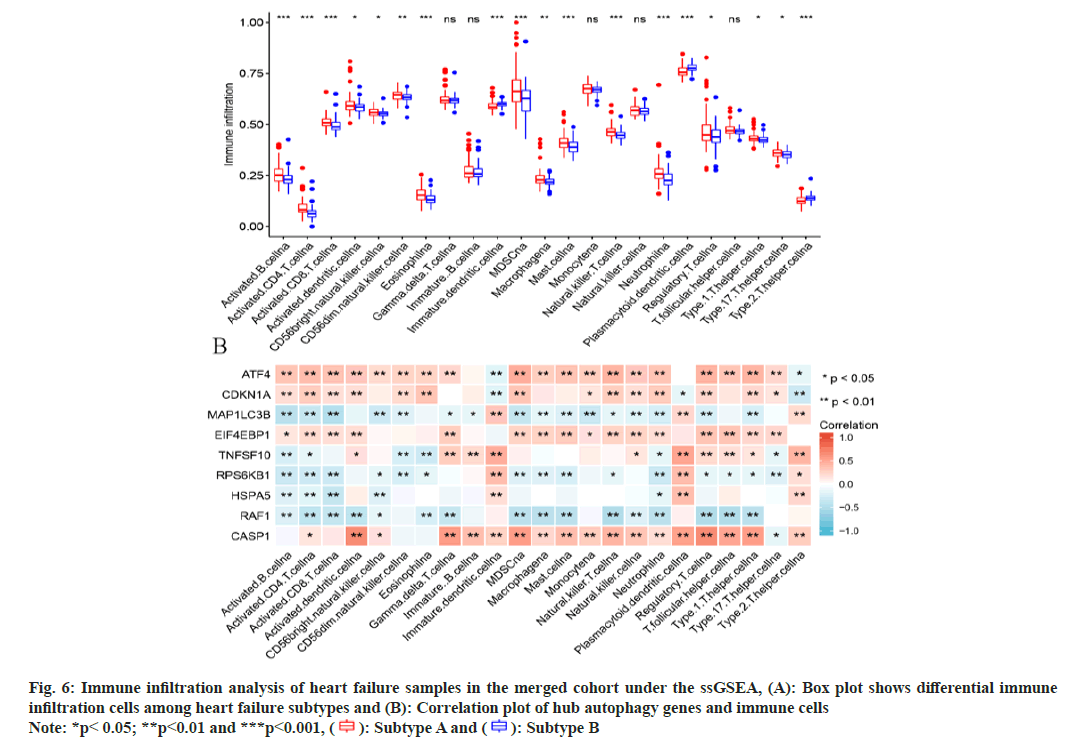
The immune infiltration status of HF disease samples was assessed using the ssGSEA. The results showed that most immune cells were significantly higher in subtype A than in subtype B, such as activated B cell, activated CD4 T cell, activated CD8 T cell, and immature B cell; only immature dendritic cell, plasmacytoid dendritic cell, and Th2 cell were highly expressed in subtype B group (fig. 6A). Next, we performed correlation analysis of hub autophagy differential genes with immune infiltration score, and the results showed that ATF4, CDKN1A, EIF4EBP1, and CASP1 had a significant positive correlation with the identified immune infiltration cells, while MAP1LC3B, RAF1 showed significant negative correlation with the identified immune infiltration cells (fig. 6B). Immunoinfiltration analysis offers insights into the immune cell composition within HF, elucidating pivotal immune cell types influencing its progression. Integrating autophagy-related genes with immune cells facilitates a more comprehensive understanding of autophagy’s role in HF. Autophagyrelated genes could potentially influence the prognosis of HF by modulating immune-related cells.
