- *Corresponding Author:
- Bharathi Sharanappa Veerapur
Department of Chemistry, Vijayanagara Sri Krishnadevaraya University, Vinayaka Nagara, Ballari, Karnataka 583105, India
E-mail: veerapurbharati@gmail.com
| Date of Received | 13 July 2021 |
| Date of Revision | 27 July 2023 |
| Date of Accepted | 02 January 2024 |
| Indian J Pharm Sci 2024;86(1):253-265 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In this investigation, a novel group of compounds, 2-mercapto-benzofuro-pyrimidine derivatives, were synthesized from 2-hydroxy-5-nitro-1-benzonitrile (1) as the starting material. This was treated with various haloketones to provide the corresponding 1-(3-amino-5-nitro-1-benzofuran-2-yl) ethan-1-one (2a-d), which was then treated with ammonium thiocyanate and acetyl chloride to yield N-[(2-acetyl-5-nitro-1-benzofuran-yl) carbamothioyl]acetamide (3a-d). Further, it was reflux with sodium hydroxide yields 4-methyl/aryl-8-nitro[1] benzofuro[3,2-d]pyrimidine-2-thiol(4a-d). To obtain the appropriate acids, continue stirring compounds (4a d) with chloroacetic acid to obtain [(4-methyl-8-nitro[1]benzofuro[3,2-d]pyrimidin-2-yl)sulfanyl]acetic acid (5a-d). The structures of all the newly synthesized compounds have been established by spectral studies such as Fourier-transform infrared, Proton nuclear magnetic resonance and Mass spectra. Furthermore, all the new compounds were screened antibacterial and antifungal activity against human pathogenic viz. gram +Ve and gram -Ve organisms by agar well diffusion method. In addition, molecular docking was performed for all synthesized compounds using high inhibition organisms selected protein of both genetic and non-genetic algorithm techniques, where result are demonstrate in an excellent docking energy.
Keywords
Benzofuran, pyrimidine, in vitro, antibacterial, molecular docking
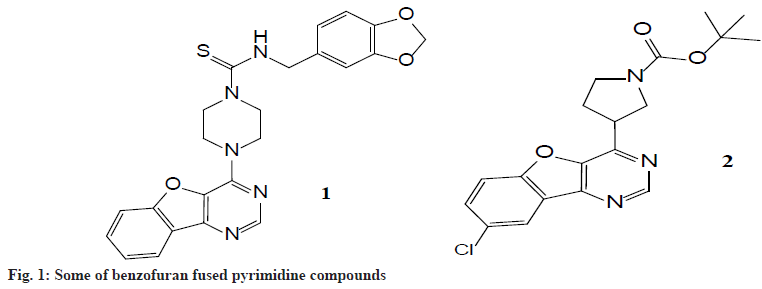
Nitrogen, oxygen and sulfur-containing heterocyclic compounds have received considerable attention due to their wide range of pharmacological applications. Pyrimidine-based heterocyclic compounds were of interest as potential bioactive molecules and exhibit analgesic, antifungal, antibacterial, and antiinfametry activities[1-3]. These are also associated with nucleic acid, antibiotics, antimalarial, and anticancer drugs. Many of the pyrimidine derivatives are reported to possess potential CNS depressant properties[4-6]. Moreover benzofurans heterocycles occur in a great number of natural products. Many natural benzofurans have physiological, pharmacological and toxic properties. There are well-known natural products having related benzofuran ring structures, which are particularly isolated from Machilus glaucescent, Ophryosporus charge, Ophryosporus Lorentz, Krameria ramosissima and Zanthoxylum ailanthoidol. The most recognized benzofurans are ailanthoidol, amiodarone, and bufuralol compounds. Ailanthoidol, a neolignan with a 2-aryl benzofuran skeleton, has been reported to possess a variety of biological activities[7-9] such as anticancer, antiviral, immunosuppressive, antioxidant, antifungal, and antifeedant activities. Amiodarone is a highly effective antiarrhythmic agent with class III activity. The benzofuran associated with pyrimidine derivatives has been shown to exhibit fluorescent sensors, oxidants, antioxidants, brightening agents, a variety of drugs, and in another field of chemistry and agriculture. In order to explore diverse biological activities, investigating various methods for synthesis and structural modification has now become an important goal of several research groups. Thus, benzofuran fused pyrimidine moiety can be taken as a lead compound for the synthesis of novel derivatives with a variety of biological activities[10-12]. Fig. 1 explains that Compound 1 is a novel drug, it is a multistage tyrosine kinase inhibitor currently in Phase I clinical trials, while compound 2 was found to be histamine H4 modulators compounds[13-15]. Benzofuran[3, 2-d] pyrimidine structure is similar to those compounds, theoretically promoting the compounds should also have good biological activity[16-18].
In view of the wide range biological activities antimicrobial resistance is the major threat to human health, growing in the field of infectious diseases mainly caused by bacteria, fungi, viruses and parasites. According to a survey, around 490 000 people have multidrug-resistant Tuberculosis worldwide and the drug resistance is commencing to complicate the battle against Human Immunodeficiency Virus (HIV) as well as Malaria. With the passage of time the need for effective antimicrobial has become essential to control the spread of “superbugs” microorganisms that develop antimicrobial resistance are sometimes referred to as superbugs. We aim to design benzofuropyrimidine hybrids with a hope for better therapeutic agents[16,19,20]. Molecular docking analysis to selected bacterial Zoanthus sp at 2.2A Resolution enzyme were adopted to aid understanding the possible binding modes and interactions between the synthesized compounds and protein target[17].
Materials and Methods
All reagents were commercially available like Sigma Aldrich, mol chem., and other standard commercial sources were used without purification, using distilled purified solvents, Progress of the reaction was monitored by Thin Layer Chromatography (TLC) using hexane and methyl acetate mobile phase. Melting points were determined with open capillary and are uncorrected. The purity of the compounds was purified by using recrystalisation methods. Infrared (IR) spectra were recorded in KBr pellets by using Shimadzu Fourier Transform Infrared Spectroscopy (FTIR) 8000 Spectrometer. Proton Nuclear Magnetic Resonance (1H NMR) was recorded in DMSO-d6 on Jeol-400 MHz Spectrometer. Chemical shifts are recorded relative to tetramethylsilane as an internal standard. Mass spectral data were obtained on Mass Spectrometer.
