- *Corresponding Author:
- N. Nagaladinne
Department of Pharmaceutical Sciences
Jawaharlal Nehru Technological University Anantapur
Anantapuramu-515001, Andhra Pradesh
E-mail: nnizamuddin1988@gmail.com
| Date of Received | 10 May 2020 |
| Date of Revision | 13 September 2020 |
| Date of Acceptance | 23 November 2020 |
| Indian J Pharm Sci 2020;82(6):984-995 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Quinazolin-4-ones connected 1, 3-thiazole were found with antibacterial action after high throughput screening. N-Methyl Anthranilic acid when treated with thiophene-2-carboxylic acid gives 1-Methyl-2- (thiophen-2-yl)-1,2-dihydro-4H-3,1-Benzoxazin-4-one which on further treatment with 4-substituted phenyl-1,3-thiazol-2-amines gives 1-Methyl-3-(4-Substituted phenyl-1,3-thiazol-2yl)-2-(thiophen-2-yl)-2,3- dihydro quinazoline-4(1H)-ones (5Fa1-5Fk11) were obtained. The structural identification was done through Infra-red, 1H- Nuclear magnetic resonance, 13C- Nuclear magnetic resonance, and Mass spectrometry. These derivatives were screened for anti-tubercular activity against Mycobacterium tuberculosis H37RV using the micro plate alamar blue assay method. Compounds 5Ff6, 5Fe5, 5Fb2, and 5Fd4 displayed better antibacterial activity at minimum inhibitory concentration 12.5 and 6.25 μg/ml. Further, 5Ff6 displayed a favourable inhibition at minimum inhibitory concentration 6.25 μg/ml, contrasted with the reference drug Isoniazide. The compounds were docked where they exhibited associations with both the vital site of penicillin-binding protein 2a and Mycobacterium tuberculosis H37Rv organism. All the derivatives showed good affinity towards the protein when compare with the standard drug. Here 5Ff6 was observed to interact with three amino acids, viz., Agn104, Lys273, and Tyr297, as signified by the great interaction energy (ΔG=-4.2 kcal/mol).
Keywords
1, 3-thiazoles, N-methyl quiazolin-4-ones, anti-tubercular activity, molecular docking studies, mycobacterium tuberculosis H37RV, and penicillin-binding protein 2a (6Q9N).
Tuberculosis is one of the major health issues with an extensive kind of manifestations caused by Mycobacterium tuberculosis (M.t). According to the recent report, it has been predicted that over onethird of the world population is infected with this microorganism. The treatment of infectious diseases has become a critical problem because of the emergence of mono drug and multidrug-resistant strains of M.t[1]. Consequently, there is a necessity for the development of new drugs with a novel mechanism other than Streptomycin (STP), Isoniazid (INH), Rifampicin (RIF) and Pyrazinamide (PZA). Since 20 y the look for new anti-tubercular agents has been ranked among the priority areas of chemotherapeutic studies. The derivatives of Quinazolin-4-one represent one of the liveliest classes of compounds owning a wide spectrum of activity used in pharmaceuticals and agrochemicals[2]. For instance, fluquinconazole controls agriculture diseases caused by pesticides[3]. Many reports had been issued on the quinazoline derivatives, which include bactericidal, herbal, and antitumor activities[4,5]. Accordingly, their synthesis attracts a lot in the vast expansion of living vital heterocyclic compounds. 1, 3-thiazoles ring system has found a wide spectrum of applications in the authority of Medicinal Chemistry (MC) because of some unique features like hydrogen bond formation, dipole-dipole, and π–π stacking interactions, stable to oxidation/reduction, and hydrolysis under acidic/ basic conditions. The current proposal for making novel bioactive molecules is about the incorporation of heterocycles and thiazole fraction in a single molecular structure[6]. This generates functionalized small molecules that were required by researchers to work in the area of MC and pharmaceutical/drug discovery.
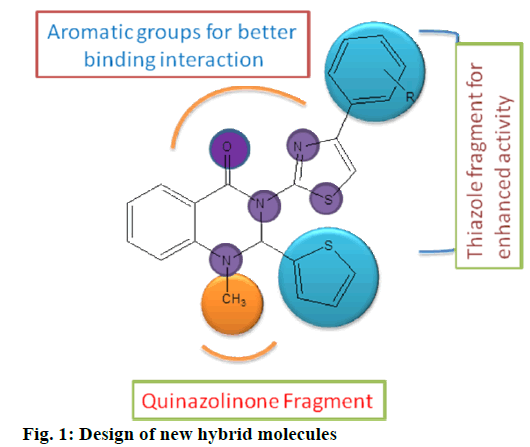
In this context, we anticipated that integration of three primary groups could be selected for the design, synthesis and SAR studies. Quinazolinone core, substituted phenyl groups and substituted 1,3-thiazole moiety in fig. 1. for the generation of small molecules as anti-tubercular agents. Molecular docking research had been performed at penicillin-binding protein 2 (PBP 2a) proteins, to study the interaction of the synthesized compounds[7]. The results of In-Silico studies indicate that quinazolinones containing 1, 3-thiazoles may serve as a potential new drug candidate in the combat against PDB code, 6Q9N and M. t H37Rv organism.
The present work is a three-fold. To synthesize a novel anti-tubercular agents based 1-Methyl-3-(4- Substituted phenyl-1, 3-thiazol-2yl)-2-(thiophen-2- yl)-2,3-dihydro quinazoline-4(1H)-ones framework, biological screening and docking studies. The later was to recognize the active binding site of the protein with respective to the compounds. These compounds were evaluated for their anti-tubercular activity by Micro plate Alamar Blue Assay (MABA)[8]. PBP 2a provides resistance required for the bacteria against various drugs. It cross links with the bacterial cell wall and also provides active site for binding. The complexity of the protein challenges the researcher to synthesize the compounds that readily bind and inhibit the protein.
Materials and Methods
All the chemicals and reagents are procured from commercial sources with analytical grade. The infrared spectra were recorded between 4000-400 cm-1 on Fourier-transform infrared spectroscopy (FTIR) Bruker 8400, Shimadzu. All 1H Nuclear magnetic resonance (NMR) and 13C NMR spectra were recorded on Bruker Avance III 500MHz NMR spectrophotometer (BIOSPIN) in CDCl3 solvent and using tetramethylsilane (TMS) as an internal standard. Mass spectra were recorded on the JEOL GCMATE II GC-MS instrument at 70 eV. Purification of compounds was done by Thin layer chromatography (TLC) where Cyclohexane and Ethyl acetate (2:1) acted as mobile phase.
