- *Corresponding Author:
- V. R. Josyula
Cell and Molecular Biology lab, Department of Pharmaceutical Biotechnology, Manipal College of Pharmaceutical Sciences, Karnataka 576104, India
E-mail: rao.josyula@gmail.com
| Date of Received | 09 July 2019 |
| Date of Revised | 01 February 2020 |
| Date of Accepted | 14 February 2020 |
| Indian J Pharm Sci 2020;82(2):329-340 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Keywords
Small interfering ribonucleic acid, folic acid, chitosan, resistance, poly lactic-co-glycolic acid, cancer and nanoparticles
Cancer can be defined as a group of diseases characterized by uncontrolled cell growth leading to invasion of other parts of the body. The major hallmarks of cancer cells include self-sufficiency of growth signals, ignoring antigrowth signals, avoidance of apoptosis, unlimited replication, angiogenesis and invasion of nearby tissues[1]. It is a bewildering condition where the global burden is very high. Breast cancer is ranked second, next to lung cancer and leading cause for deaths among women. The incidence rate was about 11.6 %, which is still rising in most countries than the last decades as per Globocan, 2018. In India, women with breast cancer burden is about 0.15 million and rising year by year[2]. Drug resistance to chemotherapeutic agents could occur in many ways. Multidrug resistance is usually caused by few mechanisms, one of the very important mechanisms is energy driven process, where drugs are effluxed out from the cytosol by an important family of transporters known as ABC (ATP binding cassette) transporters. Resistance to various anticancer agents with different mechanisms and structures is a phenomenon called as Multidrug resistance. ABC transporters are expressed widely in most of the organs. They are classified into 7 sub families. P-glycoprotein, multidrug resistance associated protein-1 (MRP-1) and breast cancer resistance protein (BCRP) are the major proteins over expressed in breast cancer. Many small molecules were developed to inhibit or modulate drug resistance. Verapamil, the first generation inhibitor, did not qualify as a promising molecule due to high toxicity. Valspodar the second generation inhibitor, too was not considered due to inappropriate pharmacokinetics[3,4]. Upregulation of MRP-1, a 190kDa protein, encoded by the ABCC1 gene is associated with development of drug resistance in breast cancer. In tumor cells, MRP- 1 transports a variety of substrates that are linked to development of resistance. This protein is responsible for the transport of drugs like doxorubicin, methotrexate, vinblastine and paclitaxel. However, MRP-1 in normal cells acts as a detoxifying factor[5].
Considering the above, it is necessary to have a different approach to tackle multidrug resistant breast cancer through small interfering RNA (siRNA), known as RNA interference. It regulates the expression of the target gene by degrading mRNA. The expression of MRP-1 can be suppressed by siRNA, by degrading the mRNA, thus preventing the protein formation responsible for effluxing. Previous studies reported that dicer-based siRNA is highly functional in degrading the mRNA efficiently, producing at least 10-100 fold more potent activity than the traditional siRNA. Previous studies reported that the dicer-based siRNA is very functional and could lead to the highest knockdown of the target gene[6,7]. Dicer acts as a bona fide sensor in detecting thermodynamically less stable ends which help in loading the guide strand. Dicer is an important component in the mechanism but has no role in integrating siRNA with the RNA-induced silencing complex (RISC). This may lead to weak binding with the target mRNA[8].
In the current study, a dicer substrate siRNA was developed for improved knockdown potential. Naked siRNA is not stable and is degraded when it encounters serum nucleases. Therefore, the strategically designed dicer substrate siRNA was packaged in the folate functionalized chitosan-coated PLGA (Fol-Ch-PLGA) nanoparticles. The physicochemical characterization of the nanoparticles was studied using dynamic light scattering technique (DLS). These nanoparticles were used to deliver the siRNA exclusively to the cancer cells. Consistently the drug/dye accumulation studies, cytotoxicity assay along with expression studies were carried out to show improved efficiency of the therapy by knocking down the ABCC1 expression using nanoformulated dicer substrate siRNA.
Materials and Methods
Poly(D,L lactide co-glycolide), (Purasorb® PDLG 5002A, MW 17 kDa, acid terminated, lactic:glycolic acid ratio 50:50) was provided as a gift sample from Purac Biomaterials (Gorinchem, Netherland). Polyvinyl alcohol (PVA; 88 % hydrolyzed, MW 22000), dimethylsulfoxide (DMSO), dichloromethane (DCM), isopropanol, glacial acetic acid (Merck), chitosan (140-220 kDa ≥ 75% deacetylated), folic acid, EDC hydrochloride and N-hydroxysuccinimide (NHS, Sigma), Quant-It Picogreen reagent, Calcein-AM (Invitrogen), PBS (Himedia) and doxorubicin (Cayman Chemicals).
Cell lines and culture media:
MCF-7, a human breast cancer cell line was purchased from National centre for cell sciences (Pune, India). Cell line was maintained in DMEM medium containing sodium pyruvate with 10 % FBS and 50 μg/ml gentamycin sulfate and incubated at 37º in 5% CO2.
Design of siRNA:
Specific siRNA for the target gene ABCC1 is designed to prevent off-target effects. Designing the siRNA sequence for silencing the gene expression includes searching for various sequences of siRNA through computational tools and then selecting the most specific sequence considering the rules for designing siRNA[6,9].
Prediction of secondary structure:
Secondary structure and free energy (ΔG) of folding for the ABCC 1 dicer-based siRNA was predicted at 310.15 K (equivalent to 37º) using RNA structure web server[10].
Calculation of T\target-siRNA binding and determination heat capacity:
RNA structure web server was used to determine the binding between target mRNA and siRNA (guide strand) in a thermodynamic manner. Determination of ensemble heat capacity of the designed siRNA as function of temperature with melting temperature Tm (Cp) was performed using the DINAMelt web Server provided by RNA Institute, University of Albany, USA.