Experimental procedure:
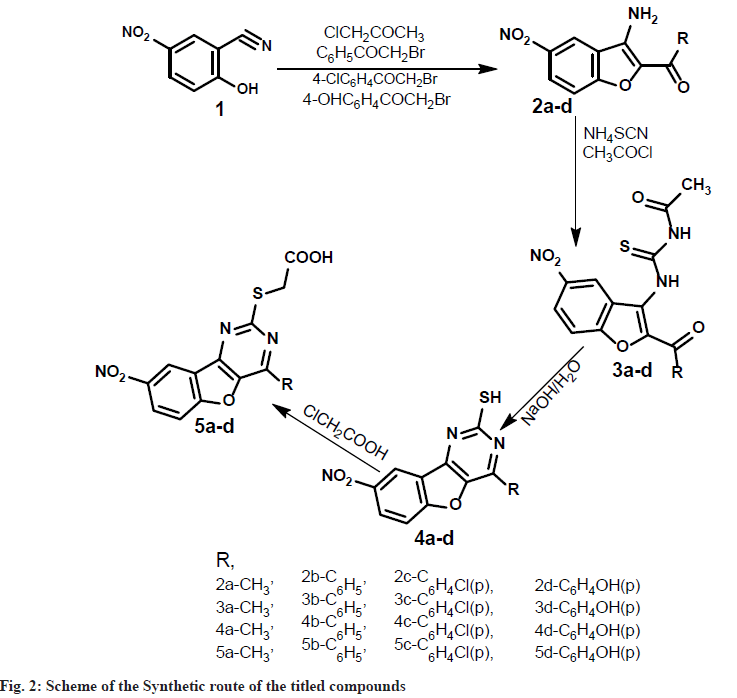
Synthesis of 2-hydroxy-5-nitro-benzonitrile (1): A mixture of 2-hydroxy-1-benzoldoxime (0.1 mol) and acetic anhydride (20 ml) was refluxed for 1 h and excess of acetic anhydride was removed by distillation under reduced pressure. The dark colored dense liquid leftover gave benzo[l,2-d] isoxazole(not isolated), to this absolute ethanol (100 ml) and freshly prepared sodium ethoxide (0.2 mol) [Prepared by adding freshly cut dry sodium (0.2 mol) into absolute ethanol (50 ml) at 0°] were added. The mixture was stirred for 1 hour at room temperature and poured into icecold water (500 ml), on careful acidification with dilute hydrochloric acid, 2-hydroxy-5- nitrobenzonitrile(1) was obtained as brown crystalline solid. It was collected by filtration and recrystallized from ethanol. Synthesis of 2-acyl- 3-amino-5-nitrobenzo [2,l-b]furan (2a-d): To a solution of 2-hydroxy-5-nitrobenzonitrile (1) (0.01 mol) in dry acetone (100 ml), chloroacetone (0.01 mol) and anhydrous potassium carbonate (0.1 mol) were added and the reaction mixture was refluxed on water bath for 8 h.
The potassium salt was filtered off and washed thoroughly with acetone. Removal of solvent from the filtrate and subsequent trisection with ethanol gave 2-acyl-3-amino-5-nitrobenzo[2, l-b]furan (2a). The compounds 2b-d was synthesized using phenacylbromide, 4-chloro-phenacylbromide, 4-hydroxyphenacylbromide, in the place of chloroacetone by the same method described for compound 2a as above.
Synthesis of N-[(2-substituted-5-nitro-1- benzofuran-3-yl)carbamothioyl] acetamide (3a-d):To a solution of ammonium thiocynate (0.01 mol) in dry acetone (25 ml), acetyl chloride (0.01 mol) was added slowly with stirring to a solution of compound 2a. To the reaction mixture dry acetone (50 ml) was added slowly so as to maintain reflux condition. After the addition was complete, the reaction mixture was stirred for 90 min at room temperature, compound 3a, which separated as precipitate on pouring into ice cold water, was collected by filtration and washed with water and cold ethanol. It was purified by crystallizing from ethanol. Similarly, the compounds 3b-d were synthesised from compounds 2b-d by following the procedure as described for compound 3a.
Synthesis of 4-substituted-8-nitro [1] benzofuro[3,2-d]pyrimidine-2-thiol (4a-d): A mixture of 3a (0.06 mol), sodium hydroxide (0.008 mol) and water (50 ml) was heated under reflex for 15 min, cooled, diluted with water and acidified with dilute hydrochloric acid. The solid that separated was filtered, washed with water, dried and recrystalised from aqueous Dimethyl Formamide (DMF) to get 2-mercapto-4- phenylbenzofuropyrimidine (4a). The compounds 4b-d was synthesized from 3b-d in a similar manner as described for compound 4a.
Synthesis of [(4-substituted-8-nitro [1] benzofuro [3, 2-d] pyrimidin-2-yl) sulfanyl] acetic acid (5ad): To a solution of chloroacetic acid (0.005 mol) and sodium carbonate (0.00275 mol) in water (10 ml) was added slowly, while stirring for 30 min, to a solution of compound 4a (0.0055 mol) in sodium hydroxide (0.015 mol) in water (10 ml). The reaction mixture was stirred overnight at room temperature, acidified with dilute hydrochloric acid and extracted with chloroform. The organic layer was dried over anhydrous sodium sulphate, the solvent was removed by distillation under reduced pressure to obtain solid, which was recrystalized from ethanol. The compounds 5b-d was synthesized from 4b-d in a similar manner as described for compound 5a.
Biological activity test method:
Antibacterial activity test method: The antibacterial activities of newly synthesized compounds were determined by well plate diffusion method in Mueller-Hinton Agar[18-20]. The antibacterial activity was carried out against 24 h old cultures of bacterial strains. The test compounds were dissolved in DMF at a concentration of 100 and 50 μg/ml. 20 ml of sterilized agar media was poured into each pre-sterilized Petri dish. Excess of the suspension was decanted and plates were dried by placing them in an incubator at 37° for 1 h. About 60 ml of 24 h old culture suspension were poured and neatly swabbed with the pre-sterilized cotton swabs. Six-millimeter diameter well was then punched carefully using a sterile cork borer and 30 ml of test solutions of different concentrations were added into each labeled well. The plates were then incubated at 37°C for 24 h. The inhibition zone that appeared after 24 h, around the well in each plate was measured as a zone of inhibition in mm. Experiments were carried out in triplicates and the standard deviation was calculated.
