- *Corresponding Author:
- H. R. Badwaik
Rungta College of Pharmaceutical Sciences and Research
Bhilai 490024,
Chhattisgarh,
India
E-mail: hemantrbadwaik@gmail.com
| Date of Received | 20 December 2020 |
| Date of Revision | 24 September 2021 |
| Date of Acceptance | 12 April 2022 |
| Indian J Pharm Sci 2022;84(2):453-464 |
Abstract
Hyperlipidemia is characterized by a rise in high-density lipoproteins and decreases in low-density lipoproteins and triglyceride level in blood serum. Numerous prescription drugs for hyperlipidemia are available, but each has significant side effects. Many works were motivated in this area by the significant pharmacological action of fused chromene. Throughout the present research, new substituted derivatives were synthesized and tested for the anti-hyperlipidemic activity for benzo[h]chromene-3 carboxylate derivatives. Molecules docking showed that compound Vf has greater energy affinity binding values (-12.898 kcal/mol) with 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase enzyme compared with atorvastatin (-11.8605 kcal/mol). All synthesized compounds (Va-l) were able to decreases total cholesterol, low-density lipoproteins and triglycerides and increased high-density lipoproteins and very low-density lipoprotein in hyperlipidemic rats. Compounds Va-l have a good affinity towards 3-hydroxy-3-methylglutaryl- coenzyme A reductase enzyme. High yield, good affinity, a nontoxic and low cardiac risk with potential antihyperlipidemic activity make these compounds the possible lead for future research.
Keywords
Anti-hyperlipidemic, benzo (h) chromene-3-carboxylate, 3-hydroxy-3-methyl-glutaryl- coenzyme-A reductase, low-density lipoproteins/high-density lipoproteins, docking
Higher levels of blood lipids may be a possible risk factor for Coronary Heart Disease (CHD) and stroke because they are closely associated with the atherosclerotic disorder. Types of lipids include High-Density Lipoprotein (HDL), Triglycerides (TG), Low-Density Lipoprotein (LDL), cholesterol esters, insulin, phospholipids and fats [1]. LDL is the primary transporter for atherogenic cholesterol, while plasma HDL passes cholesterol from the peripheral tissues to the liver. Diminished HDL and elevated LDL and TG are standard features of hyperlipidemia. Several synthetic medications for hyperlipidemia are available. However, all of them are seriously affected by the adverse effects [2]. Atorvastatin, a pioneering drug for hyperlipidemia treatment with reduced or no capacities for HDL cholesterol, which may be disadvantageous for patients with metabolism disorder or diabetes (having low HDL-Cholesterol). There was considerable exposure to the different pharmacological behaviors of synthetic coumarins. Such derived coumarins will also specifically scavenge species of reactive oxygen and influence the processes of free radical injury. The incorporation of key structural elements within the coumarins family is essential for the production and development of new analogs with the enhanced operation and for characterizing their action mechanisms and possible adverse effects [3].
Much research in this field was stimulated by the critical biological and medicinal activities of fused chromene. In fact, functionally substituted chromene has taken on an uptick in synthetic functions in the field of medicinal chemistry for pledge compounds with an antiproliferative [4], antibacterial [5], anticancer [6], antioxidant [7], aldose reductase inhibitor [8], Monoamine Oxidase (MAO) inhibitor [9], antitubercular [10], platelet anti-aggregating [11], antiinflunza [12]. By keeping these in mind, we synthesized substituted benzo [h] chromene-3-carboxylate derivatives as a potential Antihyperlipidemic agent.
Materials and Methods
All chemicals and reagents were purchased from Sigma Aldrich and Loba Chemie. Atorvastatin was obtained from Micro labs, Bangalore, India. An open capillary method has been used to determine the melting points without correcting them. The Perkins Elmer infrared-283 Fourier Transform Infrared (FT-IR) was used for recording FT-IR-spectra with the help of the Potassium Bromide (KBr) pellet. On the spectrometers of Varian 300 MHz, 1H and 13C Nuclear Magnetic Resonance (NMR) (DMSO-d6) were recorded. API 3000 Liquid Chromatography-Mass Spectrometry (LC-MS) was used to record the mass spectra.