Development of a resistant cell line:
Doxorubicin was used to induce resistance in the MCF- 7 cells. The resistant cells were selected by stepwise selection method. Initially, the cells were grown in DMEM with high glucose, containing doxorubicin at a concentration of 5 nmol. When the cells were capable of growing and reaching appropriate confluency, the cells were passaged and the concentration of doxorubicin was doubled. The procedure was continued by doubling the concentration at each step to reach a final concentration of 320 nmol. MCF-7 cells were added with doxorubicin in different concentrations as specified above and incubated for 24 h, followed by transfer to the fresh medium without drug and then incubated for another 48 h at 37º and 5 % CO2 [11].
Formulation development:
The Fol-Ch-PLGA-ABCC1 siRNA nanoparticles were designed and developed in the following manner. One hundred milligrams of PLGA was dissolved in DCM. siRNA, previously solubilized in Tris EDTA buffer was added to the PLGA solution. Then the mixture was probe sonicated at 65 % amplitude for 10 min with a pulse of 2 s to form a primary emulsion. Later to the primary emulsion, 2 ml of previously synthesized folate tagged chitosan (FS-Ch) in PVA was added and the whole mixture was sonicated for another 2 min with a pulse of 2 sec to allow the formation of double emulsion. To the double emulsion, another 2 ml of PVA was added and stirred for 3 h to evaporate DCM. The nanoparticles were collected by centrifuging at 12 000 rpm at 4º for 20 min and then the pellet was washed with RNAase free water. The resulting mixture was collected and freeze dried. The tagging of folate with chitosan was via ɣ-carboxylic group of folate moiety to amino group of chitosan in the presence of EDC and NHS to allow formation of conjugate[12].
Characterization of FA-Ch and FA-Ch-coated PLGA-siRNA nanoparticles:
The formation of the conjugate was confirmed by FT-IR spectroscopy, X-Ray diffraction technique and differential scanning calorimetry. After the confirmation of the conjugate, the nanoparticles were prepared and characterized for size, PDI and zeta potential.
Determination of transfection efficiency:
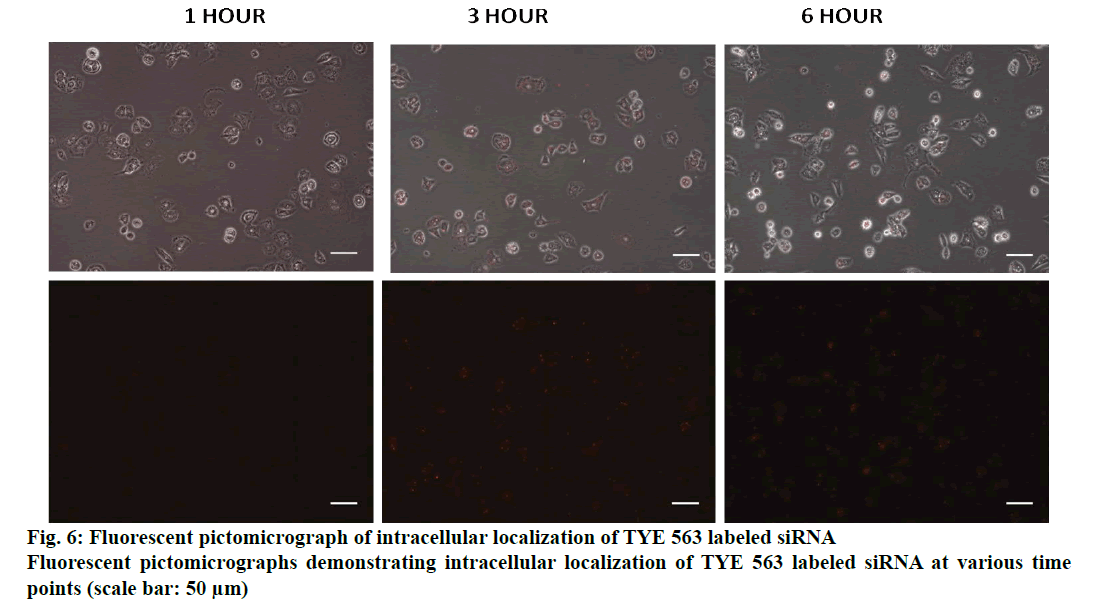
MCF-7 doxorubicin resistant cells were seeded in a 6-well plate and the nanoparticles loaded with TYE 563 labeled siRNA were used to transfect the cells. The plate was mixed by rocking back and forth and the cells were incubated for 24-72 h at 37° in a CO2 incubator. Qualitatively, the efficiency of the transfection was evaluated for 6 h under fluorescence microscopy[13].
Cellular uptake (calcein Am):
MCF 7 resistant cells were seeded (2×104 cells/well) in a 6-well plate and incubated for 24 h. The cells were pretreated with nanoformulated ABCC1 siRNA, which was then followed by incubation with florescent dye for 1 h at 37° and 5 % CO2, to evaluate the dye uptake and/or accumulation potential. Fluorescence intensity of the dye was evaluated using fluorescence microscopy[14].
Doxorubicin uptake/accumulation assay:
Cells (20×103cells/well) were seeded in 6-well culture plates and prior to the doxorubicin treatment, cells were pretreated with nanoformulated ABCC1 siRNA and incubated at 37 and 5 % CO2. After 24 h, 1 μg/ml concentration of doxorubicin was incubated in all the test groups except control and incubated for 1 hour. Then the cells were washed with PBS to remove non-internalized doxorubicin. It was then observed under Florescence microscopy[15].
Cytotoxicity determination:
To evaluate dose dependency of the cytotoxicity of doxorubicin, MCF-7 cells and MCF-7 doxorubicin resistant cells (10×103cells/well) were seeded in a 96-well plate in 100 μl of DMEM medium and incubated for 24 h prior to the test. Drug dilutions of different concentrations were prepared and added to the 96-well plate, and incubated for 48 h. Chemosensitivity was assessed using tetrazolium salt MTT assay. MTT was prepared in PBS pH 7.4 and added to each well. The plate was incubated for 4 h, and then 100 μl of DMSO was added to each well. The absorbance of each well was read on a microplate reader at a wavelength of 570 nm[16].