Synthesis:
Synthesis of 4-Substituted phenyl-1,3-thiazol-2- amines (4a1-4k11):
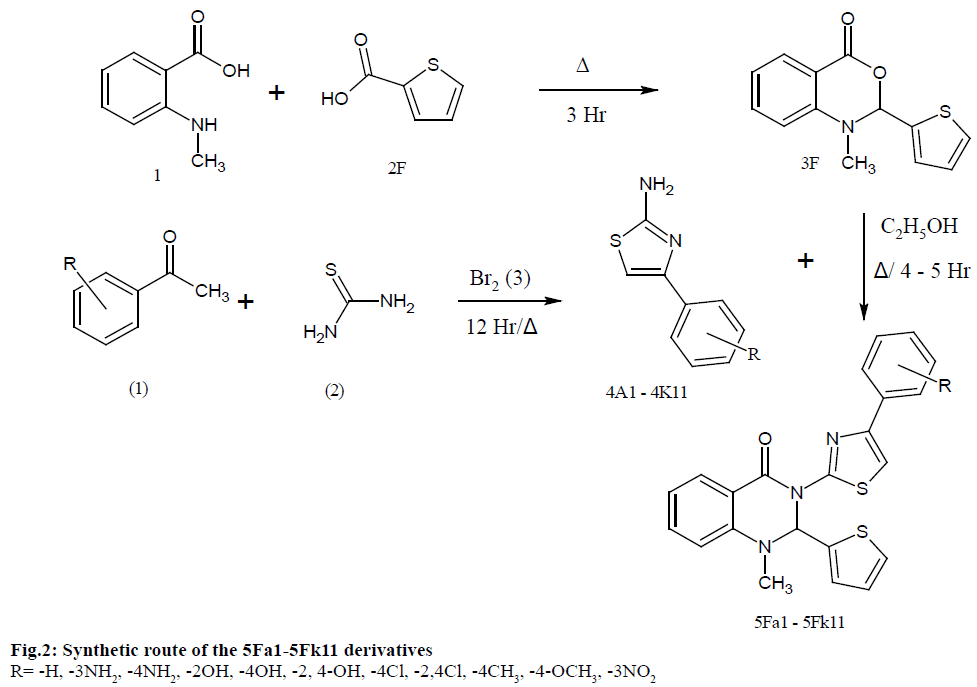
Compounds 4a1-4k11 were synthesized according to literature[9], mixture of substituted aromatic ketones (1) (0.1 mol), thiourea (2) 15 g (0.2 mol) and 100 ml of ethanol, bromine (3), 31.96 g (0.2 mol) was added drop wise. The above mixture was refluxed overnight on water bath until maximum solid has gone into the solution. The mixture was filtered. 2-amino-4- substituted phenyl-1, 3-thiazole was made alkaline using concentrated Ammonia. The obtained product was filtered, washed with a rectified spirit (rs), and warmed-over P2O5. It was recrystallized from rs, as colourless needles. The melting point (M.P.) of synthesized compounds from 4A1 to 4K11 was found to be 140, 120, 210, 140, 125, 145, 165, 150, 130, 140, and 146°, whereas the literature mp of phenyl thiazoles were 138, 126, 206, 152, 115, 139, 154, 143, 147, and 152°, respectively.
Synthesis of 1-methyl-2-(thiophen-2-yl)-1,2- dihydro-4H-3,1-benzoxazin-4-one (3F):
According to fig. 2, compound 3F was prepared in a 250 ml round bottom flask (RBF) N-Methyl anthranilic acid (1) 9.06 g (0.06 mol) was melted in 100 ml of benzene. To which 2-3 drops of pyridine and thiophen- 2-carboxylic acid (2F) 7.68 g (0.06 mol) was added under cold conditions. It was refluxed for 3 h, followed by cooling, filtration and washing using petroleum ether. Recrystallized with rs-acetone mixture (1:1) to get 1-methyl-2-(thiophen-2-yl)-1, 2-dihydro-4H-3, 1-benzoxazin-4-one (3F). The final product was determined with the aid of TLC using Cyclohexane and Ethyl acetate (2:1) as a mobile phase.
Synthesis 1-methyl-3-(4-substituted phenyl-1, 3-thiazol-2-yl)-2-(thiophen-2-yl)-2, 3-dihydro quinazolin-4(1H)-ones (5Fa1-5Fk11):
In a 100 ml RBF 1-methyl-2-(thiophen-2-yl)-1,2- dihydro-4H-3,1-benzoxazin-4-one (3F) 2.45 g (0.01 mol) was refluxed for 4-5 h with (0.01 mol) 4-substituted phenyl-1,3-thiazol-2-amine (4a1-4K11) in presence of K2CO3 in 100 ml of rs under anhydrous condition. The obtained solid was laved with hot water and recrystallized with rs-acetone mixture (1:1) and Yield was found as 79 %. (5Fa1). Remaining target quinazolinone–thiazole compounds were prepared in the same manner as above, and a series of 5Fa1-5Fk11 were synthesized and characterized by IR, 1H NMR, 13C NMR, and mass spectral analysis and its physical properties mentioned in Table 1.
| Code | R | Molecular Formula | Molecular Weight | Melting Point (°) |
Percentage Yield |
Rf Value |
|---|---|---|---|---|---|---|
| 5Fa1 | -H | C22H17N3OS2 | 403.51 | 192-195 | 68 | 0.32 |
| 5Fb2 | -3NH2 | C22H18N4OS2 | 418.53 | 180-185 | 80 | 0.23 |
| 5Fc3 | -4NH2 | C22H18N4OS2 | 418.53 | 110-115 | 86 | 0.62 |
| 5Fd4 | -2OH | C22H17N3O2S2 | 419.51 | 115-120 | 70 | 0.55 |
| 5Fe5 | -4OH | C22H17N3O2S2 | 419.51 | 152-157 | 76 | 0.52 |
| 5Ff6 | -2,4OH | C22H17N3O3S2 | 435.51 | 195-199 | 69 | 0.56 |
| 5Fg7 | -4Cl | C22H16ClN3OS2 | 437.96 | 170-175 | 68 | 0.55 |
| 5Fh8 | -2,4Cl | C22H15Cl2N3OS2 | 472.41 | 185-187 | 59 | 0.42 |
| 5Fi9 | -4CH3 | C23H19N3OS2 | 417.54 | 100-105 | 62 | 0.33 |
| 5Fj10 | -4OCH3 | C23H19N3O2S2 | 433.54 | 110-115 | 69 | 0.36 |
| 5Fk11 | -3NO2 | C22H16N4O3S2 | 448.51 | 160-165 | 70 | 0.39 |
Table 1: Physicochemical Properties of Synthesized Compounds (5FA1-5FK11)
Spectral characterization of synthesized compounds:
1-methyl-2-(thiophen-2-yl)-1, 2-dihydro-4H-3,1- benzoxazin-4-one (3F); Yield: 79 %; M.P. 152-155º; IR (cm-1): 2907 (C-H str.), 1661 (C=O str.), 1347 (C-N str.), 2991 (C-H aromatic), 1334 (C-O str. ether), 700 (C-S Str); m/z: (M+) 245.29; Anal: Calculated for C13H11NO2S.
1-methyl-3-(4-phenyl-1, 3-thiazol-2-yl)-2-(thiophen- 2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fa1); Yield: 68 %; M.P. 192-195°; IR (KBr, cm-1): 3126 (C-H Aromatic), 2926 (C-H str.), 1664 (C=O str. in ring), 1588 (C=N str.), 1268 (C-N str.), 724 (C-S str); 1H NMR (Dimethyl sulfoxide (DMSO)) δ (ppm) 2.85 (s, 3H, CH3), 6.01 (s, 1H), 7.20-7.50 (m, 5H, Ar-H), 6.6 (s, 1H), 6.55-7.99 (m, 4H, Ar-H), 6.66-6.99 (m. 3H): 13C NMR (DMSO) δ (ppm) 113.4, 115.0, 117.4, 127.4, 132.1, 148.1, 31.9, 60.2, 77.1, 122.6, 125.7, 125.9,138.6, 99.0, 147.2, 159.4, 126.5, 126.5, 127.8, 128.3, 128.3, 132.1: GCMS: m/z: (M+). 403, Anal. Calcd. For C22H17N3OS2.
3-[4-(3-aminophenyl)-1,3-thiazol-2-yl]-1-methyl- 2-(thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fb2); Yield: 80 %; M.P. 180-185ºC; IR (KBr, cm-1): 3115 (C-H Aromatic), 2930 (C-H str.), 1647 (C=O str. in ring), 1647 (C=N str.), 1328 (C-N str.), 717 (C-S str); 1H NMR (DMSO) δ (ppm) 2.95 (s, 3H, CH3), 6.01 (s, 1H), 6.32-7.17 (m, 4H, Ar-H), 6.4 (s, 1H), 6.65- 7.99 (m, 4H, Ar-H), 6.54-6.99 (m. 3H), 4.2 (s, 2H, NH2): 13C NMR (DMSO) δ (ppm) 114.4, 116.0, 118.4, 128.4, 133.1, 149.1, 32.9, 61.2, 78.1, 123.6, 126.7, 126.9,139.6, 100.0, 148.2, 160.4, 114.2, 116.3, 117.5, 130.1, 133.9, 148.1: GCMS: m/z: (M+). 418, Anal. Calcd. For C22H18N4OS2.