Fig. 6: Immune infiltration analysis of heart failure samples in the merged cohort under the ssGSEA, (A): Box plot shows differential immune
infiltration cells among heart failure subtypes and (B): Correlation plot of hub autophagy genes and immune cells
Note: *p< 0.05; **p<0.01 and ***p<0.001, ( ): Subtype A and (
): Subtype A and ( ): Subtype B
): Subtype B
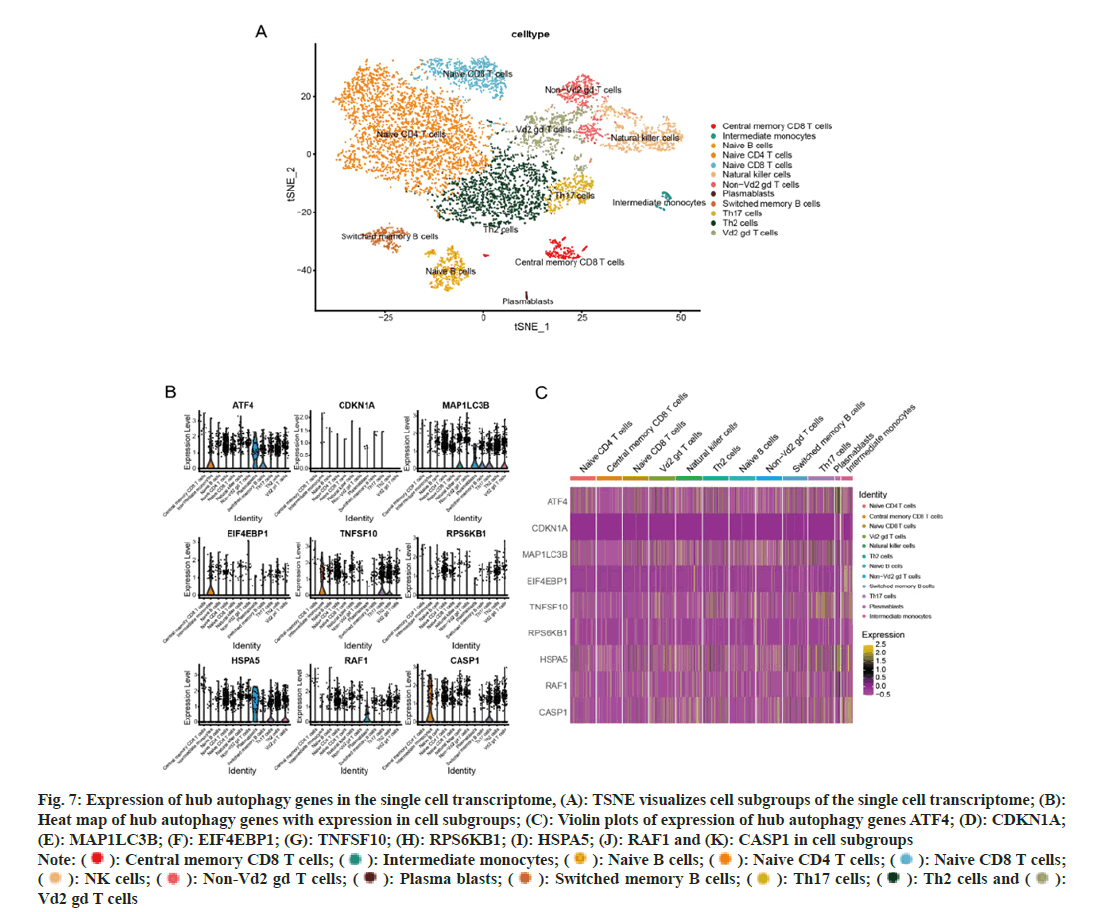
In this study, we performed scRNA-seq on samples from DCM cases, resulting in a total of 8211 cells after filtering and the removal of batch effects. Following data normalization, the top 3000 genes were selected for PCA dimensionality reduction. Subsequently, the normalized data underwent clustering of cells exhibiting analogous gene expression profiles, utilizing the first 50 principal components. T-SNE dimensionality reduction was used to visualize 13 independent clusters. We employed the human primary cell atlas data dataset within single to classify the cell types present in each cluster, resulting in the identification of 12 distinct cell types (fig. 7A).