Cellular expression studies, PCR:
Total RNA of cells was extracted with the Trizol reagent at 48 h after transfection. cDNA was generated by reverse transcription of 2 μg of total RNA using superscript first strand synthesis kit in a total reaction volume of 20 μl according to the manufacturer’s instructions. The sequences of forward and reverse oligonucleotide primers, specific to ABCC1 and housekeeping genes, were quantified. Real-time quantitative PCR (RT qPCR) was performed using Quantstudio-5 Real Time System. A 10-fold dilution of each cDNA was amplified in 20-μl volume, using the Power SYBR green with reference dye ROX. PCR cycle conditions were; hold stage 50° for 2 min and 95° for 10 min and at PCR stage 40 cycles of 95° for 15 s and 60° for 60 s. For all the target genes, the mRNA levels were normalized to β2M mRNA levels[15].
Western blot:
After the treatment, cells were washed with PBS and lysed on ice using RIPA buffer plus protease inhibitor cocktail to collect total cellular proteins. Concentration of the whole protein was estimated using Bradford method. Equal amounts (20-30 μg) of protein was loaded and electrophoresed on 12% SDS-Polyacrylamide gel and transblotted on to PVDF membrane. The membrane was blocked in PBS containing 5% non fat dry milk and Tween 20 for an hour. Then the membrane was incubated with anti MRP1 and β actin antibody at 4º overnight. Then membranes were washed with PBST, followed by incubation with horse radish peroxidase conjugated antimouse Ig-G secondary antibody for 1 h. Later it was washed thoroughly with PBST and the bands were visualized using a chemiluminescent substrate ECL in UVI-TEC system[17].
In vivo toxicity of the formulation:
Approval from the Institutional animal ethics committee (IAEC no: IAEC/KMC/09/2019) was obtained to conduct experiments on animals for the determination of acute toxicity of the formulations. This study was conducted to determine the initial dose required for the main study. siRNA nanoparticles were administered to single animal in a sequential manner from Dose FIRST to Dose LAST.
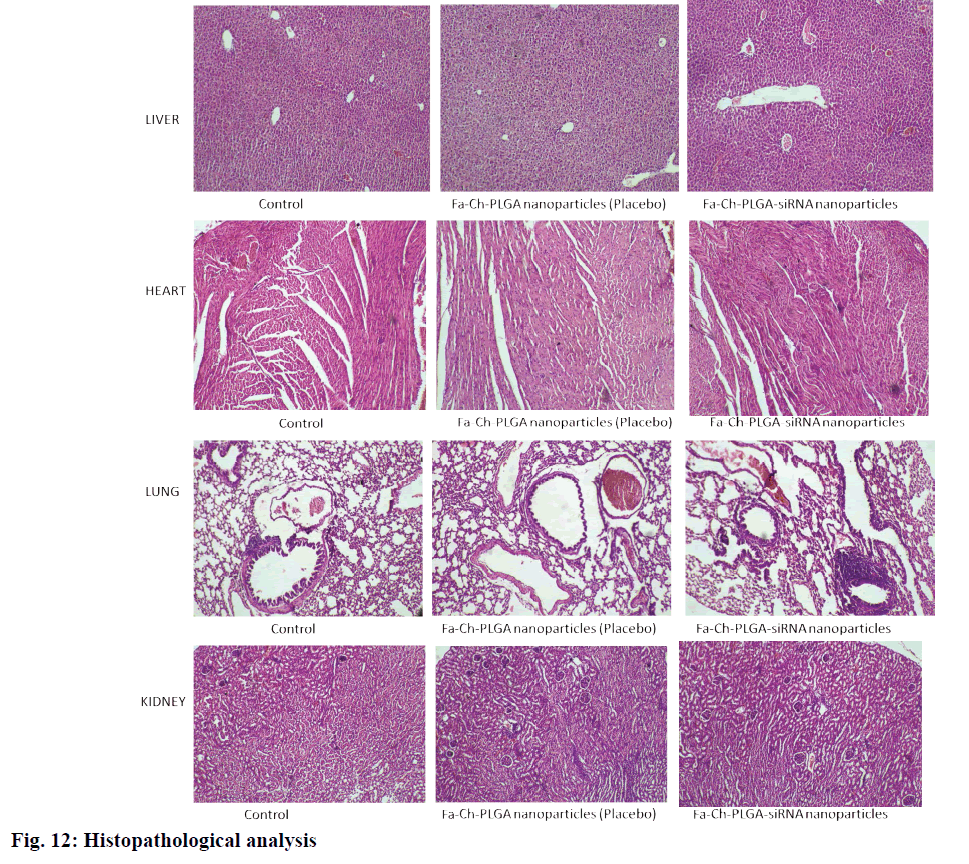
The siRNA nanoparticles were administered as a single intravenous dose through the tail vein. Animals were fasted 3-4 h prior to dosing. After completion of the fasting time, the animals were marked, grouped and then dosed. Subsequently the animals were fasted for another 1-2 h. The last dose from sighting study was considered as the initial dose in main study (siRNA 0.75 mg/kg) and (polymeric NP 150 mg/kg as placebo). A break of 24 h is given between dosing each animal. If the animal survived, then the next animal receives the higher dose. If the animal dies or appears moribund (state of dying) then it receives a lower dose. Animals were weighed before initiating the study and after completion of the study. Then all the animals were caged and observed for 14 d for any kind of toxic effects. All the animals were euthanized after the completion of the study. Visceral organs were collected to evaluate probable toxicological changes in the tissues. Organs were preserved in 10% formalin until histopathological studies were performed.
Statistical analysis:
Statistical analysis was performed using GraphPad Prism ANOVA. A value of P˂0.05 is considered as significant.