Antifungal activity test method: Antifungal studies of newly synthesized compounds were carried out against A. flavus, C. keratinophilum and C. albicans. Sabouraud’s agar media was prepared by dissolving peptone (10 g), D-glucose (40 g) and agar (20 g) in distilled water (1000 ml) and adjusting the pH to 5.7. Normal saline was used to make a suspension of spore of fungal strains for lawning. A loopful of particular fungal strain was transferred to 3 ml saline to get a suspension of the corresponding species. 20 ml of agar media was poured into each Petri dish. Excess of the suspension was decanted and plates were dried by placing them in an incubator at 37° for 1 h. Wells were made on these seeded agar plates using sterile cork borer and different concentrations of the test compounds in DMF were added into each of the labelled wells. A control was also prepared for the plates, in the same way, using DMF. Petri dishes were prepared in triplicate and maintained at 25° for 72 h. Antifungal activity was determined by measuring the diameter of inhibition zone. The activity of each compound was compared with fluconazole as standard. Zones of inhibition were determined for compounds.
Molecular docking studies method: Computational techniques offer alternatives close to reality through simulations that allow, from exhaustive studies, to analyze the different interactions that occur between the molecule under study and the protein site of interest. In the present protocol, we will address the different steps to perform molecular docking by patch dock software[15,21].
Choice of study protein: One of the key steps in molecular docking is the choice of the protein site to be studied their biological importance and the literature study of the proteins reported in the Protein Data bank database.
Preparation of protein and compound: The next step of the protocol starts with the preparation of the previously downloaded protein structure and compound saved as pdb file by using “open Babel” software.
Docking: The time has come to perform the docking calculation, it is important to note that the docking will study the different conformations that can interact in the coordinates that we have established. The calculation conducted in patch dock software will give us some values of dock score value in Kcal/mol.
Analyzing the results: The next and last step is to analyze the results for this will go to discovery studio visualize. We will go into the display ligand interactions as in fig. 1.
Results and Discussion
In the present investigation, we would like to report the synthesis of [(4-methyl-8-nitro [1] benzofuro[3,2-d]pyrimidin-2-yl)sulfanyl] acetic acid and their derivatives (5a-d) synthetic route observed in the scheme (fig. 1). The synthesis of 1-(3-amino-5-nitro-1-benzofuran-2-yl)ethan- 1-one (2a-d) were achieved by cyclisation of 2-hydroxy-5-nitro-1-benzonitrile and aryloxy ketones in the presence of alcohols. Initially, we investigated the reaction of N-[(2-acetyl-5-nitro- 1-benzofuran-3-yl)carbamothioyl]acetamide (3ad), with ammonium thiocyanate under the normal condition at room temperature. Further, we tried the cyclisation reaction of 3a-d with aqueous sodium hydroxide to produce a compound (4a-d). The cyclized product formed in this reaction was refluxed with chloroacetic acid. To our surprise, the desired product compound (5a-d) was formed in 65 %-72 % excellent yields. The versatility of the reaction was studied with different aryaloxyketones. All the synthesized compounds' physically characterized results are given in Table 1.