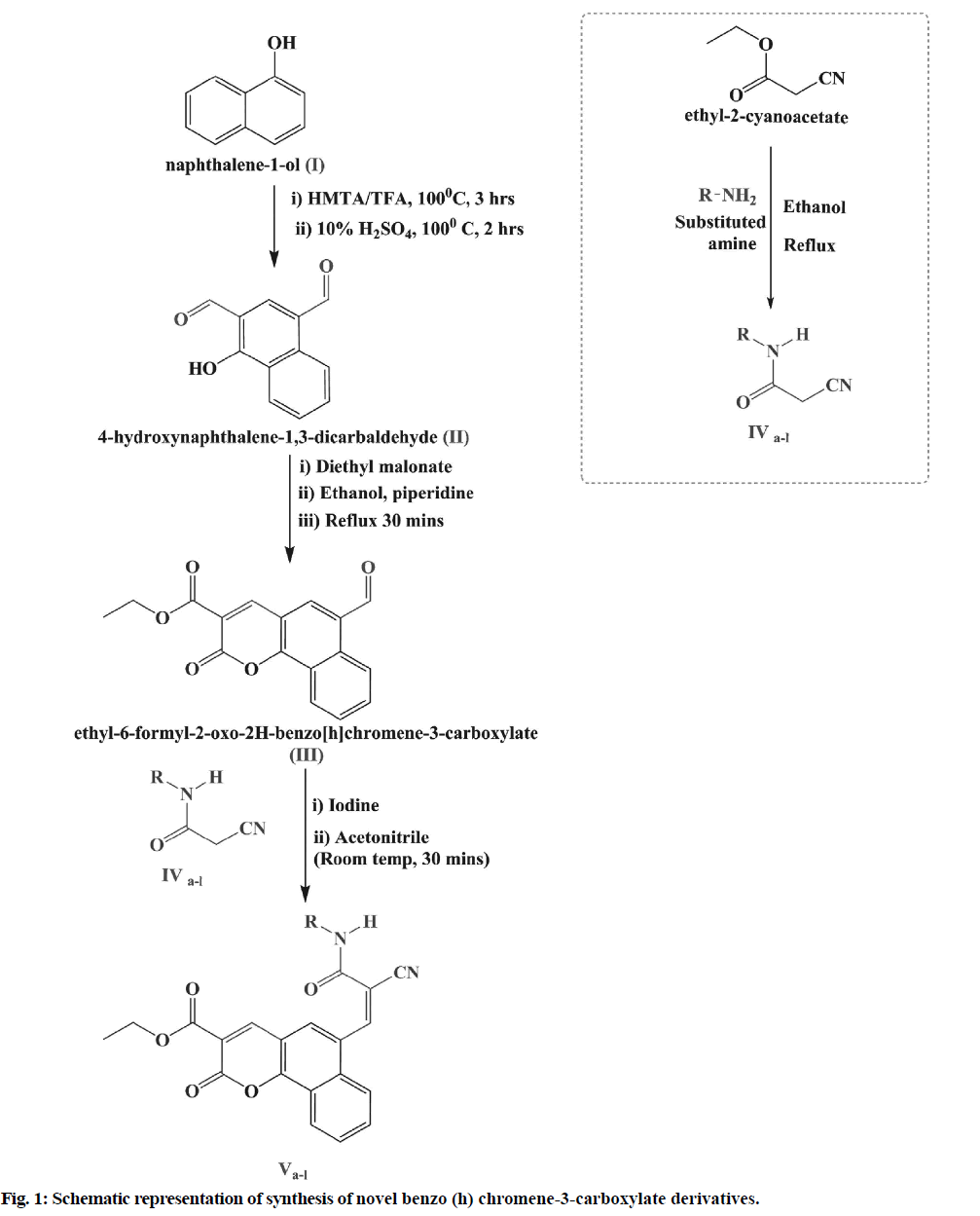
Preparation of 4-hydroxynaphthalene-1,3-dicarbaldehyde (II):
The procedure reported by Venkatraman et al. [13] was adopted for the synthesis of compound I. About 28.8 g (0.2 m) of α-naphthol (I) and 28 g of hexamine were added to 160 ml of trifluoroacetic acid in 500 ml Round- Bottom (R.B) flask fitted with a reflux condenser. The reaction mixture was maintained at 100° for 5 h. The reaction mixture was enriched with 10 % sulphuric acid after completion of 3 h and the reaction continues. The reddish-brown 4-hydroxynaphthalene-1, 3-dicarbaldehyde (II) has been isolated from the reaction mixture by using ether.