Results and Discussion
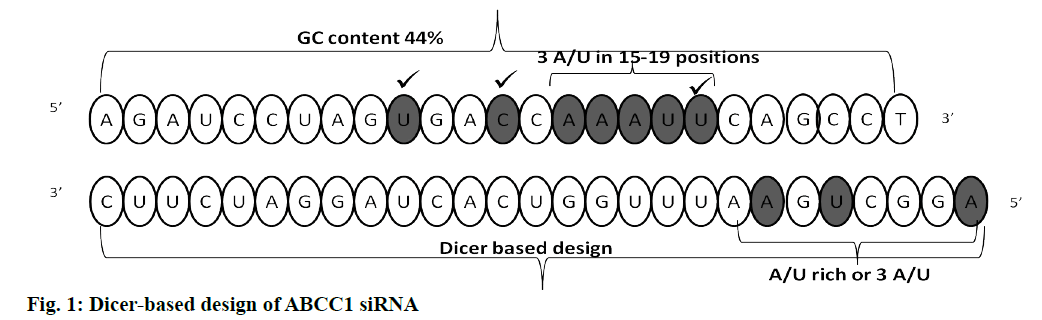
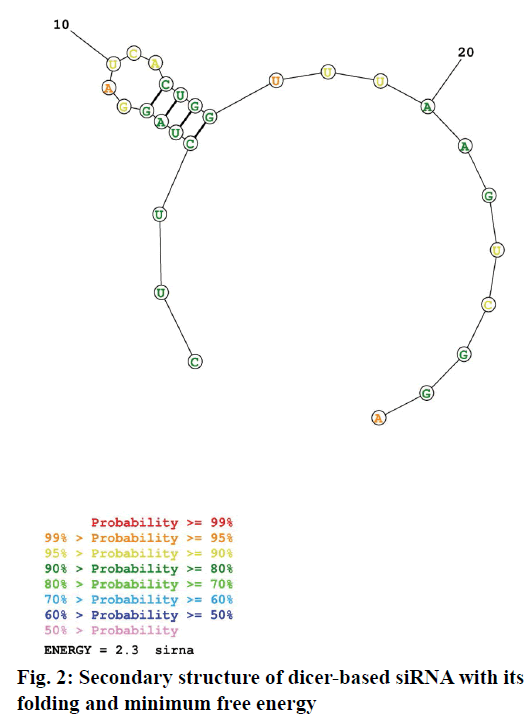
Dicer substrate siRNA was designed (27mer RNA duplex) using IDT software. For the uptake of the guide strand by RISC, maintaining a less stable 5’ end is critical. Asymmetric designing of siRNA by generating a 2 nt on 3’ overhang for an antisense strand can be effective for dicing. While designing siRNA, the following rules were considered; GC content, thermodynamic parameters, usage of appropriate bases and avoidance of some bases at specific positions. Especially inclusion of U at 10th position of sense strand, absence of G at 13th position of sense strand, inclusion of U at 19th position of sense strand, maintenance of A/U richness in 15-19 of sense strand, similarly on 5’ end of anti sense strand and satisfying the rule of GC content (44 %). Finally BLAST analysis was done to rule out off target binding. Fig. 1 depicts the siRNA sequence generated and it was synthesized by IDT. The folding pattern led to minimum free energy which is useful in determination of secondary structure of dicer substrate siRNA and was found to be 2.3. The predicted secondary structure was depicted in fig. 2.
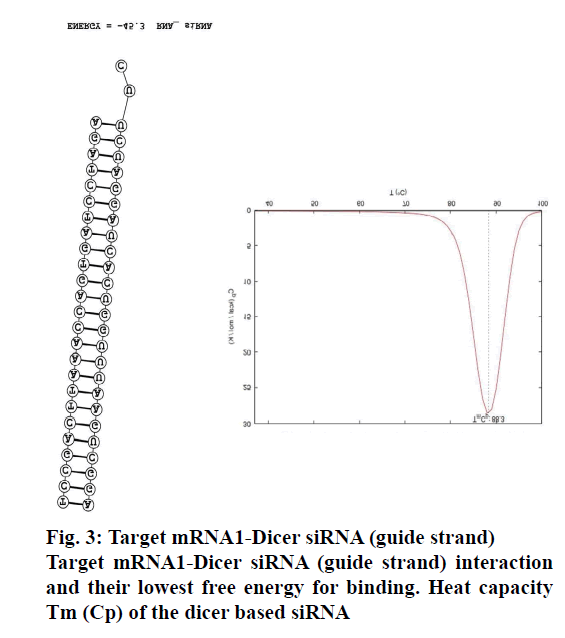
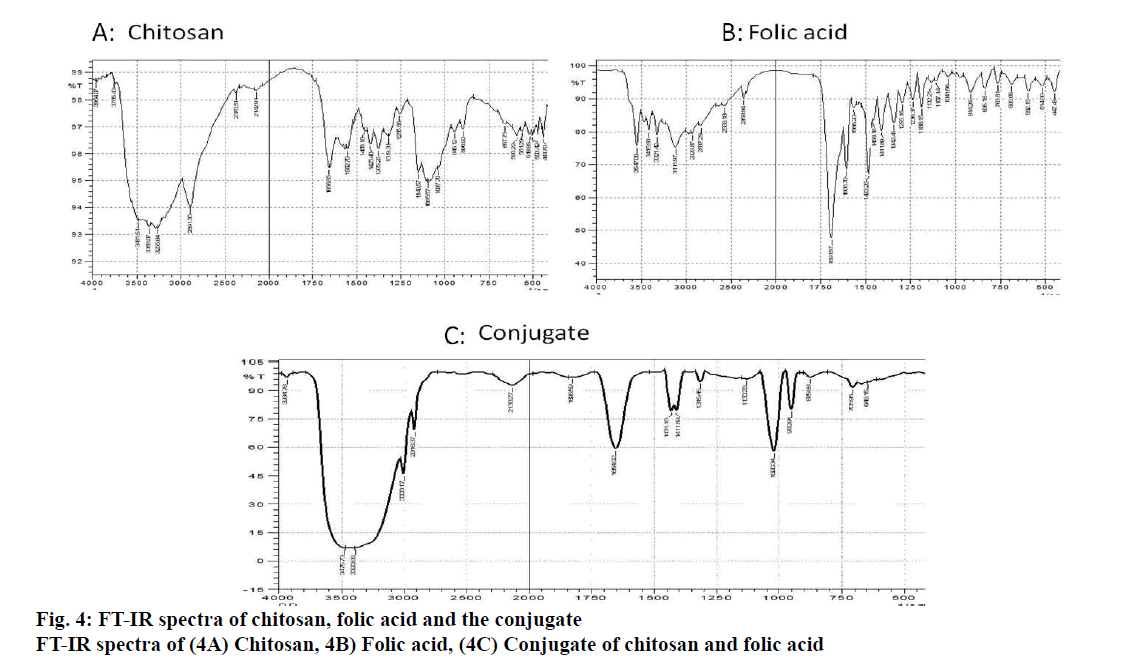
The binding of the designed siRNA with the target mRNA was represented in fig. 3 and the free energy was mentioned. The binding energy was -45.3. Heat capacity Tm (Cp), a function of temperature and concentration, was observed to be 88.3º. FA-Ch conjugate was synthesized and characterized with FTIR and XRD. From FTIR, it could be inferred that there are spectral intensity variations in the amide region (1654-1657). Also, there are multiple differences in the 3500-2800 region when pure chitosan compared with the conjugate indicating that conjugation must have occurred between FA-Ch. The intensity of the signals coming from the conjugate was masked due to the resonance of protons of the chitosan in XRD. Though there were no strong signals, it could be deduced that the conjugation of ɣ-carboxylic group of folate moiety to amino group of chitosan occurred due to steric hindrances and the reaction conditions. Further, the nanoparticles were characterized for their size and zeta potential. The zeta potential of nanoparticles was +11.57±5.9 mV, PDI was 0.247±0.085. The particle size was observed using DLS technique and was found to be 246.83±39.13 nm. Qualitative determination of transfection efficiency of TYE 563 labeled siRNA in the doxorubicin-resistant MCF-7 cells were measured up to 6 h. From fig. 4, it was observed that in the first hout, a very less amount of TYE 563 labeled siRNA got transfected compared to third hour and then sixth h.