| Compound | R | M.P | Yield (%) | Solvent | Molecular formula |
|---|---|---|---|---|---|
| 3a | CH3 | 58 | 75 | Ethanol | C13H11N3O5S |
| 3b | C6H5 | 126 | 73 | Ethanol | C18H13N3O5S |
| 3c | 4-Cl C6H5 | 143 | 68 | Ethanol | C18H12ClN3O5S |
| 3d | 4-OHC6H5 | 136 | 65 | Ethanol | C18H13N3O6S |
| 4a | CH3 | 96 | 67 | DMF | C11H7N3O3S |
| 4b | C6H5 | 110 | 70 | DMF | C16H9N3O3S |
| 4c | 4-Cl C6H5 | 159 | 62 | Aq DMF | C16H9ClN3O3S |
| 4d | 4-OHC6H5 | 133 | 68 | Aq DMF | C16H9N3O4S |
| 5a | CH3 | 118 | 78 | Ethanol | C13H11N3O5S |
| 5b | C6H5 | 69 | 83 | Ethanol | C18H11N3O5S |
| 5c | 4-Cl C6H5 | 84 | 73 | Ethanol | C18H10ClN3O5S |
| 5d | 4-OH C6H5 | 95 | 64 | Ethanol | C18H11N3O6S |
Table 1: Analytical Data of the Synthesized Compounds
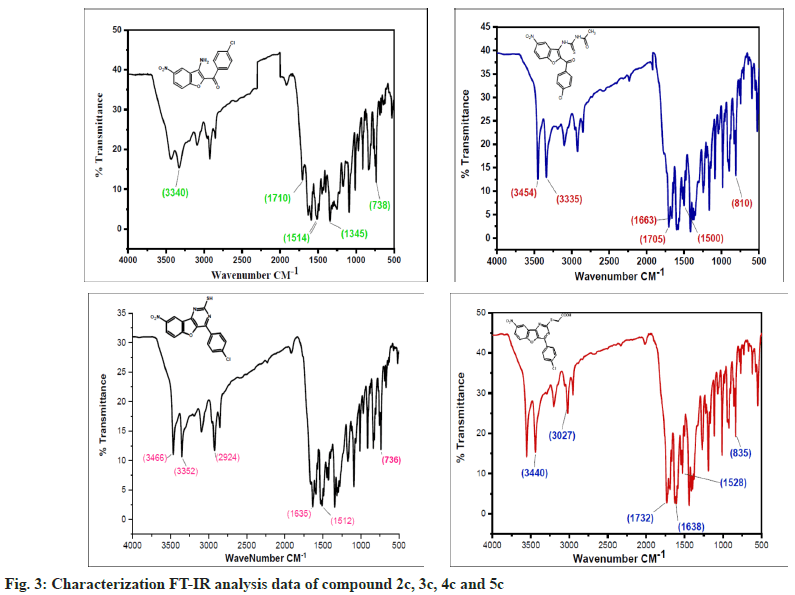
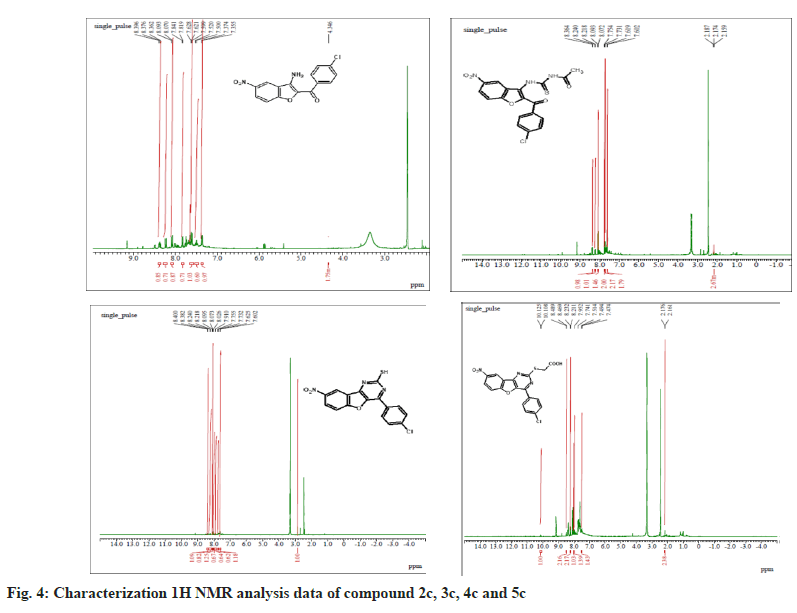
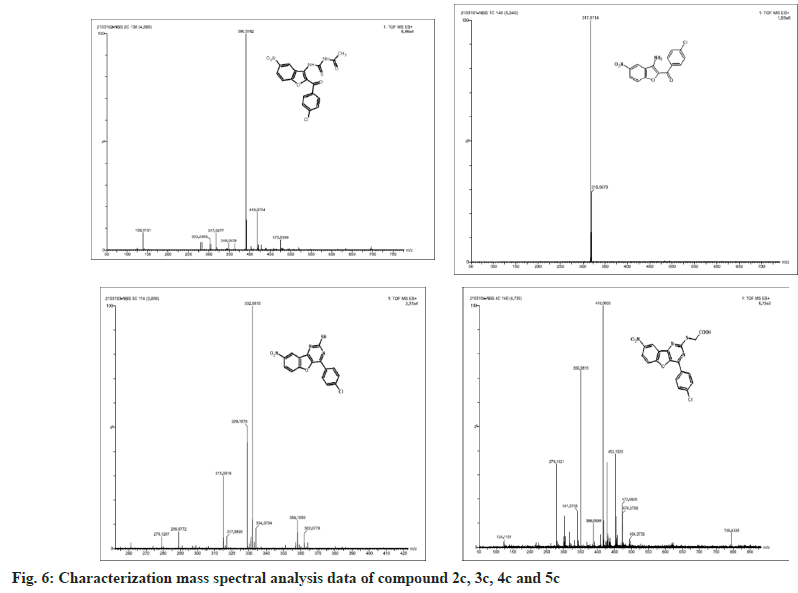
The synthesized compounds were confirmed by FT-IR, 1H NMR, and Mass spectral data. The IR spectrum of the final or desired compound 5c sowed a strong absorption band for the stretching peak of the carboxyl group in the region 1715-1685 cm-1, and the imine group in the region 1690-1630 cm-1. These results are observed in IR spectral graph (fig. 2). The formations of final products were evidenced by proton NMR, the appearance of a singlet for the exocyclic (>CH) proton in the region δ 2.85-3.04 ppm, the aromatic group of multiples (Ar-H) in the region δ 7.70–7.80 ppm, and another hydroxyl group of singlet (-OH) in the region δ 8.94-9.05 ppm, this results depicted in their 1H NMR spectral graph (fig. 3). This deshielded singlet further confirms the stereochemistry of the exocyclic double bond. These two results are confirmed by the mass spectral data. It shows the synthesized compounds' molecular weight is 415 on the bases of their M+2 fragment ion peak is 417 due to the presence of halogen element, this was observed in the mass spectral graph (fig. 4).
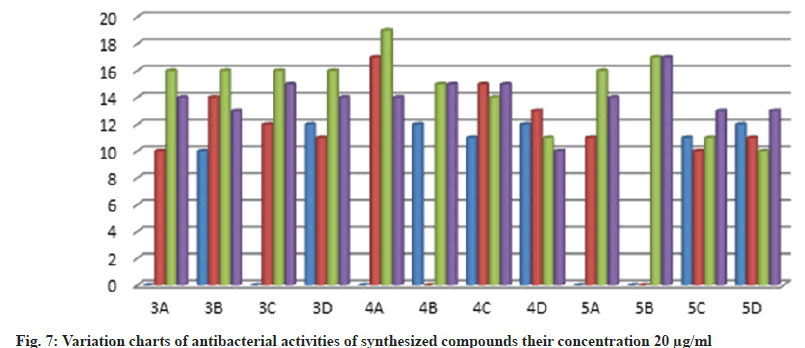
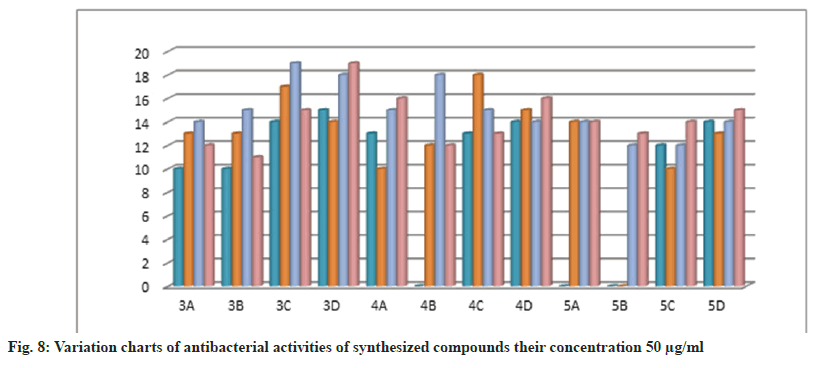
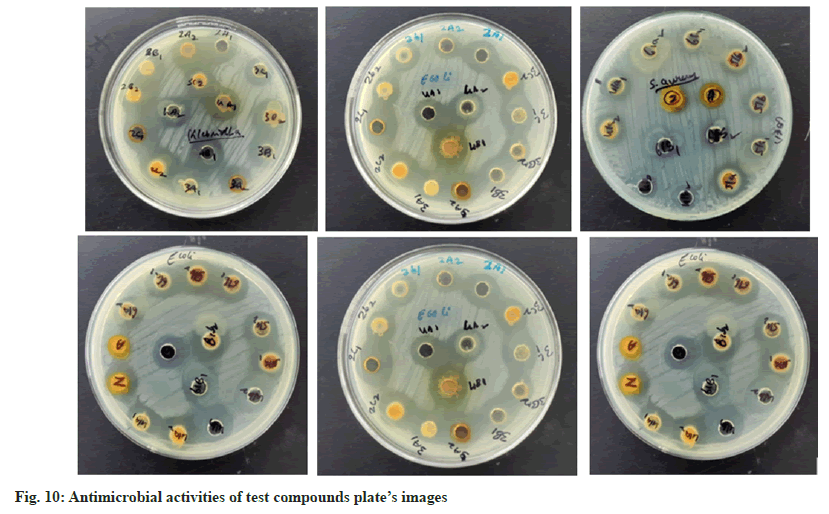
Further, the confirmed desired synthesized compounds were screened for biological activity like antibacterial and antifungal activity tests in two concentrations 20 μg/ml and 50 μg/ml. The compound shows excellent antibacterial activity against the four organisms Staphylococcus aureus, Staphylococcus epidermidis, Klebsiella pneumoniae, and Escherichia coli, which are compared with the standard drugs penicillin and streptomycin. This activity result of the zone of inhabitation was observed in their activity plate’s images in fig. 5 and this result was studied with bar graphs in fig. 6 and fig. 7.