3-[4-(4-aminophenyl)-1,3-thiazol-2-yl]-1-methyl- 2-(thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fc3); Yield: 86 %; M.P. 110-115º; IR (KBr, cm-1): 2931 (C-H str.), 3157 (C-H Aromatic), 1661 (C=O str. in ring), 1661 (C=N str.), 665 (C-S str), 1331 (C-N str.); 1H NMR (DMSO) δ (ppm) 2.65 (s, 3H, CH3), 6.11 (s, 1H), 6.32-7.43 (m, 4H, Ar-H), 6.8 (s, 1H), 6.77-7.99 (m, 4H, Ar-H), 6.70-6.95 (m. 3H), 4.4 (s, 2H, NH2) : 13C NMR (DMSO) δ (ppm) 115.4, 117.0, 119.4, 129.4, 134.1, 150.1, 33.9, 62.2, 79.1, 124.6, 127.7, 127.9,140.6, 100.0, 148.2, 160.4, 116.6, 116.8, 123.1, 128.3, 128.3, 148.4: GCMS: m/z: (M+). 418, Anal. Calcd. For C22H18N4OS2.
3-[4-(2-hydroxyphenyl)-1,3-thiazol-2-yl]-1-methyl- 2-(thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fd4); Yield: 70 %; M.P. 115-120ºC; IR (KBr, cm-1): 2925 (C-H str.), 2985 (C-H Aromatic), 1655 (C=O str. in ring), 1688 (C=N str.), 1247 (C-N str.), 745 (C-S str); 1H NMR (DMSO) δ (ppm) 2.85 (s, 3H, CH3), 6.10 (s, 1H), 6.68-7.31 (m, 4H, Ar-H), 6.9 (s, 1H), 6.55-7.99 (m, 4H, Ar-H), 6.70-6.75 (m. 3H), 5.2 (s, 1H, OH): 13C NMR (DMSO) δ (ppm) 113.2, 116.4, 115.2, 128.4, 131.1, 149.1, 34.9, 61.2, 77.1, 123.6, 124.7, 126.9,139.6, 101.0, 148.2, 160.4, 116.4, 120.6, 121.9, 128.9, 130.2, 155.3: GCMS: m/z: (M+). 419, Anal. Calcd. For C22H17N3O2S2.
3-[4-(4-hydroxyphenyl)-1,3-thiazol-2-yl]-1-methyl- 2-(thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fe5); Yield: 76 %; M.P. 152-157°; IR (KBr, cm-1): 2925 (C-H str.), 3073 (C-H Aromatic), 1630 (C=O str. in ring), 1680 (C=N str.), 1260 (C-N str.), 719 (C-S str); 1H NMR (DMSO) δ (ppm) 2.25 (s, 3H, CH3), 6.21 (s, 1H), 6.65-7.45 (m, 4H, Ar-H), 6.6 (s, 1H), 6.55- 7.79 (m, 4H, Ar-H), 6.50-6.85 (m. 3H), 5.2, (s, 1H, OH): 13C NMR (DMSO) δ (ppm) 114.4, 116.0, 118.4, 129.4, 133.1, 146.1, 32.9, 61.2, 78.1, 125.6, 126.7, 126.9,139.6, 100.0, 148.2, 160.4, 116.4, 116.4, 125.9, 128.9, 128.9, 158.5: GCMS: m/z: (M+). 419, Anal. Calcd. For C22H17N3O2S2.
3-[4-(2,4-dihydroxyphenyl)-1,3-thiazol-2-yl]-1- methyl-2-(thiophen-2-yl)-2,3-dihydroquinazolin- 4(1H)-one (5Ff6); Yield: 69 %; M.P. 195-199º; IR (KBr, cm-1): 2822 (C-H str.), 3074 (C-H Aromatic), 1634 (C=O str. in ring), 1590 (C=N str.), 1322 (C-N str.), 689 (C-S str); 1H NMR (DMSO) δ (ppm) 2.85 (s, 3H, CH3), 6.25 (s, 1H), 6.20-7.20 (t, 3H, Ar-H), 6.4 (s, 1H), 6.70-7.79 (m, 4H, Ar-H), 6.72-6.95 (m. 3H), 5.2, 5.0 (s, 1H, 1H, OH): 13C NMR (DMSO) δ (ppm) 114.4, 113.0, 118.4, 128.4, 133.1, 147.1, 33.9, 61.2, 78.1, 123.6, 126.7, 126.9,139.6, 100.0, 148.2, 160.4, 104.1, 109.0, 130.3, 132.2, 156.7, 159.9: GCMS: m/z: (M+). 435, Anal. Calcd. For C22H17N3O3S2.
3-[4-(4-chlorophenyl)-1,3-thiazol-2-yl]-1-methyl- 2-(thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fg7); Yield: 68 %; M.P. 170-175º; IR (KBr, cm-1): 2927 (C-H str.), 3113 (C-H Aromatic), 1634 (C=O str. in ring), 1529 (C=N str.), 1335 (C-N str.), 665 (C-S str); 1H NMR (DMSO) δ (ppm) 2.65 (s, 3H, CH3), 6.21 (s, 1H), 6.4 (s, 1H), 7.25-7.50 (m, 4H, Ar-H), 6.55-7.99 (m, 4H, Ar-H), 6.70-6.75 (m. 3H): 13C NMR (DMSO) δ (ppm) 112.4, 116.0, 118.4, 126.4, 133.1, 149.1, 32.9, 61.2, 78.1, 123.6, 126.7, 126.9,139.6, 100.0, 148.2, 160.4, 128.9, 128.9, 129.4, 129.4, 131.2, 134.3: GCMS: m/z: (M+). 437, Anal. Calcd. For C22H16ClN3OS2.
3-[4-(2,4-dichlorophenyl)-1,3-thiazol-2-yl]-1-methyl- 2-(thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fh8); Yield: 59 %; M.P. 185-187º; IR (KBr, cm-1): 2644 (C-H str.), 3143 (C-H Aromatic), 1660 (C=O str. in ring), 1588 (C=N str.), 1252 (C-N str.), 706 (C-S str); 1H NMR (DMSO) δ (ppm) 2.80 (s, 3H, CH3), 6.21 (s, 1H), 6.4 (s, 1H), 7.10-7.36 (m, 3H, Ar-H), 6.59-7.99 (m, 4H, Ar-H), 6.50-6.78 (m. 3H): 13C NMR (DMSO) δ (ppm) 114.4, 116.0, 118.4, 128.4, 133.1, 149.1, 32.9, 61.2, 78.1, 123.6, 126.7, 126.9,139.6, 100.0, 148.2, 160.4, 127.5, 128.1, 130.3, 130.9, 133.7, 135.7: GCMS: m/z: (M+). 472, Anal. Calcd. For C22H15Cl2N3OS2.
1-methyl-3-[4-(4-methylphenyl)-1,3-thiazol-2-yl]- 2-(thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fi9); Yield: 62 %; M.P. 100-105º; IR (KBr, cm-1): 2922 (C-H str.), 3125 (C-H Aromatic), 1651 (C=O str. in ring), 1629 (C=N str.), 1336 (C-N str.), 708 (C-S str); 1H NMR (DMSO) δ (ppm) 2.80 (s, 3H, CH3), 2.45 (s, 3H, CH3), 6.20 (s, 1H), 6.3 (s, 1H), 7.10-7.36 (m, 4H, Ar-H), 6.59-7.99 (m, 4H, Ar-H), 6.70-6.85 (m. 3H): 13C NMR (DMSO) δ (ppm) 114.4, 116.0, 118.4, 128.4, 133.1, 149.1, 32.9, 61.2, 78.1, 123.6, 126.7, 126.9,139.6, 100.0, 148.2, 160.4, 127.4, 127.4, 129.6, 129.6, 130.1, 138.4, 24.3: GCMS: m/z: (M+). 417, Anal. Calcd. For C23H19N3OS2.