Next, the expression of hub autophagy genes in cell subgroups was visualized using heat maps with violin plots. The results showed that ATF4 was highly expressed in intermediate monocytes and plasma blasts; CDKN1A was not found to be expressed in the identified cell groups; MAP1LC3B was highly expressed in NK cells, plasma blasts, switched memory B cells, Th17 cells, and Vd2 gd T cells; EIF4EBP1 was expressed in intermediate monocytes; TNFSF10 was expressed in intermediate monocytes, Th17 cells and Th2 cells; RPS6KN1 was not found to be expressed in the identified cell groups; HSPA5 was highly expressed in plasma blasts, Th17 cells and Vd2 gd T cells; RAF1 was expressed in plasma blasts; CASP1 was expressed in intermediate monocytes and Th17 cells (fig. 7B-fig. 7K).
Fig. 7: Expression of hub autophagy genes in the single cell transcriptome, (A): TSNE visualizes cell subgroups of the single cell transcriptome; (B):
Heat map of hub autophagy genes with expression in cell subgroups; (C): Violin plots of expression of hub autophagy genes ATF4; (D): CDKN1A;
(E): MAP1LC3B; (F): EIF4EBP1; (G): TNFSF10; (H): RPS6KB1; (I): HSPA5; (J): RAF1 and (K): CASP1 in cell subgroups
Note: ( ): Central memory CD8 T cells; (
): Central memory CD8 T cells; ( ): Intermediate monocytes; (
): Intermediate monocytes; ( ): Naive B cells; (
): Naive B cells; (  ): Naive CD4 T cells; (
): Naive CD4 T cells; (  ): Naive CD8 T cells;
(
): Naive CD8 T cells;
( ): NK cells; (
): NK cells; ( ): Non-Vd2 gd T cells; (
): Non-Vd2 gd T cells; ( ): Plasma blasts; (
): Plasma blasts; ( ): Switched memory B cells; (
): Switched memory B cells; (  ): Th17 cells; (
): Th17 cells; ( ): Th2 cells and (
): Th2 cells and ( ):
Vd2 gd T cells
):
Vd2 gd T cells
In summary, we have indicated the expression patterns of differentially expressed autophagy-related genes within distinct cell populations associated with HF at the single-cell level.
HF is a serious and terminal stage resulting from a spectrum of heart diseases; it is characterized by significant morbidity and mortality rates. The condition can induce fatigue, fluid overload (marked by occurrences like pulmonary edema or ankle edema), and lead to structural and functional abnormalities within the heart[2]. DCM is the 2nd most common cause of HF (after coronary artery disease), accounting for approximately 36 % of all HF cases, and has an estimated prevalence of >0.4 % in the general population[29]. However, there is no specific target and therapeutic mechanism for addressing this direction of the disease, and drug development is hampered, at least in part, by the lack of consensus on appropriate standards for HF.
Autophagy, which is the self-eating activity involved in the maintenance of cellular homeostasis, is currently a therapeutic target in several diseases, including HF[30]. The present study has provided evidence regarding the importance of autophagy in the failing hearts of patients with DCM. Of note, the quantification of autophagy vacuoles and the expression levels of cathepsin D emerged as independent predictors of Left Ventricular Reverse Remodeling (LVRR) in HF patients. These findings strongly advocate for the consideration of autophagy as a promising therapeutic target in the management of HF among patients with DCM. Leveraging data from the GEO database, we conducted an analysis to discern the transcriptional signature of autophagy genes in both the normal group and HF groups. Remarkably, RPS6KB1 emerged as a pivotal player in the interaction of autophagy genes, while ATF4 and CDKN1A exhibited significant negative correlations with several other genes. To gain further insights into the functional implications of differentially expressed autophagy genes, we performed GO and KEGG enrichment analysis. These analysis allowed us to predict the potential functions and pathways associated with the identified genes, indicating their roles in the context of HF and autophagy regulation.