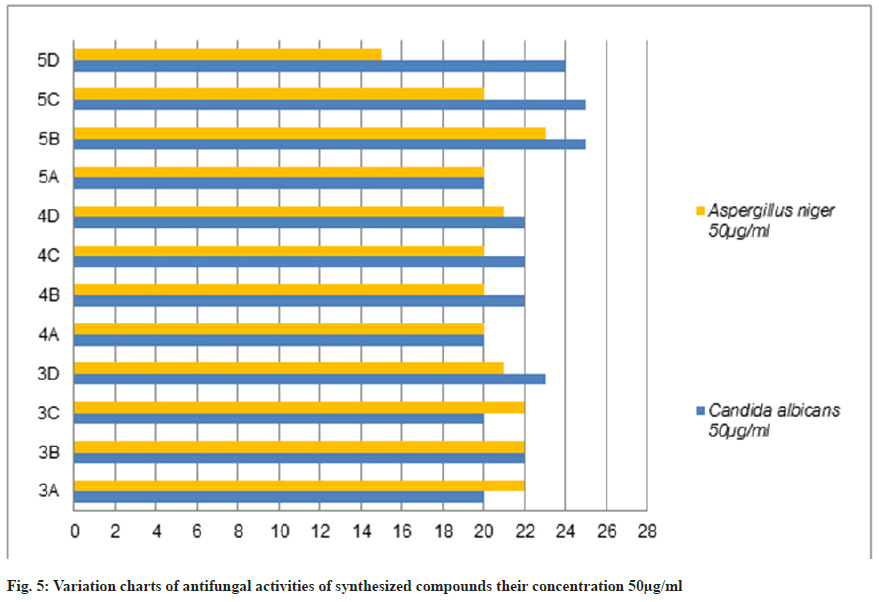
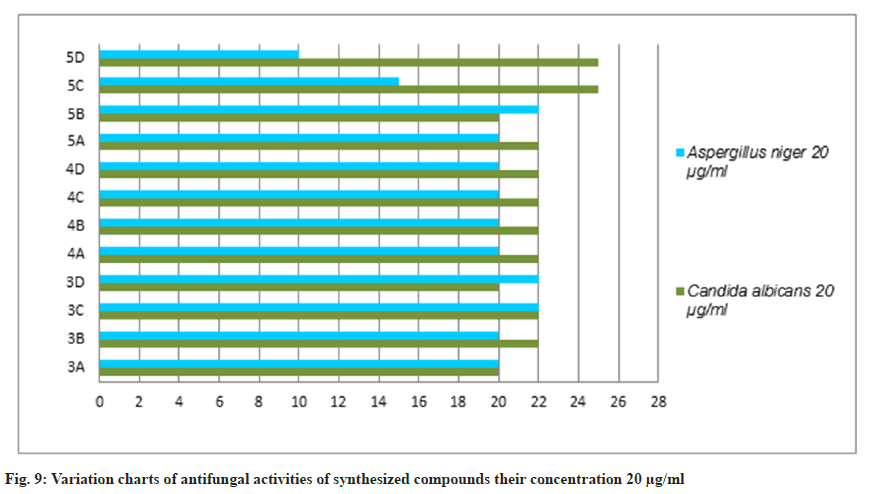
The antifungal activity of all the synthetic compounds exhibits predominant activity against the two organisms, Candida albicans and Aspergillus niger with concentrations of 20 μg/ml and 50 μg/ml, these synthesized compounds were compared with the standard drug Griseofulvin, which is depicted the activity results are the zone of inhabitation plating images in fig. 5 and this result was shown with bar graph in fig. 8 and fig. 9.
Compound 2c: (3-amino-5-nitro-1-benzofuran-2- yl)(4-chlorophenyl)methanone: IR in cm-1 using KBr (-NH2) 3340, (-C=O) 1710, (-NO2) 1514, (-C-N) 1345, (-C-Cl stretching) 738; 1H NMR in δ ppm (DMSO d6) 7.46-7.75 (d, 4H, Ar-H), 4.34- 4.38 (s, 2H, -NH2), 8.37-8.39 (s, 1H, Ar-H), 7.72- 8.09 (d, 2H, Ar-H); Mass Spectra of ion peak at (m/z) 317 (may be M+2 peak).
Compound 3c: N-{[2-(4-chlorobenzoyl)-5-nitro-1- benzofuran-3-yl]carbamothioyl} acetamide: IR in cm-1 using KBr: (–NH) 3454, (NH) 3335, (>C=O) 1705, (>C=O) 1663, (-NO2) 1500, (-NO2) 810; 1H NMR in δ ppm (DMSO d6): 8.07-8.36 (m, 3H, Ar- H), 4.6-4.65 (s, 1H, -NH), 9.10-9.15 (s, 1H, NH), 2.15-2.18(s, 3H, -CH3), 7.60-7.75(d, 4H, Ar-H); Mass Spectra in M+ (m/z) 418 (may be M+1 peak).