3-[4-(4-methoxyphenyl)-1,3-thiazol-2-yl]-1-methyl- 2-(thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)- one (5Fj10); Yield: 69 %; M.P. 110-115ºC; IR (KBr, cm-1): 2927 (C-H str.), 3157 (C-H Aromatic), 1651 (C=O str. in ring), 1608 (C=N str.), 1255 (C-N str.), 727 (C-S str); 1H NMR (DMSO) δ (ppm) 2.60 (s, 3H, CH3), 3.93 (s, 3H, OCH3), 5.80 (s, 1H), 6.1 (s, 1H), 6.70-7.52 (m, 4H, Ar-H), 6.77-7.99 (m, 4H, Ar-H), 6.71-6.95 (m. 3H): 13C NMR (DMSO) δ (ppm) 114.4, 116.0, 118.4, 128.4, 133.1, 149.1, 32.9, 61.2, 78.1, 123.6, 126.7, 126.9,139.6, 100.0, 148.2, 160.4, 114.8, 114.8, 125.4, 128.4, 128.4, 160.7, 55.9: GCMS: m/z: (M+). 433, Anal. Calcd. For C23H19N3O2S2.
1-methyl-3-[4-(3-nitrophenyl)-1,3-thiazol-2-yl]-2- (thiophen-2-yl)-2,3-dihydroquinazolin-4(1H)-one (5Fk11); Yield: 70%; M.P. 160-165ºC; IR (KBr, cm- 1): 2925 (C-H str.), 3108 (C-H Aromatic), 1653 (C=O str. in ring), 1653 (C=N str.), 1256 (C-N str.), 676 (C-S str); 1H NMR (DMSO) δ (ppm) 2.89 (s, 3H, CH3), 6.29 (s, 1H), 6.9 (s, 1H), 7.50-8.55 (m, 4H, Ar-H), 6.77-7.89 (m, 4H, Ar-H), 6.70-6.75 (m. 3H): 13C NMR (DMSO) δ (ppm) 114.4, 116.0, 118.4, 128.4, 133.1, 149.1, 32.9, 61.2, 78.1, 123.6, 126.7, 126.9,139.6, 100.0, 148.2, 160.4, 121.6, 121.6, 128.4, 128.4, 139.2, 148.4: GCMS: m/z: (M+). 448, Anal. Calcd. For C22H16N4O3S2.
Anti-tubercular activity:
The stock solution was made as per the Centres for Disease Control and Prevention (CDC 1985) recommendations. The successive concentrations had been used for the screening of standard antimicrobial agents STP and INH: 3.25 and 6.25 μg/ml. The standard drugs were stored as a stock suspension (1 %) using distilled water or else methanol and stored at -25°[10]. 100 μg/ml stock solution was prepared with DMSO (1 %), from which 50, 25, and 12.5 to 0.2μg/ml concentrations were obtained using serial dilution technique.
Middlebrook 7H9 Broth medium preparation:
It is a liquid growth medium, mainly used for the culture of Mycobacterium species (Ms). 7H9 improves the growth of Ms, as it posses nutrients such as glycerol, oleic acid, albumin, and dextrose (GOAD). Cultures need to be studied between 5th and 7th d of a w, which is continued for 8 w after inoculation. Numerous developments were found using Middlebrook broth viz. antimicrobial assays, biochemical test (arylsulfatase and tellurite reduction), and maintenance of stock strains[11].
Random Screening for anti-tubercular activity (Alamar –Blue assay):
The anti-mycobacterial activity of compounds was assessed for Mycobacterium, sensitive to STP and INH-H37RV, using the MABA[12,13]. This nontoxic approach uses a thermal-stable reagent, and it suits for screening the anti-mycobacterial activity randomly. During incubation, 200 μl of sterile deionised water was added to the outer perimeter of a pure plate of 96 wells. This minimizes the evaporation of medium in the test wells. Later, these 96 well plates were filled with 100 μl of Middlebrook 7H9 broth[14] (with loopful inoculum of bacteria-108 colony forming units (CFU)), suitable compounds and standards with different dilutions. The plates were wrapped with parafilm, and then incubated at 37° for 120 h. In addition to this, add 25 μl of freshly prepared 1:1 mixture of Alamar blue reagent and 10 % tween 80 and it was incubated for one day. The well with blue colour indicates no mycobacterial growth and pink colour for mycobacterial growth.
Molecular Docking studies:
Selection of protein:
In structural biology, biomolecular docking (Deoxyribonucleic acid (DNA)-Ligand docking) is becoming a popular approach to understand the biomolecular interactions responsible for affinity to the target. In this study, we selected the DNA target (PDB ID: 6Q9N) from the Protein Data Bank (PDB)[15,16]. Protein is selected based on the various factors, i.e., the resolution should be 1.5-2.5 Ao, it should contain co-crystallized Ligand, and the structure should be evaluated by X-ray diffraction. Selected protein should not contain breaks in their three dimensional structure.
Preparation of ligand:
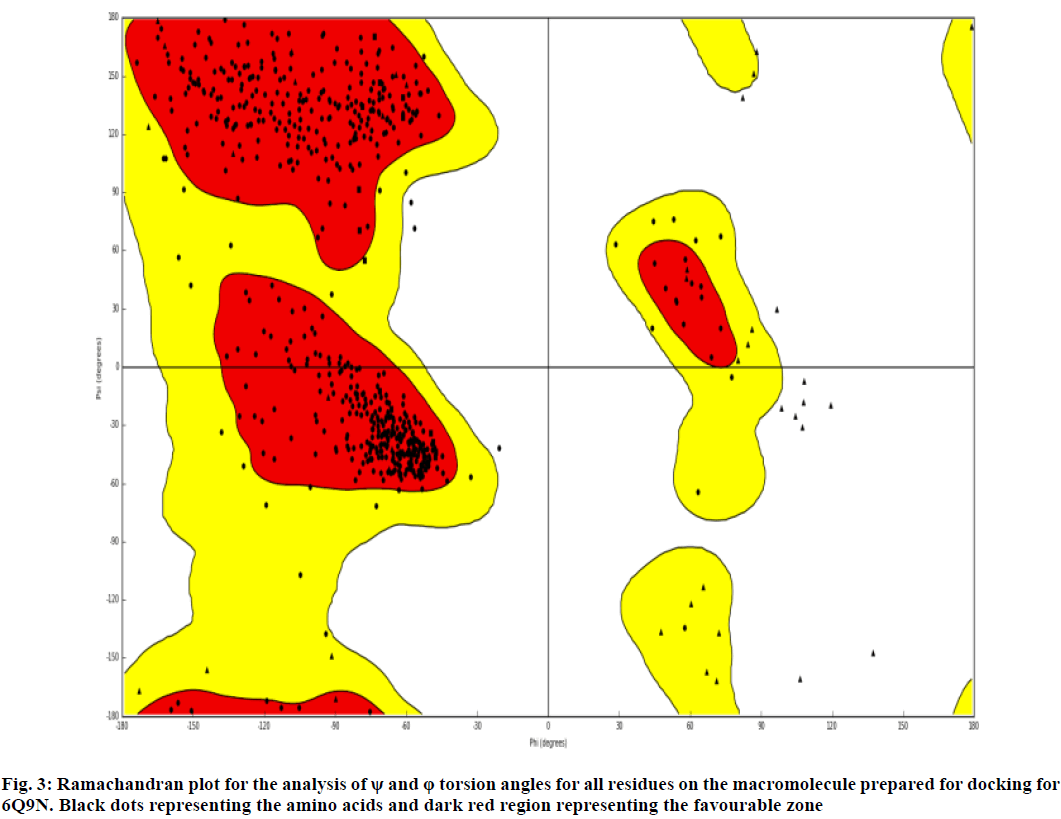
The generated structures were imported into maestro Schrödinger software (Version 2020_1) by using SMILES notation in two dimensional sketcher. The protein structure imported by giving the code 6Q9N in protein preparation wizard (it uses Protein Data Bank (http://www.rcsb.org) online database to import the protein). Then the protein is pre-processed by assigning bond orders, adding hydrogen’s, create disulfide bonds, etc. The co-crystal ligand 6Q9N was kept as such. The two-dimensional Ramachandran plot represents the allowed and disfavoured values of phi (φ) and psi (ψ). This plot shows the torsional angles (φ) and (ψ) of the residues (amino acids) contained in a peptide represented in fig. 3.