Preparation of ethyl-6-formyl-2-oxo-2H-benzo [h] chromene-3-carboxylate (III):
About 20 g of (0.1 m) of 4-hydroxynaphthalene-1, 3-dicarbaldehyde (II) was added to a mixture of 16 ml of diethyl malonate (0.1 m), 10 ml ethanol and 10 ml of piperidine in a reaction flask. The whole mixture was shaken and refluxed for 30-40 min. The yellowish- orange coumarin, namely ethyl-6-formyl-2-oxo-2H- benzo (h) chromene-3-carboxylate (III), was isolated and re-crystallized according to the procedure stated by Venkatraman et al. [13]
Preparation of 3-oxo-3-pyrrolidin-1-yl-propionitrile (IVa):
A compound 3-pyrrolidine-1-yl-propionitrile was synthesized as per procedure reported by Darla et al. [14] A required mass of ethyl cyanoacetate (2.72 g, 0.024 mol) was added with stirring to a solution of pyrrolidine (1.42 g, 0.02 mol) in 100 ml ethanol. The resulting mixture was then refluxed for the duration of 5 h. The resulting product was then cooled, filtered to obtain 3-oxo-3-pyrrolidine-1-yl propionitrile (IVa). Alike process was also adopted for the production of compounds (IVb-l).
Preparation of ethyl 6-((Z)-2-cyano-3-oxo-3- (pyrrolidin-1-yl) prop-1-enyl)-2-oxo-2H-benzo (h) chromene-3-carboxylate (Va) by knoevenagel condensation:
A catalytic volume of piperidine and ethyl-6-formyl-2- oxo-2H-benzo [h]chromene-3-carboxylate (3.8 g, 0.02 mol) was dissolved in ethanol in a round bottom flask, fitted with a reflux condenser. The 3-oxo-3-pyrrolidin- 1-yl-propionitrile (IVa, 2.78 g, 0.002 mol) compound was introduced into the reaction mixture and heated for 2 h. The reaction mixture was then cooled to 0°-5° and separated. The resulting product was crystallized using solvent dioxane: Ethanol to get ethyl 6-((Z)-2-cyano-3- oxo-3-(pyrrolidin-1-yl) prop-1-enyl)-2-oxo-2H-benzo (h) chromene-3-carboxylate (Va). For the preparation of compounds (Vb-l), a similar procedure was adopted.
Anti-hyperlipidemic studies:
Male albino strains of Wistar rats were randomly divided into fifteen groups of six animals. The experimental protocol of the current study was approved by the Institutional Animal Ethics Committee (1189/PO/a/08/ CPCSEA) of Rungta College of Pharmaceutical Sciences and Research, Bhilai, India. Animals with ad libitum food and water were housed in the standard laboratory conditions (temperature, 22±3°; relative moisture, 30 %-70 %; time frame of 12:12 h dark- light). Group I rats are used as normal control. Group II to XV rats was given 400 mg/kg body weight dose of cholesterol for 30 d. Group II was served as cholesterol control. Group III received standard drug atorvastatin 05 mg/kg body weight from 15 d to 30 d. At the same time, Group IV to XV received test compounds (Va-l), respectively, at a dose of 05 mg/kg body weight from 15 d to 30 d.
Biochemical analysis:
All of the rats were slaughtered at the end of 30 d, blood was gathered and serum was permitted to pool after centrifugation. Serum was tested with standard protocol procedures for total cholesterol, TG, HDL, LDL, Very Low-Density Lipoproteins (VLDL) content.