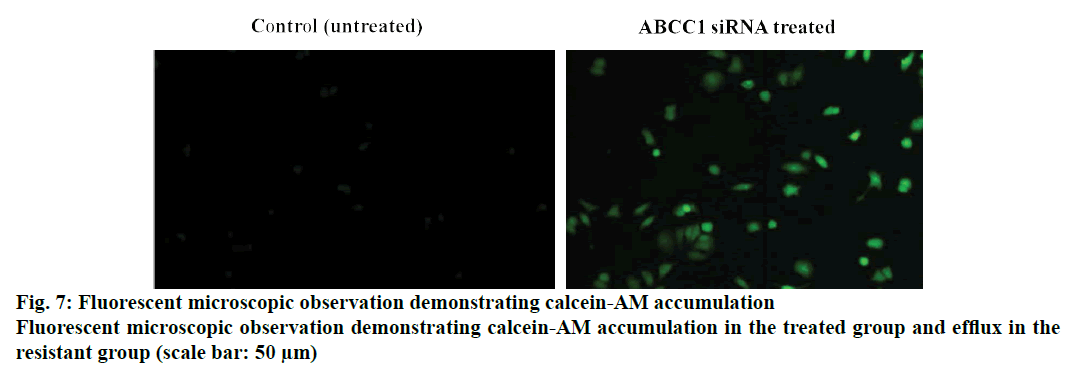
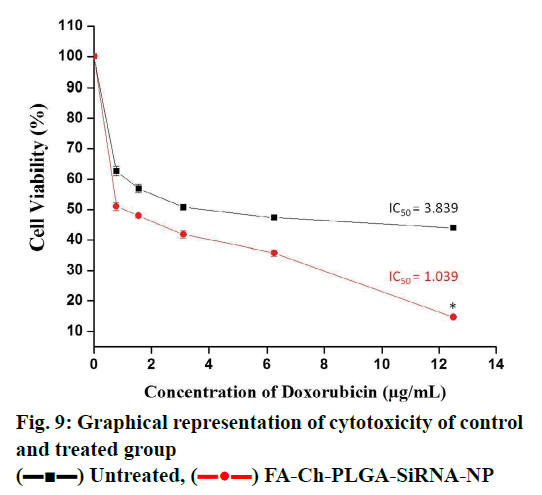
Cellular uptake or dye accumulation in the cells is a direct correlation of the retention capacity of the dye in the cells. The florescence of the green dye calcein was qualitatively measured under the florescent microscope in untreated and treated resistant cell groups. In control untreated group, there was a minimal amount of observed florescence as it remains untreated but in the cells treated with siRNA that targets ABCC1, an intense green florescence of calcein-AM was observed indicating the efficiency/knockdown potential of the ABCC1 siRNA (figs. 5 and 6). The retention capacity indicating the amount of florescence in untreated and treated groups was depicted in fig. 7. Doxorubicin uptake or dye accumulation of the cells is a direct correlation of the retention capacity of doxorubicin in the cells. The red florescence produced by doxorubicin was qualitatively measured under the microscope in untreated and treated resistant cell groups. In the untreated group, insignificant amount of florescence was observed but in the group treated with siRNA that targets ABCC1, the red florescence of doxorubicin was observed as shown in the fig. 8. This signified the reduction of ABCC1efflux of doxorubicin. The impact of ABCC1 knockdown was gauged in doxorubicinresistant MCF-7 cells. This phenomenon, which was produced by the dicer-based siRNA, reduced the IC50 of doxorubicin from 7.04 μmol to 1.89 μmol. Therefore, by downregulating ABCC1 mRNA expression using nanoformulated siRNA, it is clearly evident that the resistant breast cancer cells became sensitive to the effect of doxorubicin as shown in figs. 7 and 8.