In maestro task, select receptor grid generation suitable for peptide docking by choosing any one atom of the ligand molecule on screen; it shows grid box with X, Y, and Z coordinates -17.81, -25.5 and 69.87 respectively. Then the other options: site, constraints, and rotatable groups had been enabled. By clicking run, the grid box started its execution[17,18].
In Silico Absorption, Distribution, Metabolism, Excretion and Toxicity (ADMET) prediction by Swiss Absorption, Distribution, Metabolism, Excretion (ADME) tool:
After designing and studying the physicochemical properties about the INH and RIF, which is mandatory to match the pharmacokinetics properties, i.e., absorption in the body, distribution into the different compartments, metabolism by organs, and elimination through the body[19]. It was mandatory to perform a computational study to divine the ADME parameters of the designed molecules to prioritize the molecules for synthesis[20]. Hence, the ADME study is an essential step for checking the drug-likeness. ADME study of the selected whole docked library was carried out using the QikProp[21]. The present study, properties like molecular weight predicted central nervous system activity, octanol/water partition coefficient, aqueous solubility, inhibitory concentration (IC)-50 value for the blockage of MAPK channels, cell permeability, binding of a drug to human serum albumin, no. of hydrogen bond donors and acceptors was calculated for each compound. The drug-likeness was evaluated[22]. Moreover, the properties of the Ligand to noxious and carcinogenicity were analyzed online using the ADMET SAR and Swiss ADME[23]. QSAR studies and drug-likeness are also predicted to know the octanol/ water partition coefficient (log Po/w), Topological polar surface area (TPSA), Hydrogen bond Acceptor (HBA), Hydrogen Bond Donor (HBD), Lipinski Rule and synthetic accessibility which are tabulated in Table 2.
| Code | RB | HBA | HBD | MR | Log S & Class |
GI absorption & Blood brain barrier permeant |
log Kp (cm/s) | Synthetic Accessibility | Log S | TPSA | Lipinski Rule |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 5Fa1 | 3 | 2 | 0 | 122.19 | -6.9 & Poorly soluble |
High & No | -5.07 | 3.78 | -6.9 | 92.92 | 0 |
| 5Fb2 | 3 | 2 | 1 | 126.59 | -6.74 & . Poorly soluble |
High & No | -5.64 | 3.88 | -6.74 | 118.94 | 0 |
| 5Fc3 | 3 | 2 | 1 | 126.59 | -6.74 & Poorly soluble |
High & No | -5.64 | 3.86 | -6.74 | 118.94 | 0 |
| 5Fd4 | 3 | 3 | 1 | 124.21 | -6.96 & Poorly soluble |
High & No | -5.42 | 3.84 | -6.96 | 113.15 | 0 |
| 5Fe5 | 3 | 3 | 1 | 124.21 | -6.96 & Poorly soluble |
High & No | -5.42 | 3.79 | -6.96 | 113.15 | 0 |
| 5Ff6 | 3 | 4 | 2 | 126.23 | -7.01 & Poorly soluble |
Low & No | -5.77 | 3.88 | -7.01 | 133.38 | 0 |
| 5Fg7 | 3 | 2 | 0 | 127.2 | -7.55 & Poorly soluble |
High & No | -4.83 | 3.78 | -7.55 | 92.92 | 0 |
| 5Fh8 | 3 | 2 | 0 | 132.21 | -8.21 & Poorly soluble |
High & No | -4.6 | 3.85 | -8.21 | 92.92 | 0 |
| 5Fi9 | 3 | 2 | 0 | 127.15 | -7.28 & Poorly soluble |
High & No | -4.89 | 3.9 | -7.28 | 92.92 | 0 |
| 5Fj10 | 4 | 3 | 0 | 128.68 | -7.07 & Poorly soluble |
High & No | -5.27 | 3.89 | -7.07 | 102.15 | 0 |
| 5Fk11 | 4 | 4 | 0 | 131.01 | -7.68 & Poorly soluble |
Low& No | -5.46 | 3.89 | -7.68 | 138.74 | 0 |
| INH | 2 | 3 | 2 | 35.13 | -0.25 & Very soluble |
High & No | -7.63 | 1.24 | -0.25 | 68.01 | 0 |
| RIF | 5 | 14 | 6 | 234.22 | -9.84 & Poorly soluble |
Low& No | -7.44 | 9.23 | -9.84 | 220.15 | 3 |
R.B: Rotatable Bonds, HBA: Hydrogen Bond Acceptors, HBD: Hydrogen Bond Donors, M.R: Molar Refractive, TPSA: Topologocal Polar Surface Area
Table 2: In Silico Admet Properties of the Synthesised Compounds 5FA1-5FK11
Lipinski’s rule of five:
Given by Lipinski[24] in 1997, the rule of five is based on certain criteria to estimate drug-likeness of a molecule having a pharmacological activity. These criteria are log P lower than 5, number of HBD<5, HBA<10 and M.W not exceeding 500 Da. The rule is used in drug design to preselect molecules presenting good absorption, distribution, metabolism, and excretion (ADME) properties that must have a medicament in the organism. We have used the ADME property calculator (http://www.SwissADME.com) to calculate the four parameters of Lipinski’s rule in addition to the number of rotatable bonds that have to be inferior to 10 to have a good oral bioavailability[25].
Results and Discussion
The synthesis of 1-methyl-3-(4-substituted phenyl-1,3- thiazol-2-yl)-2-(thiophen-2-yl)-2,3-dihydro quinazolin- 4(1H)-one (5Fa1-5Fk11) were achieved through the steps outlined in Scheme 1. The intermediate 1-methyl- 2-(thiophen-2-yl)-1, 2-dihydro-4H-3, 1-benzoxazin-4- one (3F) was obtained by reacting N-Methyl Anthranilic acid (1) with thiophen-2-carboxylic acid (2F). Aromatic aldehydes (1), thiourea (2) were mixed in ethanol along with bromine (3) to obtain second intermediate 4-Substituted phenyl-1, 3-thiazol-2-amine (4a1-4k11). Finally, the desired title compounds were obtained by reaction of the intermediates 1-methyl-2-(thiophen-2- yl)-1,2-dihydro-4H-3,1-benzoxazin-4-one with various 4-Substituted phenyl-1,3-thiazol-2-amines. All the compounds obtained were at good yields, characterized by spectral analysis, and examine the anti-tubercular activity to find a potent molecule.