Histopathological evaluation:
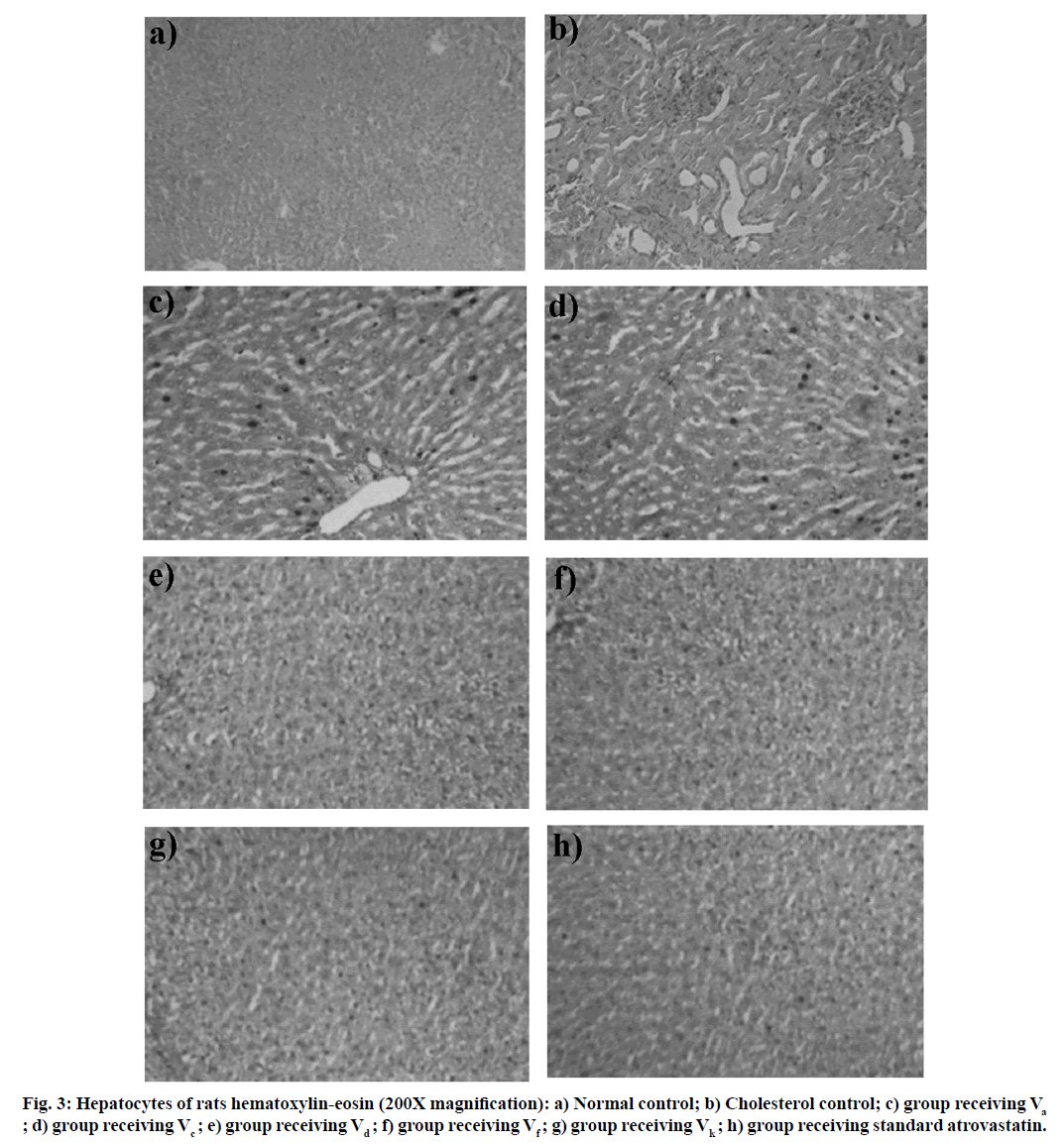
Under the influence of anesthetic (ether), animals were euthanized and their liver was subsequently examined after dissection and weight were measured. The liver and adipose tissue are set for histopathological examination at room temperature in 10 percent formalin. The tissue was coated in paraffin, split up into 3-4 μm size and then mounted using standard histological techniques on glass microscopes slides. The pieces were drenched with hematoxylin-eosin for staining and analyzed at 200X magnitudes using light microscopy. An image analyzer assessed these light microscopic fields in each section.