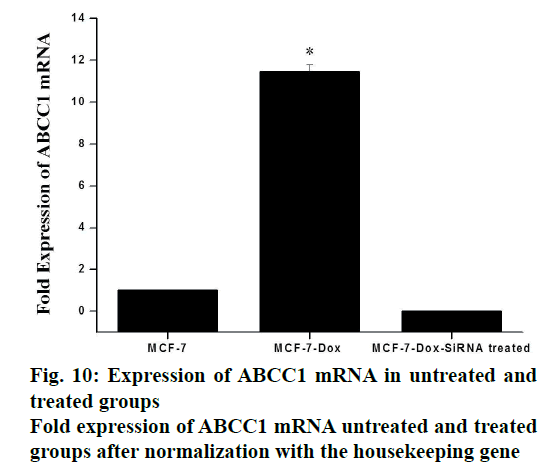
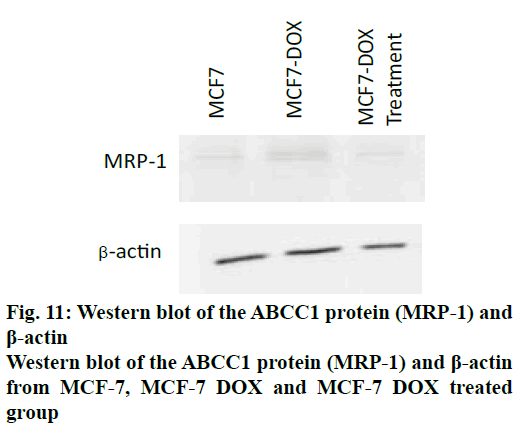
The cellular expression was quantified using gene expression studies with RT qPCR and protein expression studies by Western blot. The expression profile of the gene ABCC1 was normalized with β2M (housekeeping gene) in MCF-7. The primer sequences for ABCC1 gene (sense strand) 5’-GTGTTTCTGGTCAGCCCAACT-3’ and (antisense strand) 5’-TTGGATCTCAGGATGGCTAGG-3’ and β2M (sense) 5’-GTCTTTCAGCAAGGACTGGTC 3’ and (antisense) 5’-CAAATGCGGCATCTTCAAACC-3’. SiRNA, targeting ABCC1 gene, was found to be very effective in down regulating the expression of ABCC1 gene. It was observed that the expression levels of ABCC1 gene in resistant cells were increased by 11.44 fold, compared to the normal cells (figs. 9 and 10). But in the presence of siRNA, 0.01 fold expression is observed. which was insignificant. It appeared that the potency of siRNA was very high such that the expression was reduced by greater than 95 %. A graphical representation of ABCC1 mRNA expression was shown in fig. 11). Western blot was done to study the protein expression levels. Three groups of cells, MCF-7, doxorubicin-resistant MCF-7 cells without siRNA treatment (control group) and with siRNA treatment (treatment group) were used for this study. The results, depicted in fig. 12, indicated that the protein corresponding to ABCC1 gene was expressed in significant amounts by the control group of cells, whereas it was insignificant with respect to normal and treatment group of cells. Protein corresponding to β-actin housekeeping gene was expressed at same level by all groups of cells.
Although, the cancer therapeutics has undergone significant advancements, there is still a challenge to improve the patient survival due to the development of MDR. MDR is a phenomenon where in the tumors are responsive to the drugs in the initial stages, but gain resistance with time. However, 80 % of the breast tumors are said to be sensitive in their initial stages. These tumors do not respond and develop crossresistance to structurally and functionally unrelated compounds leading to ineffectiveness of the drug. This hurdle is majorly a result of elevated expression levels of ABC pumps/transporters, causing efflux of the drug. Several studies evaluated the expression of these efflux pumps and suggested that P-gp encoded by ABCB1and MRP1 encoded by ABCC1 genes are mainly responsible for the efflux of drugs. In this regard, understanding and coming up with new strategies like RNAi could overcome the problem of resistance[18,19]. In the present study, ABCC1-mediated resistance in doxorubicin-resistant MCF-7 cells were used to observe the upregulation of ABCC1 through qPCR experiment and the same was evidenced by decreased intracellular concentration of calcein-AM and doxorubicin through calcein-AM and doxorubicin accumulation assays. Therefore, an siRNA strategy was devised, to specifically target and down-regulat the overexpressing ABCC1 gene in order to sensitize the drug resistant cancer cells to doxorubicin.
To design the siRNA that specifically targets the desired gene of interest without leading any off-target effects, a strategic way of designing was followed. Sufficient number of studies experimentally proved that the impact of potency and functionality of siRNA were significantly improved, if a dicer substrate siRNA was used[6]. Therefore, this study was designed to develop a dicer substrate siRNA (27mer siRNA) that efficiently targets and produces a strong knock down potential with no off target effects. To design the dicer substrate siRNA that targets ABCC1, mRNA sequence of the gene was retrieved from NCBI database and that was used to generate siRNA specific to the gene of interest. Another key criterion is the generation of highly potent 27mer siRNA with the aid of the IDT siRNA designing tool, which also satisfies the basic criteria of presence/ absence of specific nucleotides at specific positions. Including A or U at 3rd, 10th and 19th of sense strand and excluding G at 13th and 19th of sense strand might have increased the functionality of siRNA to a great extent[20]. In this regard, the presence of U nucleotide at 10th position is helpful in cleaving the mRNA by the activated RISC. The GC content is considered to be first important feature as per Reynolds rule, siRNA with a GC content of 30-52% is said to be potent and functional. We considered it as important parameter that can influence the functionality. Therefore the GC content of our designed siRNA was 44%. We also maintained asymmetric ends (where 27 nucleotides in the guide strand and 25 nucleotides in the passenger strand) keeping 2 nucleotides on 3’ overhangs of antisense strand to make the siRNA more potent, as this terminal end structure helps in proper loading of siRNA into RISC. The presence of 2 nucleotides on 3’ overhangs of antisense strand makes the site as an excellent site for the RISC processing to initiate while the blunt end gives asymmetry[9]. This approach also serves as an advantage in preventing the consideration of passenger strand as guide due to its thermodynamically unstable ends. In this effort of producing a highly functional siRNA, features that facilitate the guide strand selection by the RISC along with an additional parameter to predict the secondary structure of siRNA was also employed, as these factors impact potency and stability[10]. In this regard, minimum free energy (ΔG) of the siRNA secondary structure was the determinant, and the folding energy for the designed siRNA was 2.3 at 37.0º. Off targeting is a major hurdle in generating a successful siRNA candidate. If the designed siRNA is not precise enough, it can cross match with multiple genes and may have higher chances to induce the off target effects. Therefore BLAST was performed to evaluate cross match/homology match, the result showed no off target match. Another determinant of efficacy of the knockdown depends on the interaction of siRNA with target mRNA. It is predicted using a tool Bifold, where it hybridizes target mRNA and siRNA to determine the lowest binding free energy[10,21]. In our study, the free energy with our designed siRNA was -45.3, which is strong enough to bind with target mRNA, thereby preventing translation as depicted in fig. 3. Heat capacity (Cp), as a function of temperature and concentration, is used to determine melting temperature (Tm) of the hybridized nucleic acids. siRNA, with higher Tm, can be more functional. As in our study also, the Tm (Cp) was observed to be ˃85º.