The anti-mycobacterium activity of the synthesized derivatives had been evaluated at different concentrations, time intervals and minimum inhibitory concentration (MIC) was investigated and is postulated in Table 3. All the compounds were evaluated for mycobacteria inhibitory action at different strengths (100 to 0.2 μg/ml); STP and INH were taken as standard/control. All the compounds exhibited >90 % growth inhibition at the strength of 6.25 and 12.5 μg/ml. Screening of compounds 5Fa1-5Fk11 towards tuberculosis showed that some compounds had mild to good inhibitory activity which is evident from their MIC values. Compounds 5Ff6, 5Fe5 and 5Fb2 show prominent activity, compared to other synthesized compounds.
| Code | MIC (mm) | 100 | 50 | 25 | 12.5 | 6.25 | 3.125 | 1.6 | 0.8 | 0.4 | 0.2 (µg/ml) |
||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5Fa1 | 22 | S | S | S | R | R | R | R | R | R | R | ||
| 5Fb2 | 25 | S | S | S | S | S | R | R | R | R | R | ||
| 5Fc3 | 21 | S | S | S | R | R | R | R | R | R | R | ||
| 5Fd4 | 22 | S | S | S | R | R | R | R | R | R | R | ||
| 5Fe5 | 28 | S | S | S | S | R | R | R | R | R | R | ||
| 5Ff6 | 30 | S | S | S | S | S | R | R | R | R | R | ||
| 5Fg7 | 18 | S | S | R | R | R | R | R | R | R | R | ||
| 5Fh8 | 10 | S | R | R | R | R | R | R | R | R | R | ||
| 5Fi9 | 7 | S | R | R | R | R | R | R | R | R | R | ||
| 5Fj10 | 28 | S | S | S | R | R | R | R | R | R | R | ||
| 5Fk11 | 28 | S | S | S | R | R | R | R | R | R | R | ||
| PZA | 32 | 3.125 µg/ml | |||||||||||
| STR | 36 | 6.25 µg/ml | |||||||||||
| CIP | 37 | 3.125 µg/ml | |||||||||||
Strain Used: M. Tuberculosis (H37 RV Strain)
R: Resistance, S: Sensitive; MIC: Minimum Inhibitory Concentration
Table 3: MIC Data for the Anti-Mycobacterial Activity of Target Compounds Obtained by the Microdilution Method toward Micobacterium Tuberculosis H37RV
The integration of three primary groups selected for the design, synthesis, and SAR studies are quinazolinone core, substitution on second and third positions, and presence of 1, 3-thiazole moiety. Observation of Table 3 (anti-TB investigations): it is clear that compound 5Ff6 having 2, 4-dihydroxy phenyl group on 1, 3-thiazole moiety exhibit potent activity followed by 5Fb2, 5Fe5. Based on the Lipinski,s rule criteria, all the molecules fulfill predictions, and it shows LogP, TPSA, HBA, and HBD are within the range, which is tabulated in Table 4. Suggesting their good pharmacokinetic permeability and their oral bioavailability, which may allow them to constitute lead compounds for cancer treatment.
| Code | Interacting amino acids |
Binding energy (Kcal/mol) |
Glide energy |
|---|---|---|---|
| 5Fa1 | Tyr105, Lys273, Lys316 | -3.391 | -31.521 |
| 5Fb2 | Tyr105, Lys273, Gly271, Lys316 | -4.048 | -34.226 |
| 5Fc3 | Agn104 | -3.8 | -33.226 |
| 5Fd4 | Tyr297, Asp275 | -3.919 | -33.901 |
| 5Fe5 | Asn104, Tyr297 | -4.022 | -36.228 |
| 5Ff6 | Agn104, Lys273, Tyr297 | -4.2 | -37.965 |
| 5Fg7 | Tyr105, Lys273, Tyr297 | -2.916 | -33.599 |
| 5Fh8 | Lys273, Tyr297 | -3.11 | -37.26 |
| 5Fi9 | Tyr105, Lys273, Tyr297 | -3.031 | -32.625 |
| 5Fj10 | Tyr105, Lys273, Tyr297 | -3.092 | -33.587 |
| 5Fk11 | Tyr105, Lys273, Lys316 | -3.797 | -35.577 |
| 6Q9N | Asn104, Tyr297 | -3.899 | -50.289 |
| STP | Tyr105, Asn146, Lys273, Asp275, Gly395, Asp295,Lys316 |
-5.169 | -19.985 |
| INH | Lys273, Asp295 | -4.743 | -35.056 |
Table 4: Docking Score (KCAL/MOL) of the Compounds 5FA1-5FK11 with Protein Binding Protein 2A (6Q9N)
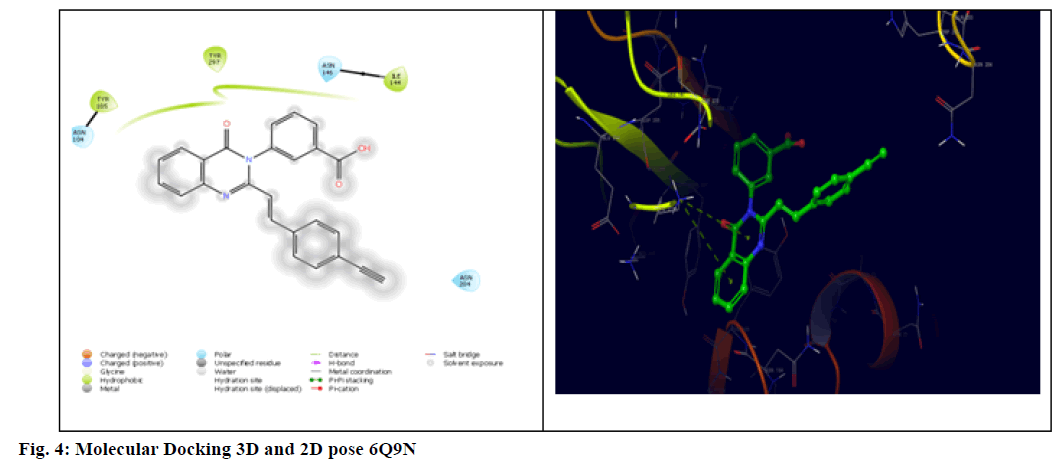
The three-dimensional crystal structure of penicillin-binding protein 2a complex with 3-[2-[({E})-2- (4-ethynyl phenyl)ethynyl]-4-oxidanylidenequinazolin- 3-yl] benzoic acid (6Q9N) was taken from protein data bank[26]. All the derivatives were docked against PBP 2a protein by using Schrödinger software (Version 2020_1). Docking score and Binding energies were measured for all the compounds. Good docking score was obtained due to the presence of H-bond interactions, and Vander Waal forces. The docking score of each ligand with respective to interaction energy were calculated and mentioned in Table 4 along with accompanying amino acids.