Autophagy has been extensively studied as a foundation for understanding HF in various articles[5,31]. However, investigations from the standpoint of scRNA-seq analyses have been lacking, which has resulted in research content that is limited in scope and lacks a comprehensive systematic approach. Therefore, our study is specifically oriented towards filling this gap by concentrating on the collaborative insights derived from scRNA-seq analysis. The utility of scRNA-seq helps us to better understand HF and may bring promising prospects for clinical diagnosis and therapy. A growing body of studies has identified that scRNA-seq can be utilized to explore genetic alterations in HF. In this study, we performed a comprehensive analysis of singlecell genome datasets sourced from patients with HF that are available in the GEO database, thereby elucidating distinct cell subsets and their expression patterns of autophagy-related genes in the context of HF. Notably, our correlation analysis between hub autophagy genes and immune infiltration scores revealed significant positive associations between ATF4, CDKN1A, EIF4EBP1, and CASP1 and the identified immune infiltrating cells. Conversely, MAP1LC3B and RAF1 exhibited clear negative correlations with certain immune cell populations.
The previously published literature has highlighted the significance of the interplay between autophagy and the immune system. Given this recognition, there is a need for further exploration into the intricate relationship between autophagy and immune cells[32]. Both autophagy and immune cells exhibit a notable association with subtype A HF, thereby potentially establishing a connection between autophagy and immune cells. This integrated approach not only augments our comprehensive understanding of autophagy’s contribution within the HF framework but also suggests a plausible scenario where autophagy-related genes could impact the prognosis of HF by influencing immune-related cells. Further investigation allowed us to identify specific cell types that showed elevated expression of these hub autophagy genes. We identified high expression of ATF4 in intermediate monocytes and plasma blasts. A previous study has demonstrated that ATF4 expression in cardiomyocytes confers protection against HF through its anti-oxidative stress properties[33]. The presence of ATF4 in nonmyocardial cells and its involvement in autophagy could potentially introduce a novel perspective on its impact on HF prognosis. We notably detected substantial MAP1LC3B expression in NK cells, plasma blasts, switched memory B cells, Th17 cells, and Vd2 gd T cells. While prior research has elucidated its involvement in autophagy within renal cell carcinoma[34], its relevance to cardiovascular diseases remains unexplored. Therefore, the potential correlation between MAP1LC3B and autophagy in the context of HF holds promise for further investigation. The remaining autophagy-related hub genes identified through the single-cell method have not been previously linked to cardiovascular diseases. This study could potentially contribute new insights into the roles of these genes in HF. However, the absence of existing reports on these genes presents a challenge in further elucidating their underlying mechanisms.
In this study, we have observed significant transcriptional alterations between healthy controls and samples from individuals with DCM. Collectively, our findings point to a potential link between autophagy and HF within noncardiomyocyte populations using single-cell methodologies. These investigations also offer a foundation for delving into the intricate interplay between autophagy and HF. However, it is important to acknowledge the presence of several limitations in this study that warrant careful consideration. In our exploration of autophagy-related genes through single-cell strategies, it is noteworthy that the HF data exclusively originated from the GEO database. The augmentation of our finding’s robustness and applicability could be achieved through the integration of data from additional sources and experimental validations. Furthermore, considering the retrospective nature of this study, prospective investigations and experiments are imperative to validate the stability and precision of the identified gene signature in future clinical scenarios. While this study indicated the possible correlation between autophagy-related genes and HF, it is essential to delve into the underlying molecular mechanisms. A comprehensive exploration of the regulatory pathways and interactions involving these genes would provide a profound comprehension of their roles in the pathogenesis of HF. This, in turn, could uncover novel therapeutic targets, addressing the complexities of this multifaceted cardiovascular disorder.