Even after designing a successful siRNA candidate, another key element has to be resolved i.e. delivery of the siRNA using a suitable carrier. Delivery of the siRNA with high specificity to targeted location and keeping the siRNA intact by preventing the degradation mediated by nucleases is a major obstacle. Also the designed dicer based siRNA is highly polyanionic and will not diffuse in the cell membranes easily. Naked siRNA may not produce sufficient knockdown potential yet causing some toxic issues at higher doses[22]. To address these critical issues like specificity and stability, we utilized polymeric substances that are considered to be biologically and genomically compatible without having a compromise on specificity aspect. PLGA, a polymer that is approved by FDA for its use in delivery[23] and another coating polymer, chitosan were used in this study. Chitosan coating improves the surface charge properties by modifying the overall charge of the particles. Usage of chitosan can also have a favorable impact in the incorporation of negatively charged siRNA[24]. Using only PLGA has a problem as the negative charge influences the drug leakage and affects the activity of the drug incorporated. To overcome this problem, chitosan coating is used. Folate has a specific advantage of targeting the cancer cells, as folic acid expression is very high on the cancer cells and in contrast an insignificant level of expression on normal cells[25]. In the present study, a strategy was devised to deliver the designed siRNA in an active targeting manner, using folic acid. Therefore, functionalizing the nanoparticles with folate can target cancer cells with exclusivity. Improved transfection efficiency is the major determinant of gene silencing potential. PLGA is considered to have poor transfection efficiency for siRNA. Therefore, layering the PLGA nanoparticles with cationic polymer can greatly improve the transfection efficiency. Considering this, chitosan was used to layer over the PLGA nanoparticles and folate as targeting entity[26]. The transfection efficiency was qualitatively determined for over a period of 6 h using a TYE 563-labeled siRNA to ensure that maximum amount of siRNA was delivered using this formulation. Then, the similar pattern was used to develop a formulation for ABCC1 siRNA. The mean particle size of the developed formulation was 246.83±39.13 nm and the zeta potential was observed to be 11.57±5.9mV. The Zeta potential of the coated formulation was found to be more towards a positive value, which implied that chitosan was layered on the negatively charged PLGA core due to its electrostatic attraction and a positively charged nanoparticle is regarded to be stable. The size of the nanoparticles can influence the intracellular uptake of the nanoparticles; there is a slight increase in the size of the nanoparticles due to functionalization of folate moiety and it is in accordance with PDI, producing a mono-disperse particles[12]. The unprotected or unmodified siRNA delivered through bloodstream is prone to degradation by nucleases. Hickerson et al. performed degradation studies under various conditions and found that siRNA is neither degraded at incubating in high temperatures nor after repeated freeze/thaw cycles but it is immediately degraded in serum[27]. Similar pattern resulted in the current study where naked siRNA underwent instantaneous degradation but the folate functionalized nanoparticles resisted degradation by serum nuclease. This nanoformulated protected ABCC1 siRNA, appeared to have multiple layers of polymer protection. This elevated expression of ABCC1, as depicted in doxorubicin accumulation assay, led to the efflux of the doxorubicin resulting in its diminished efficiency. In the siRNA-treated group, doxorubicin efflux was decreased thereby making the resistant cells sensitive to doxorubicin as shown in fig. 7. A similar phenomenon was noticed with calcein- AM accumulation (fig. 8).
The knockdown potential of the designed dicer-based siRNA depended on the ability to silence the expression of ABCC1 mRNA that blocks the expression of protein levels. In the present study, the designed dicer-based siRNA produced a knockdown potential ˃95%. This is due to the dicer-induced knockdown. The dicer is a large protein with multiple domains like Piwi, Argonaute, and Zwille domains (PAZ), dsRNA binding domains (RBD) and RNA-III domains. The RNA-III like domains serve as an excellent site for the cleavage of the dicer based siRNA confers to the generation of 21-23 nucleotide sequence. The exogenously entered 27mer siRNA initiates the silencing by endogenous RNAi pathway[28,29]. The 27mer siRNA serves as a substrate for the dicer, where it integrates along with other binding proteins and introduces 27mer siRNA into RISC in proper orientation that is essential for strong knockdown potential[9]. Also, as observed in many other studies, dicer-based siRNA is 10-100 times more potent than traditional siRNA[6,7]. An additional advantage with this design is that the presence of 2 nt on 3’ overhang of guide strand produces homogenous dicer products rather than blunt 27mer siRNA which reduces the off targeting[22,29].
Nano formulations (placebo and siRNA) were administered intravenously through the tail vein. A maximum concentration of 0.75 mg/kg ABCC1 siRNA was used. Further increase in dose was not attempted, as this would be sufficient to attain therapeutic efficacy. As per the histopathological studies and observations from the study, shown in fig. 12, the formulation did not show any signs or toxic symptoms or mortality even at a very high polymer concentration (polymeric NP 300 mg/kg).
In this study, all essential rules were followed for designing an efficient dicer substrate siRNA with better knockdown efficiency. The folate functionalized nanoparticles delivered the siRNA exclusively to the cancer cells. The designed siRNA could target ABCC1, significantly reduced its expression, sensitized the resistant cells to doxorubicin and improved the cytotoxic effect. Thus the results demonstrated that the dicer-based design could be a promising approach in improving the specificity and functionality of siRNA.
Acknowledgements
The authors acknowledge Nano Mission Council of the Department of Science and Technology, the Govt. of India, New Delhi (SR/NM/NS-1090/2012 (G)) for funding the project, the authorities of the Manipal Academy of Higher Education (MAHE) for providing all facilities, Daphne and G. Karthik for their help in this study.
References
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell 2000;100(1):57-70.
- Mallath MK, Taylor DG, Badwe RA, Rath GK, Shanta V, Pramesh C, et al. The growing burden of cancer in India: epidemiology and social context. Lancet Oncol 2014;15(6):e205-e12.