Statistical analysis:
All values have been represented as mean±Standard Error of the Mean (SEM). Results have been evaluated in one way Analysis of Variance (ANOVA), accompanied by multiple range tests by Newman keul's. Statistically significant values were those, which have the p values <0.05.
Docking studies:
Molecular docking is carried out in order to provide a binding site with a population of probable ligand orientations and conformations [15,16]. 3-Hydroxy-3-Methyl-Glutaryl-Coenzyme A (HMG-CoA) reductase (1HWK) crystal structure was obtained from the protein data bank Research Collaboratory for Structural Bioinformatics (RCSB). The Two Dimensional (2D) structure of the ligands (i.e., synthesized molecules (Va-l) and standard drug (Atorvastatin) was designed with the help of chem draw professional 16.0 software. Molecular mechanics optimization of the 2D structures of ligands was done by converting them into Three Dimensional (3D) structures with the help of chem 3D 16.0 application of the same software (Chem draw professional 16.0). Argus Lab 4.0.1 was used for the docking [17]. The grid with dimension X=70, Y=90 and Z=60 Å are assigned to cover entire 3-dimensional active site of HMG-CoA reductase. Upon testing the docking technique, dynamic molecular docking was done on the active site of proteins. Finally, the software retrieved the result of binding energy. The protein- ligand interaction was visualized by using discovery studio 2016 software.
Results and Discussion
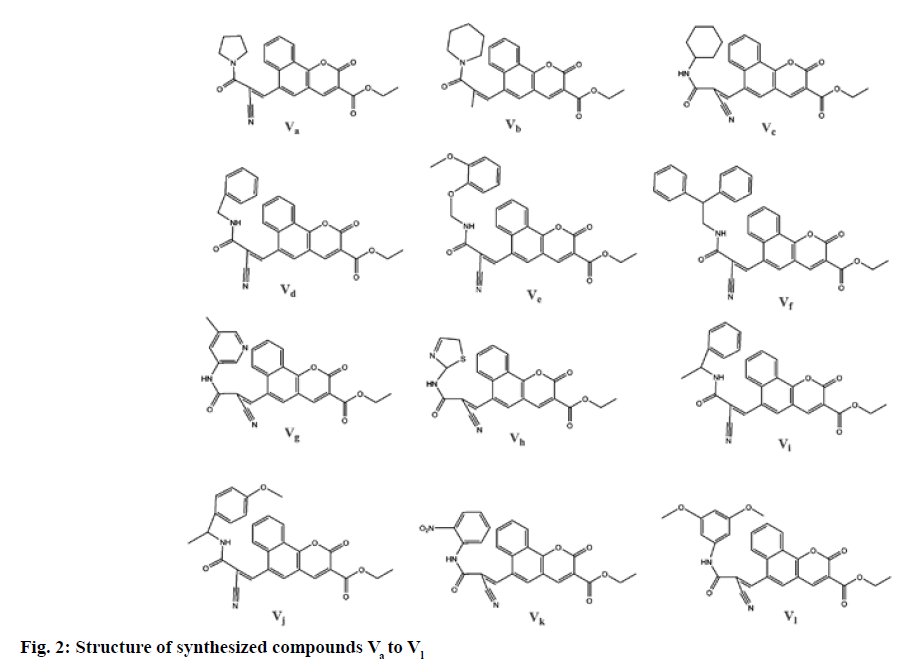
In the present study, we have reported the synthesis and antihyperlipidemic screening of novel benzo [h] chromene-3-carboxylate derivatives. The conformation of synthesis of benzo [h]chromene-3-carboxylate derivatives (Va-l) was ascertained using Thin Layer Chromatography (TLC), combustion analysis, FT-IR, NMR and Mass Spectrometry (MS). The synthetic method used in the preparation of these derivatives is shown in fig. 1. Total 12 different benzo (h) chromene-3-carboxylate derivatives were prepared (fig. 2) by Knoevenagel condensation of ethyl-6-formyl- 2-oxo-2H-benzo [h]chromene-3-carboxylate (III) with compounds IVa-l. Chemical structure, melting point, combustion analysis, and other physical data of synthesized compounds are mentioned in Table 1. The conformation of synthesis of different derivatives ascertained by I.R., 1H NMR and 13C NMR and mass analysis.