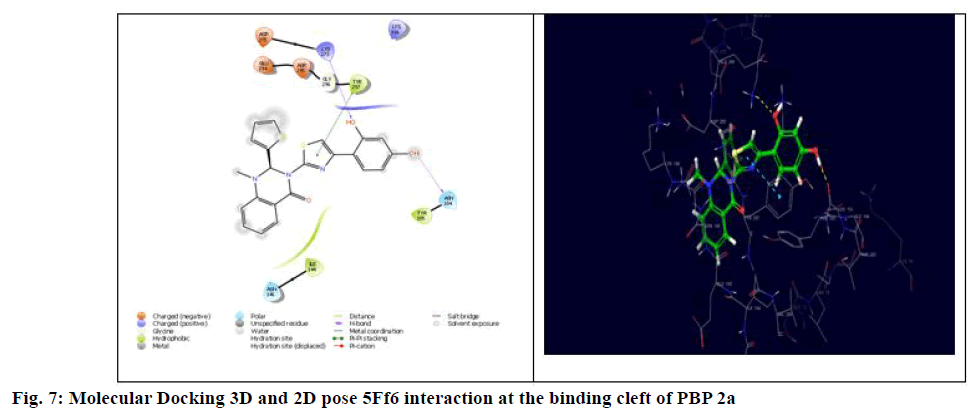
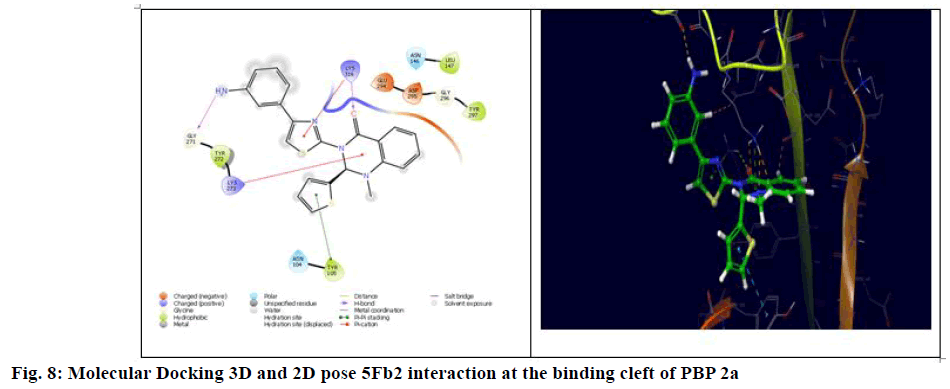
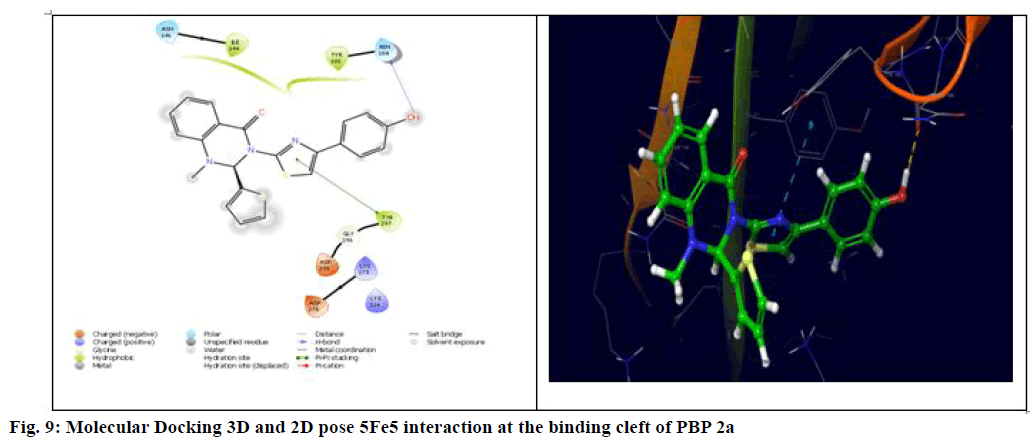
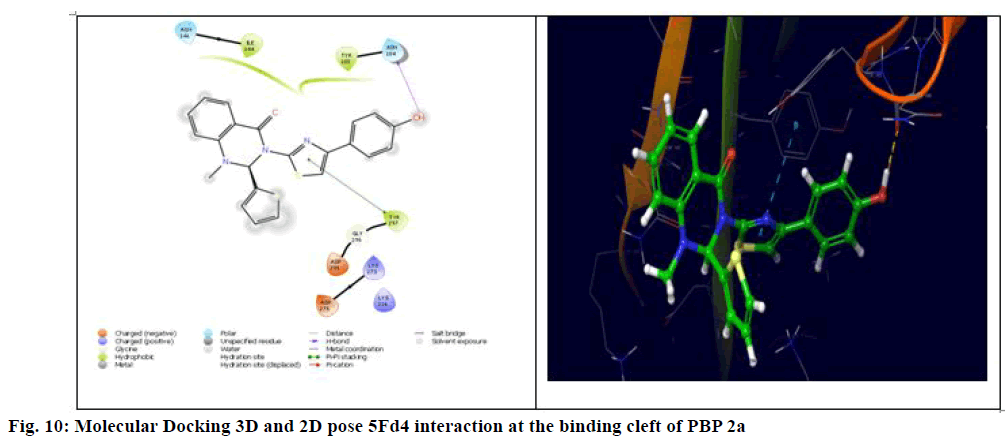
The generalized structure of 6Q9N and docking ligand were shown in fig. 4. From the data, it is observed that except 5Fg7 (the interaction energy of -2.916 kcal/mol), all other synthesized compounds showed interaction energy similar to standard and Lys273 and Tyr297 as common interacting amino acids. Compound 5Ff6 showed interaction energy of -4.2 kcal/mol with Agn104, Lys273, and Tyr297 were shown in fig. 7. Compound 5Fb2 showed interaction energy of −4.048 kcal/mol with Tyr104, Gly271, Lys273, and Lys316 was shown in fig. 8. Compound 5Fe5 showed binding energy of -4.022 kcal/mol with Asn104 and Tyr297 was shown in fig. 9. Compound 5Fd4 showed interactions with Asn275 and Tyr297 and has the binding power of −3.919 kcal/mol was shown in fig. 10.
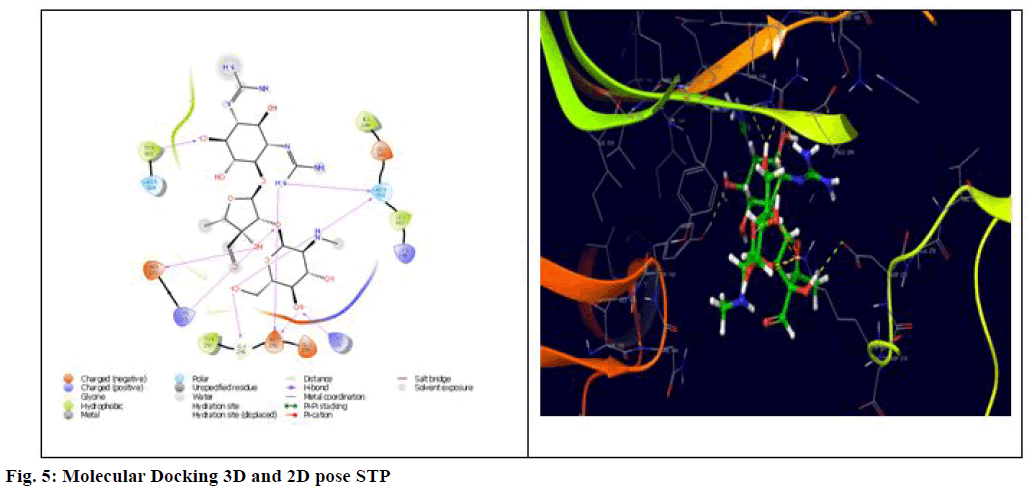
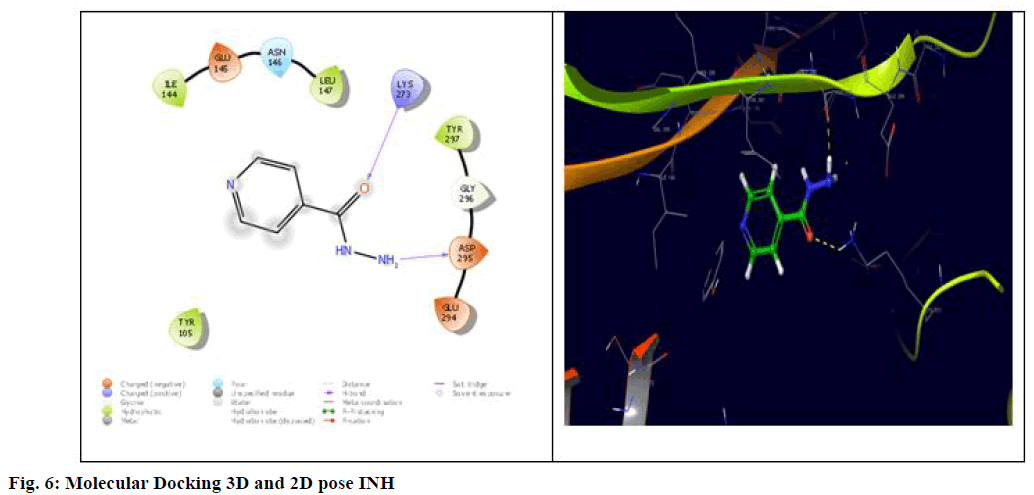
Finally, these interaction energies were compared to the standard drug molecules STP and INH, where it showed binding energy of -5.169 and -4.743 with Lys273, Asp295 and Tyr 105, Asn146, Asp275, Lys273, Tyr294, Gly296, Asp295, Lys316 respectively shown in fig. 5 and 6. This indeed suggests that except 5Fc3, the remained compounds showed equivalent interaction energies compared to STP and INH. Overall, docking studies reveal that the interaction with Asn104, Lys273 and Tyr297 is very crucial in the design of new chemical entities for potent inhibition of PBP 2a protein and suggested that 5Ff6 (R=2,4-OH), 5Fb2 (R=-3 NH2), 5Fe5 (R=4-OH) and 5Fd4 (R=2-OH) can be potent candidatures for the inhibition against the standard.