- Li YT, Chua MJ, Kunnath AP, Chowdhury EH. Reversing multidrug resistance in breast cancer cells by silencing ABC transporter genes with nanoparticle-facilitated delivery of target siRNAs. Int J Nanomed 2012;7:2473.
- Ughachukwu P, Unekwe P. Efflux Pump. Mediated Resistance in Chemotherapy. Annals Med Health Sci Res 2013;2(2):191-8.
- Wind N, Holen I. Multidrug resistance in breast cancer: from in vitro models to clinical studies. Int J Breast Cancer 2011;2011.
- Kim DH, Behlke MA, Rose SD, Chang MS, Choi S, Rossi JJ. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat Biotechnol 2005;23(2):222-6. Epub 2004/12/28.
- Dudek H, Wong DH, Arvan R, Shah A, Wortham K, Ying B, et al. Knockdown of β-catenin with dicer-substrate siRNAs reduces liver tumor burden in vivo. Mol Ther 2014;22(1):92-101.
- Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell 2011;43(1):110-21.
- Gatta AK, Hariharapura RC, Udupa N, Reddy MS, Josyula VR. Strategies for improving the specificity of siRNAs for enhanced therapeutic potential. Expert Opin Drug Discov 2018;13(8):709-25.
- Sharif Shohan MU, Paul A, Hossain M. Computational design of potential siRNA molecules for silencing nucleoprotein gene of rabies virus. Future Virol 2018;13(3):159-70.
- Kars MD, İşeri ÖD, Gündüz U, Ural AU, Arpaci F, Molnar J. Development of rational in vitro models for drug resistance in breast cancer and modulation of MDR by selected compounds. Anticancer Research 2006;26(6B):4559-68.
- Ji J, Zuo P, Wang Y-L. Enhanced antiproliferative effect of carboplatin in cervical cancer cells utilizing folate-grafted polymeric nanoparticles. Nanoscale Res Lett 2015;10(1):453.
- Feldmann DP, Cheng Y, Kandil R, Xie Y, Mohammadi M, Harz H, et al. In vitro and in vivo delivery of siRNA via VIPER polymer system to lung cells. J Control Release. 2018;276:50-8.
- Dogan AL, Legrand O, Faussat A-M, Perrot J-Y, Marie J-P. Evaluation and comparison of MRP1 activity with three fluorescent dyes and three modulators in leukemic cell lines. Leukemia Res 2004;28(6):619-22.
- Dönmez Y, Akhmetova L, İşeri ÖD, Kars MD, Gündüz U. Effect of MDR modulators verapamil and promethazine on gene expression levels of MDR1 and MRP1 in doxorubicin-resistant MCF-7 cells. Cancer Chemother Pharmacol 2011;67(4):823-8.
- Tabatabaei-Mirakabad FS, Akbarzadeh A, Milani M, Zarghami N, Taheri-Anganeh M, Zeighamian V, et al. A Comparison between the cytotoxic effects of pure curcumin and curcumin-loaded PLGA-PEG nanoparticles on the MCF-7 human breast cancer cell line. Artificial cells, Nanomed Biotechnol 2016;44(1):423-30.
- Kang KW, Chun M-K, Kim O, Subedi RK, Ahn S-G, Yoon J-H, et al. Doxorubicin-loaded solid lipid nanoparticles to overcome multidrug resistance in cancer therapy. Nanomed Nanotechnol Biol Med 2010;6(2):210-3.
- Kim DW, Kim K-O, Shin MJ, Ha JH, Seo SW, Yang J, et al. siRNA-based targeting of antiapoptotic genes can reverse chemoresistance in P-glycoprotein expressing chondrosarcoma cells. Mol Cancer 2009;8(1):28.
- Chang G. Multidrug resistance ABC transporters. FEBS Lett 2003;555(1):102-5.
- Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol 2004;22(3):326-30.
- Shawan M, Hossain MM, Hasan MA, Hasan MM, Parvin A, Akter S, et al. Design and prediction of potential RNAi (siRNA) molecules for 3'UTR PTGS of different strains of zika virus: a computational approach. Nat Sci 2015;13(2):37-50.
- Bégin-Lavallée V, Midavaine É, Dansereau M-A, Tétreault P, Longpré J-M, Jacobi AM, et al. Functional inhibition of chemokine receptor CCR2 by dicer-substrate-siRNA prevents pain development. Mol Pain 2016;12:1744806916653969.
- Danhier F, Ansorena E, Silva JM, Coco R, Le Breton A, Préat V. PLGA-based nanoparticles: an overview of biomedical applications. J Control Release 2012;161(2):505-22.
- Nafee N, Taetz S, Schneider M, Schaefer UF, Lehr C-M. Chitosan-coated PLGA nanoparticles for DNA/RNA delivery: effect of the formulation parameters on complexation and transfection of antisense oligonucleotides. Nanomed Nanotechnol, Biol Med 2007;3(3):173-83.
- Cheng L, Ma H, Shao M, Fan Q, Lv H, Peng J, et al. Synthesis of folate‑chitosan nanoparticles loaded with ligustrazine to target folate receptor positive cancer cells. Mol Med Rep 2017;16(2):1101-8.
- Chen J, Tian B, Yin X, Zhang Y, Hu D, Hu Z, et al. Preparation, characterization and transfection efficiency of cationic PEGylated PLA nanoparticles as gene delivery systems. J Biotechnol 2007;130(2):107-13.
- Hickerson RP, Vlassov AV, Wang Q, Leake D, Ilves H, Gonzalez-Gonzalez E, et al. Stability study of unmodified siRNA and relevance to clinical use. Oligonucleotides 2008;18(4):345-54.
- Song M-S, Rossi JJ. Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem J 2017;474(10):1603-18.
- Sarret P, Doré-Savard L, Tétreault P, Bégin-Lavallée V, Beaudet N. Application of dicer-substrate siRNA in pain research. RNA technologies and their applications: Springer;2010. p. 161-90.

 ) FA-Ch-PLGA-SiRNA-NP
) FA-Ch-PLGA-SiRNA-NP