Compound 4c: 4-(4-chlorophenyl)-8-nitro [1] benzofuro [3,2-d]pyrimidine-2-thiol: IR in cm-1 using KBr: 3466 (-NH), 3352(-NH), 2924(-SH), 1635(-C=N), 1512(-NO2), 736(-C-Cl); 1H NMR in δ ppm (DMSO d6) 8.38-8.40 (s, 1H, Ar-H), 7.42-8.24 (d, 6H, Ar-H), 2.84-2.91 (s, 1H, SH); Mass Spectra in M+ (m/z) 358 (may be M+1 peak).
Compound 5c: {[4-(4-chlorophenyl)-8-nitro[1] benzofuro[3,2-d]pyrimidin-2 yl]sulfanyl}acetic acid: IR in cm-1 using KBr 3440(-CH2), 3027(- OH), 1732(-C=O), 1638 (-C=N), 1528 (-NO2), 835 (-C-Cl); 1H NMR in δ ppm (DMSO d6) 7.49-7.95 (d, 4H, Ar-H), 8.21-8.46 (d, 3H, Ar-H), 2.16-2.19 (s, 2H, CH2), 10.10-10.12 (s, 1H, O-H); Mass Spectra in M+ (m/z) 417 (may be M+2 peak).
The antimicrobial activity of newly synthesized compounds was determined by well diffusion method. The antimicrobial activities of different extracts and fractions were compared with standard antibacterial drug agent Penicillin and Streptomycin and antifungal activity were compared with slandered drug fluconozole (fig. 10).
The bacteria were used Gram-positive Staphylococcus aureus, Staphylococcus epidermidis, Gram-negative Klebsiella pneumoniae, and Escherichia coli. The compounds were tested at a concentration of 0.001 mg/ml in Doderma MF against both organisms. The standard drug streptomycin was used for comparison with the compounds' antibacterial activities. The zone of inhibition results are recorded in Table 2.
| Synthetic Compounds (Label) | Comp. zone of inhibition in mm | |||||||
|---|---|---|---|---|---|---|---|---|
| Gram-positive | Gram-negative | |||||||
| Staphylococcus aureus | Staphylococcus epidermidis | Klebsiella pneumoniae | Escherichia coli | |||||
| 20 µg/ml | 50µg/ml | 20 µg/ml | 50µg/ml | 20 µg/ml | 50µg/ml | 20 µg/ml | 50µg/ml | |
| 3a | - | 10 | 10 | 13 | 16 | 14 | 14 | 12 |
| 3b | 10 | 10 | 14 | 13 | 16 | 15 | 13 | 11 |
| 3c | - | 14 | 12 | 17 | 16 | 19 | 15 | 15 |
| 3d | 12 | 15 | 11 | 14 | 16 | 18 | 14 | 19 |
| 4a | - | 13 | 17 | 10 | 19 | 15 | 14 | 16 |
| 4b | 12 | - | - | 12 | 15 | 18 | 15 | 12 |
| 4c | 11 | 13 | 15 | 18 | 14 | 15 | 15 | 13 |
| 4d | 12 | 14 | 13 | 15 | 11 | 14 | 10 | 16 |
| 5a | - | - | 11 | 14 | 16 | 14 | 14 | 14 |
| 5b | - | - | - | - | 17 | 12 | 17 | 13 |
| 5c | 11 | 12 | 10 | 10 | 11 | 12 | 13 | 14 |
| 5d | 12 | 14 | 11 | 13 | 10 | 14 | 13 | 15 |
Table 2: Antibacterial Activities of Synthesized Compounds (3a-D), (4a-D) and (5a-D)
The antifungal activities of the synthesized compounds were performed against standard fungal strains, Aspergillus niger, and Candida albicans. The compounds were tested at a concentration of 0.001 mg/ml in Doderma MF against both organisms. Fluconazole was used as a standard antifungal drug. The zone of inhibition was compared with the standard drug 72 h at incubation 25° for antifungal activity. The results are recorded in Table 3.
| Synthetic Compounds (Label) | Comp. Zone of inhibition in mm | |||
|---|---|---|---|---|
| Gram-positive | Gram-negative | |||
| Candida albicans | Aspergillus niger | |||
| 20 µg/ml | 50 µg/ml | 20 µg/ml | 50 µg/ml | |
| 3a | 20 | 20 | 20 | 22 |
| 3b | 22 | 22 | 20 | 22 |
| 3c | 22 | 20 | 22 | 22 |
| 3d | 20 | 23 | 22 | 21 |
| 4a | 22 | 20 | 20 | 20 |
| 4b | 22 | 22 | 20 | 20 |
| 4c | 22 | 22 | 20 | 20 |
| 4d | 22 | 22 | 20 | 21 |
| 5a | 22 | 20 | 20 | 20 |
| 5b | 20 | 25 | 22 | 23 |
| 5c | 25 | 25 | 15 | 20 |
| 5d | 25 | 24 | 10 | 15 |
Table 3: Antifungal Activities of Synthesized Compounds (3a-D), (4a-D) and (5a-D)
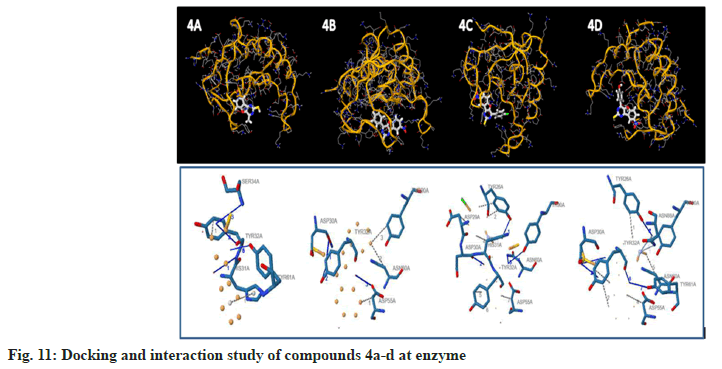
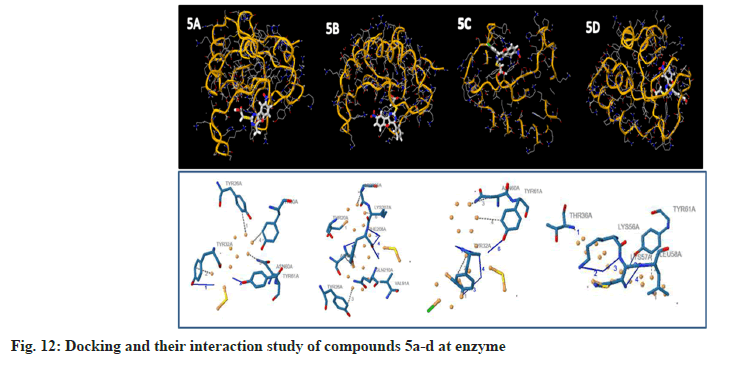