We conducted an investigation into autophagyrelated genes using bioinformatics analysis to construct and validate the transcriptional characteristics of autophagy genes in HF. Furthermore, our study identified nine hub autophagyrelated genes associated with HF occurrence. We also detected the expression of these genes in cell subsets using single-cell strategies for prognostic recognition in HF and their potential immunotherapy response. For future research, it is significant to delve deeper into the underlying mechanisms through which the nine hub genes influence the immune microenvironment in HF. Such investigations will serve as a fundamental basis for the development of potential immunotherapeutic strategies aimed at effectively managing HF effectively.
Funding:
This work was supported by the National Science Foundation of China (No: 82170503).
Conflict of interests:
The authors declared no conflict of interests.
References
- Murphy SP, Ibrahim NE, Januzzi JL. Heart failure with reduced ejection fraction: A review. JAMA 2020;324(5):488-504.
[Crossref] [Google Scholar] [PubMed]
- Bueno H, Moura B, Lancellotti P, Bauersachs J. The year in cardiovascular medicine 2020: Heart failure and cardiomyopathies. Eur Heart J 2021;42(6):657-70.
[Crossref] [Google Scholar] [PubMed]
- Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res 2017;120(11):1812-24.
[Crossref] [Google Scholar] [PubMed]
- Liao M, Xie Q, Zhao Y, Yang C, Lin C, Wang G, et al. Main active components of Si-Miao-Yong-An decoction (SMYAD) attenuate autophagy and apoptosis via the PDE5A-AKT and TLR4-NOX4 pathways in isoproterenol (ISO)-induced heart failure models. Pharmacol Res 2022;176:106077.
[Crossref] [Google Scholar] [PubMed]
- Kanamori H, Yoshida A, Naruse G, Endo S, Minatoguchi S, Watanabe T, et al. Impact of autophagy on prognosis of patients with dilated cardiomyopathy. J Am Coll Cardiol 2022;79(8):789-801.
[Crossref] [Google Scholar] [PubMed]
- Tammaro A, Kers J, Scantlebery AM, Florquin S. Metabolic flexibility and innate immunity in renal ischemia reperfusion injury: The fine balance between adaptive repair and tissue degeneration. Front Immunol 2020;11:1346.
[Crossref] [Google Scholar] [PubMed]
- Muroya Y, He X, Fan L, Wang S, Xu R, Fan F, et al. Enhanced renal ischemia-reperfusion injury in aging and diabetes. Am J Physiol Renal Physiol 2018;315(6):F1843-54.
[Crossref] [Google Scholar] [PubMed]
- Hannenhalli S, Putt ME, Gilmore JM, Wang J, Parmacek MS, Epstein JA, et al. Transcriptional genomics associates FOX transcription factors with human heart failure. Circulation 2006;114(12):1269-76.
[Crossref] [Google Scholar] [PubMed]
- Cappuzzello C, Napolitano M, Arcelli D, Melillo G, Melchionna R, di Vito L, et al. Gene expression profiles in peripheral blood mononuclear cells of chronic heart failure patients. Physiol Genom 2009;38(3):233-40.
[Crossref] [Google Scholar] [PubMed]
- Muller C, Schillert A, Rothemeier C, Trégouet DA, Proust C, Binder H, et al. Removing batch effects from longitudinal gene expression-quantile normalization plus ComBat as best approach for microarray transcriptome data. PloS One 2016;11(6):e0156594.
[Crossref] [Google Scholar] [PubMed]
- Rao M, Wang X, Guo G, Wang L, Chen S, Yin P, et al. Resolving the intertwining of inflammation and fibrosis in human heart failure at single-cell level. Basic Res Cardiol 2021;116(1):55.
[Crossref] [Google Scholar] [PubMed]
- Wang NN, Dong J, Zhang L, Ouyang D, Cheng Y, Chen AF, et al. HAMdb: A database of human autophagy modulators with specific pathway and disease information. J Cheminform 2018;10(1):34.
[Crossref] [Google Scholar] [PubMed]
- von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, et al. STRING: Known and predicted protein–protein associations, integrated and transferred across organisms. Nucl Acids Res 2005;33(1):D433-7.