A sequence of novel 1.3-thiazole hybrids linked to quinazolin-4-ones were synthesized, characterized and evaluated for anti-tubercular activity. The majority of synthesized derivatives (5Fa1-5Fk11) displayed moderate to promising action against the tested M. tuberculosis H37Rv organism. Compounds 5Ff6, 5Fe5 and 5Fb2 showed significant activity with MIC between 12.5 and 6.25 μg/ml. Furthermore, molecular modelling study indicated that the interactions of compounds 5Ff6, 5Fb2, 5Fe5 and 5Fd4 with Asn104, Lys273, Asp275, Asp295 and Tyr297 amino acids of PBP 2a were responsible for biological activity they exhibited dock score which was close enough when compared with standard. Moreover Swiss ADME results ensure the similarity of compounds with standard in safety and efficacy.
Acknowledgments:
Thankful to DST-SERB facility of molecular modelling and Drug Discovery division RERDS – CPR, RIPER, Anantapur – 515721, Andhra Pradesh, India for supporting in Docking studies.
Conflict of interest:
The authors declared no conflict of interest.
References
- Cek M, Lenk S, Naber KG, Bishop MC, Johansen TE, Botto H, et al. EAU guidelines for the management of genitourinary tuberculosis. Eur Urol 2005;48(3):353-62.
- Tobe M, Isobe Y, Tomizawa H, Nagasaki T, Obara F, Matsumoto M, et al. Structure-Activity Relationships of 6-Nitroquinazolines: Dual-Acting Compounds with Inhibitory Activities toward both TNF-α Production and T cell Proliferation. Chem Pharm Bull 2002;50(8):1073-80.
- Xu GF, Song BA, Bhadury PS, Yang S, Zhang PQ, Jin LH, et al. Synthesis and antifungal activity of novel s-substituted 6-fluoro-4-alkyl (aryl) thioquinazoline derivatives. Bioorg Med Chem 2007;15(11):3768-74.
- Raffa D, Daidone G, Schillaci D, Maggio B, Plescia F. Synthesis of new 3-(3-phenyl-isoxazol-5-yl) or 3-[(3-phenyl-isoxazol-5-yl)-amino] substituted 4 (3H)-quinazolinone derivatives with antineoplastic activity. Die Pharmazie 1999;54(4):251-4.
- Chenard BL, Welch WM, Blake JF, Butler TW, Reinhold A, Ewing FE, et al. Quinazolin-4-one α-Amino-3-hydroxy-5-methyl-4-isoxazole propionic Acid (AMPA) Receptor Antagonists: Structure-Activity Relationship of the C-2 Side Chain Tether. J Med Chem 2001;44(11):1710-7.
- Hosseinzadeh L, Aliabadi A, Rahnama M, Sadeghi HM, Khajouei MR. Synthesis and cytotoxic evaluation of some new 3-(2-(2-phenylthiazol-4-yl) ethyl)-quinazolin-4 (3H) one derivatives with potential anticancer effects. Res Pharm Sci 2017;12(4):290.
- Fedarovich A, Djordjevic KA, Swanson SM, Peterson YK, Nicholas RA, Davies C. High-throughput screening for novel inhibitors of Neisseria gonorrhoeae penicillin-binding protein 2. PLoS One 2012;7(9):e44918.
- Vanitha JD, Paramasivan CN. Evaluation of microplate Alamar blue assay for drug susceptibility testing of Mycobacterium avium complex isolates. Diagn Microbiol Infect Dis 2004;49(3):179-82.
- Saravanan G, Alagarsamy V, Pavitra TG, Kumar GC, Savithri Y, Naresh L, et al. Synthesis, characterization and anti-microbial activities of novel thiazole derivatives. Int J Pharma Biosci 2010;1:6-11.
- Leite CQ, Beretta AL, Anno IS, Telles MA. Standardization of broth microdilution method for Mycobacterium tuberculosis. Mem Inst Oswaldo Cruz 2000;95(1):127-9.
- Larsen MH, Biermann K, Jacobs Jr WR. Laboratory maintenance of Mycobacterium tuberculosis. Curr Protoc Microbiol 2007;6(1):10A-1.
- Zandhaghighi M, Ghazvini K, Meshkat Z, Rezaee SA, Derakhshan M, Soleimanpour S, et al. Screening of the anti-mycobacterial activity of novel lipophilic agents by the modified broth-based method. J Clin Tuberc Other Mycobact Dis 2016;3:1-5.
- Rajasekhar KK, Nizamuddin ND, Surur AS, Mekonnen YT. Synthesis, characterization, anti-tubercular and antibacterial activity, and molecular docking of 2, 3-disubstituted quinazolinone derivatives. Res Rep Med Chem 2016; 6:15-26.
- Cho S, Lee HS, Franzblau S. Microplate Alamar blue assay (MABA), and low oxygen recovery assay (LORA) for Mycobacterium tuberculosis. Methods Mol Biol 2015;1285:281-92.
- Patel SB, Cameron PM, O'Keefe SJ, Frantz-Wattley B, Thompson J, O'Neill EA, et al. The three-dimensional structure of MAP kinase p38β: different features of the ATP-binding site in p38β compared with p38α. Acta Crys 2009;65(8):777-85.
- Lungu CN, Diudea MV. Binding site and potency prediction of Teixobactin and other Lipid II ligands by statistical base scoring of conformational space maps. Cur Comp Aided Drug Des 2018;14(1):29-34.
- Sharma D, Kumar S, Narasimhan B, Ramasamy K, Lim SM, Shah SA, et al. 4-(4-Bromophenyl)-thiazol-2-amine derivatives: synthesis, biological activity and molecular docking study with ADME profile. BMC Chem 2019;13(1):60.
- McTigue M, Murray BW, Chen JH, Deng YL, Solowiej J, Kania RS. Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc Natl Acad Sci 2012;109(45):18281-9.
- Winiwarter S, Ahlberg E, Watson E, Oprisiu I, Mogemark M, Noeske T, et al. In silico ADME in drug design-enhancing the impact. ADMET and DMPK. 2018;6(1):15-33.
- Aswathy L, Jisha RS, Masand VH, Gajbhiye JM, Shibi IG. Design of novel amyloid β aggregation inhibitors using QSAR, pharmacophore modeling, molecular docking and ADME prediction. In-silico Pharmcol 2018;6(1):12.
- Cheng F, Li W, Zhou Y, Shen J, Wu Z, Liu G, et al. admetSAR: a comprehensive source and free tool for assessment of chemical ADMET properties.
- Zhang X, Perez-Sanchez H, C Lightstone F. A comprehensive docking and MM/GBSA rescoring study of ligand recognition upon binding antithrombin. Cur Top Med Chem 2017;17(14):1631-9.
- Daina A, Michielin O, Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 2017;7:42717.
- Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Del Rev 1997;23(1-3):3-25.
- Veber DF, Johnson SR, Cheng HY, Smith BR, Ward KW, Kopple KD. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem 2002;45(12):2615-23.
- Janardhanan J, Bouley R, Martínez-Caballero S, Peng Z, Batuecas-Mordillo M, Meisel JE, et al. The quinazolinone allosteric inhibitor of PBP 2a synergizes with piperacillin and tazobactam against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 2019;63(5):e02637-18.