To examine the complete intermolecular interactions between the molecule and the object protein, a program patch-dock was used to determine the orientation of inhibitors bound in the active site of the enzyme staphylococcal enterotoxin c2 from Staphylococcus aureus, chosen on the basis of inhibition of the antibacterial activity. The protein structure file (PDB ID: 1STE) was taken from PDB (www.rcsb.org/pdb). The ligand molecules were designed, and the structure was analysed using Chem-Draw Ultra 6.0 3D coordinates. This protein and legend molecule Docking studies give a fair idea of the analysed drug-receptor interactions. The docking study was given the docking score and Atomic Contact Energy (ACE) on PDB 1STE to sustain the interaction study of docking results with the PLIP Programme and estimate the bond angle and bond distance of amino acid residue modes of compounds 4a-d and 5a-d. Docking scores and their interaction study results are depicted in Table 4. The compounds 4a–d make hydrogen bonding interactions at the active site of the enzyme Staphylococcal enterotoxin C2 from Staphylococcus aureus (PDB ID: 1STE), among them the 4c compound, which makes seven hydrogen bonding interactions increase from nitrogen and oxygen present on the pyrimidine ring. The bond angles of legend and amino acid hydrogens are TYR30A (119, 145), ASP30A (105, 144, 101), TYR32A (167), and TYR61A (148), as depicted in Fig 10. The compounds 5a-d make hydrogen bonding interactions at the enzyme (PDB ID: 1STE), among them, 5c and 5d have good docking scores, their bonding interactions are raised by oxygen atoms from the carboxy group present on the benzofuran derivatives of the perimidine ring with hydrogens of TYR32A, TYR61A, THR36A, and LYS57A. As depicted in fig. 11 and fig. 12.
| Sample Code | Protein Receptor (PDB Id) | Patch dock Score | ACE | No. of H interaction | Bond Distance | Bond Angle | AA Residue |
|---|---|---|---|---|---|---|---|
| 4a | 1STE | 3954 | -171.3 | 5 | 2.66 | 142 | ASP30A |
| 3.72 | 144 | TYR32A | |||||
| 3.83 | 108 | SER34A | |||||
| 2.57 | 123 | TYR61A | |||||
| 2.57 | 153 | TYR61A | |||||
| 4b | 4806 | -149.97 | 3 | 4.02 | 105 | TYR32A | |
| 4.02 | 150 | TYR32A | |||||
| 3.93 | 122 | ASP55A | |||||
| 4c | 5212 | -186.61 | 4 | 3.37 | 148 | TYR26A | |
| 2.52 | 118 | ASP30A | |||||
| 2.69 | 178 | ASN60A | |||||
| 3.21 | 137 | TYR90A | |||||
| 4d | 5034 | -179.84 | 7 | 3.16 | 119 | TYR26A | |
| 3.16 | 145 | TYR26A | |||||
| 3.34 | 105 | ASP30A | |||||
| 3.85 | 144 | ASP30A | |||||
| 3.9 | 101 | ASP30A | |||||
| 3.2 | 167 | TYR32A | |||||
| 3.3 | 148 | TYR61A | |||||
| 5a | 4336 | -171.66 | 3 | 3.81 | 116 | TYR32A | |
| 2.86 | 145 | TYR61A | |||||
| 2.86 | 144 | TYR61A | |||||
| 5b | 5118 | -154.47 | 4 | 2.11 | 107 | ASN23A | |
| 3.38 | 130 | PHE208A | |||||
| 3.67 | 137 | PHE208A | |||||
| 3.12 | 155 | PHE208A | |||||
| 5c | 5250 | -209.22 | 6 | 2.98 | 139 | TYR32A | |
| 3.9 | 112 | TYR32A | |||||
| 3.62 | 162 | TYR32A | |||||
| 3.45 | 113 | TYR32A | |||||
| 3.01 | 126 | TYR61A | |||||
| 3.01 | 156 | TYR61A | |||||
| 5d | 5360 | -146.17 | 6 | 3.72 | 126 | THR36A | |
| 3.58 | 117 | LYS56A | |||||
| 2.84 | 128 | LYS57A | |||||
| 3.08 | 121 | LYS57A | |||||
| 2.93 | 120 | TYR61A | |||||
| 2.93 | 139 | TYR61A |
Table 4: Molecular Docking Study and their Interaction Results of the Compounds 4a-D and 5a-D (Pdb Id: 1ste)
In conclusion, we have synthesised substituted benzofuranyl pyrimidines with a simple experimental method in a short reaction time. And these newly synthesised compounds are concurrent with assigned structures purified by TLC and confirmed by IR, 1H NMR, and mass spectral data. Most substituted benzofuranyl pyrimidine compounds exhibit moderate to high activity against antibacterial organisms of S. aureus, S. epidermidis, Pseudomonas, E. coli, and antifungal organisms of Candida, and Aspergillus. Some of the compounds with 4a-d and 5a-d activity are supported by molecular docking studies of staphylococcal enterotoxin C2 from Staphylococcus aureus proteins.
Acknowledgements:
The authors thanks to the Chairman, Department of Chemistry, the Department of Microbiology Vijayanagar Sri Krishanadevaraya University Bellary for providing laboratory facilities; Sophisticated Instrumentation Facility, (SAIF) Karnataka University, Dharawad for providing NMR spectral data; University of Mysore, for providing Mass spectral Data; and Siri Laboratory Microbiology, Ballari.
Conflict of interests:
The authors declared no conflict of interests.
References
- Basavaraja KM, Vaidya VP, Agasimundin YS. Synthesis and biological activity of 4-arylbenzofuro[3,2-d]pyrimidine-3-N-oxides and 3-(3’-aryl-1,2,4-triazol-yl)benzofurans. Indian J Heterocycl Chem 2005;15:1-6.