[Crossref] [Google Scholar] [PubMed]
- Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, et al. STRING 8-A global view on proteins and their functional interactions in 630 organisms. Nucl Acids Res 2009;37(1):D412-6.
[Crossref] [Google Scholar] [PubMed]
- Gene Ontology Consortium. Gene ontology consortium: Going forward. Nucl Acids Res 2015;43(D1):D1049-56.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acids Res 2000;28(1):27-30.
[Crossref] [Google Scholar] [PubMed]
- Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012;16(5):284-7.
[Crossref] [Google Scholar] [PubMed]
- Guerriero S, Pascual M, Ajossa S, Neri M, Musa E, Graupera B, et al. Artificial intelligence (AI) in the detection of rectosigmoid deep endometriosis. Eur J Obstet Gynecol Reprod Biol 2021;261:29-33.
[Crossref] [Google Scholar] [PubMed]
- Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med 2013;19(5):619-25.
[Crossref] [Google Scholar] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26(12):1572-3.
[Crossref] [Google Scholar] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 2005;102(43):15545-50.
[Crossref] [Google Scholar] [PubMed]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst 2015;1(6):417-25.
[Crossref] [Google Scholar] [PubMed]
- Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011;27(12):1739-40.
[Crossref] [Google Scholar] [PubMed]
- Jin Y, Wang Z, He D, Zhu Y, Chen X, Cao K. Identification of novel subtypes based on ssGSEA in immune-related prognostic signature for tongue squamous cell carcinoma. Cancer Med 2021;10(23):8693-707.
[Crossref] [Google Scholar] [PubMed]
- Hänzelmann S, Castelo R, Guinney J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:1-5.
[Crossref] [Google Scholar] [PubMed]
- Huang Q, Liu Y, Du Y, Garmire LX. Evaluation of cell type annotation R packages on single-cell RNA-seq data. Genom Proteom Bioinform 2021;19(2):267-81.
[Crossref] [Google Scholar] [PubMed]
- Pezzotti N, Lelieveldt BP, van Der Maaten L, Hollt T, Eisemann E, Vilanova A. Approximated and user steerable tSNE for progressive visual analytics. IEEE Trans Vis Comput Graph 2016;23(7):1739-52.
- Yao Z, Liu H, Xie F, Fischer S, Adkins RS, Aldridge AI, et al. A transcriptomic and epigenomic cell atlas of the mouse primary motor cortex. Nature 2021;598(7879):103-10.
- Redfield MM, Jacobsen SJ, Burnett JC, Mahoney DW, Bailey KR, Rodeheffer RJ. Burden of systolic and diastolic ventricular dysfunction in the community: Appreciating the scope of the heart failure epidemic. JAMA 2003;289(2):194-202.
[Crossref] [Google Scholar] [PubMed]
- Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. New Engl J Med 2013;368(7):651-62.
[Crossref] [Google Scholar] [PubMed]
- Vacek TP, Vacek JC, Tyagi N, Tyagi SC. Autophagy and heart failure: A possible role for homocysteine. Cell Biochem Biophys 2012;62:1-11.
[Crossref] [Google Scholar] [PubMed]
- Wang H, Zhang Y, Chen L, Liu Y, Xu C, Jiang D, et al. Identification of clinical prognostic features of esophageal cancer based on m6A regulators. Front Immunol 2022;13:950365.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Zhang G, Dasgupta S, Niewold EL, Li C, Li Q, et al. ATF4 protects the heart from failure by antagonizing oxidative stress. Circ Res 2022;131(1):91-105.
[Crossref] [Google Scholar] [PubMed]
- Kang HM, Noh KH, Chang TK, Park D, Cho HS, Lim JH, et al. Ubiquitination of MAP1LC3B by pVHL is associated with autophagy and cell death in renal cell carcinoma. Cell Death Dis 2019;10(4):279.
[Crossref] [Google Scholar] [PubMed]