- Naveenakumari HM, Manjunatha Harihara Mathada, Shivakumar H, Basavaraja KM. Synthesis, characterization and antimicrobial screening of 5-bromo-benzofuranyl aryl triazoles. J Appl Chem 2019;8(4):1704-10.
- Giri SK, Hanumanagoud H, Basavaraja KM. Synthesis of biologically active 3-amino-2-(1,3,4-oxadiazolyl,1,3,4-thiadiazolyland1,2,4-triazolyl)naphtha[2,1-b]furans. Indian J Heterocycl Chem 2010;20:113-6.
- Latha, KP, Vaidya VP, Keshavayya J, Vijay Kumar ML, Shreedhara CS. Synthesis, characterization and biological studies of metal complexes of 2-acetylnaphtho [2,1-b] furan. Natl Acad Sci lett2002;25(5):153-8. [Crossref]
[Google Scholar] [PubMed]
- Ramesh D, Vaidya VP, Kumaraswamy MN, Chandrashekhar C. Synthesis of 2-(8-bromonaphtho [2, 1-b] furan-2-yl)-5-aryl-1, 3, 4-oxadiazoles as potential antimicrobial agents. Int J Pharm Chem Biol Sci 2014;4(2):298.
- Shruthi, E, Vaidya VP, Girija CR, Shalini S. Synthesis and structural studies of 1-[(8-nitronaphtho [2, 1-b] furan-2-yl) carbonyl] piperidine. European Scientific Journal 2013;9(33):1-12.
- Shaikh F, Shastri SL, Naik NS, Kulkarni R, Madar JM, Shastri LA, et al. Synthesis, antitubercular and antimicrobial activity of 1, 2, 4-triazolidine-3-thione functionalized coumarin and phenyl derivatives and molecular docking studies. Chemistry Select,2019;4(1):105-15.
- Mathew V, Keshavayya J, Vaidya VP, Moinuddin Khan MH. Triazoles as complexing agents: Synthesis, characterization and pharmacological activities of copper complexes of 4-amino-3-mercapto-5-substituted aryl-1, 2, 4-triazoles. J Coord Chem 2008;61(16):2629-38.
- Gaikwad SS, Suryawanshi VS, Lohar KS, Jadhav DV, Shinde ND. Synthesis and biological study of some new thiazolidinone derivatives containing naphthofuran moiety. E-Journal of Chemistry 2012;9(1):175-80.
- Nagaraja GK, Prakash GK, Satyanarayan ND, Vaidya VP, Mahadevanc KM. Microwave assisted synthesis of novel 5-aryl-1, 2, 4-triazolo [3, 4-b][1,3,4] thiadiazepino [3, 2-f] quinolines as potent antimicrobial agents. Arkivoc 2006;15(1):142-52.
- Prakash MS, Vaidya VP, Mahadevan KM, Shivananda MK, Suchetan PA, Nirmala B, et al. Synthesis, characterization and comparative antimicrobial studies of some novel chalcones and pyrazolines containing naphthofuryl substituents. J Chem Pharm2012;4(2):1179-84.
- Nagashree AS, Chandrashekhar C, Vaidya VP. Synthesis of 1-N-phenyl-3-{3-nitronaphtho [2,1-b] fur-2-yl}-5-aryl-2-pyrazolines and their biological activities. Indian J Heterocycl Chem2010; 20(1):65-8.
- Veena K, Ramaiah M, Shashikaladevi K, Avinash TS, Vaidya VP. Synthesis and antimicrobial activity of asymmetrical azines derived from naphtho [2,1-b] furan. J Chem Pharm Res2011;3(5):130-5.
- Rajashekhar H, Ramesh D, Chandrashekhar C, Mahadevan KM, Vaidya VP. Synthesis of some naphtho (2,1-b) furo (3,2-d) pyrimidines, naphtho (2,1-b) furo (3',2':4,5) pyrimido (1,2-b) benzo (d) thiazole and their antimicrobial activity. Indian J Heterocycl Chem2009;18(3):205.
- Kumaraswamy MN, Mathias DP, Vaidya VP. Synthesis and pharmacological evaluation of 2-mercapto-4-substituted-naphtho [2, 1-b] furo [3, 2-d] pyrimidines. Indian J Pharm Sci 2006; 68(6): 731.
- Harshita PS, Supriya CH, Sravanthi S, Jyothi V, Peddi SR, Manga V, et al. Design synthesis and biological evaluation of dithiocarbamate substituted 2-aminobenzothiazole derivatives as proviral integration site of moloney murine leukaemia virus 1 kinase inhibitors. Indian J Pharm Sci 2020;82(6):1015-24.
- Devi KS, Ramaiah M, Roopa DL, Vaidya VP. Synthesis and investigation of antimicrobial and antioxidant activity of 3-nitro-n-(3-chloro-2-oxo-substituted-phenyl-azetidin-1-yl) naphtho [2,1-b] furan-2-carboxamides. J Chem 2010;7(S1):S358-62.
- Govindaiah S, Sreenivasa S, Ramakrishna RA, Rao TM, Nagabhushana H. Regioselective synthesis, antibacterial, molecular docking and fingerprint applications of 1-benzhydrylpiperazine derivatized 1, 4-disubstituted 1, 2, 3-triazoles. Chemistry Select2018;3(28):8111-7.
- Savanur HM, Naik KN, Ganapathi SM, Kim KM, Kalkhambkar RG. Click chemistry inspired design, synthesis and molecular docking studies of coumarin, quinolinone linked 1,2,3triazoles as promising anti‐microbial agents. Chemistry Select 2018;3(19):5296-303.
- Hanumanagouda H, Basavaraja KM. Synthesis, antibacterial, antifungal activity and DNA cleavage study of 3-(7-methoxybenzofuran-2-yl)-5-yl-4H-[1,2,4]triazoles. J Chem Pharm Res 2012;4(12):5165-71. [Crossref]
[Google Scholar] [PubMed]
- Manjuraj T, Yuvaraj TCM, Jayanna ND, Shreedhara SH, Saravajith MS. Spectral, DFT, molecular docking and antibacterial activity studies of Schiff base derived from furan-2-carbaldehyde and their metal (II) complexes. J Turk Chem 2020;7(2):449-62.