| Compound ID | Molecular formula | Molecular weight | Melting point (°) | Rf Value | % Yield | Chemical name | Elemental Analysis | |
|---|---|---|---|---|---|---|---|---|
| Theoretical | Observed | |||||||
| Va | C24H20N2O5 | 416.4 | 225-227 | 0.8 | 72 | Ethyl 6-((Z)-2-cyano-3-oxo-3-(pyrrolidin-1-yl)prop-1-enyl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 69.22; H, 4.84; N, 6.73; O, 19.21 | C, 69.20; H, 4.86; N, 6.70; O, 19.24 |
| Vb | C25H22N2O5 | 430.5 | 215-217 | 0.6 | 62 | Ethyl (Z)-6-(2-cyano-3-oxo-3-(piperidin-1-yl)prop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 69.76; H, 5.15; N, 6.51; O, 18.58 | C, 69.75; H, 5.17; N, 6.54; O, 18.54 |
| Vc | C26H24N2O5 | 444.5 | 219-221 | 0.7 | 59 | Ethyl (Z)-6-(2-cyano-3-(cyclohexylamino)-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 70.26; H, 5.44; N, 6.30; O, 18.00 | C, 70.23; H, 5.43; N, 6.32; O, 18.02 |
| Vd | C27H20N2O5 | 452.5 | 225-227 | 0.7 | 60 | Ethyl (Z)-6-(3-(benzylamino)-2-cyano-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 71.67; H, 4.46; N, 6.19; O, 17.68 | C, 71.64; H, 4.48; N, 6.21; O, 17.67 |
| Ve | C29H24N2O7 | 512.5 | 235-237 | 0.7 | 62 | Ethyl (Z)-6-(2-cyano-3-((2-(2-methoxyphenoxy)ethyl)amino)-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 67.96; H, 4.72; N, 5.47; O, 21.85 | C, 67.94; H, 4.74; N, 5.45; O, 21.87 |
| Vf | C33H24N2O5 | 528.6 | 222-224 | 0.7 | 73 | Ethyl (E)-6-(3-(benzhydrylamino)-2-cyano-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 74.99; H, 4.58; N, 5.30; O, 15.13 | C, 74.97; H, 4.56; N, 5.33; O, 15.14 |
| Vg | C26H19N3O5 | 453.5 | 228-241 | 0.7 | 65 | Ethyl (E)-6-(2-cyano-3-((5-methylpyridin-3-yl)amino)-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 68.87; H, 4.22; N, 9.27; O, 17.64 | C, 68.84; H, 4.25; N, 9.24; O, 17.67 |
| Vh | C23H17N3O5S | 447.1 | 184-187 | 0.7 | 70 | Ethyl (Z)-6-(2-cyano-3-((2,5-dihydrothiazol-2-yl)amino)-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 61.74; H, 3.83; N, 9.39; O, 17.88; S, 7.16 | C, 61.72; H, 3.85; N, 9.37; O, 17.86; S, 7.20 |
| Vi | C28H22N2O5 | 466.5 | 210-212 | 0.8 | 61 | Ethyl (Z)-6-(2-cyano-3-oxo-3-((1-phenylethyl)amino)prop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 72.09; H, 4.75; N, 6.01; O, 17.15 | C, 72.05; H,4.87 ; N, 5.98; O, 17.18 |
| Vj | C29H24N2O6 | 496.5 | 258-260 | 0.7 | 55 | Ethyl (Z)-6-(2-cyano-3-((1-(4-methoxyphenyl)ethyl)amino)-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 70.15; H, 4.87; N, 5.64; O, 19.33 | C, 70.18; H, 4.87; N, 5.69; O, 19.26 |
| Vk | C26H17N3O7 | 483.4 | 238-240 | 0.8 | 54 | Ethyl (Z)-6-(2-cyano-3-((2-nitrophenyl)amino)-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 64.60; H, 3.54; N, 8.69; O, 23.17 | C, 64.64; H, 3.52; N, 8.65; O, 23.19 |
| Vl | C29H24N2O7 | 512.5 | 247-249 | 0.7 | 70 | Ethyl (Z)-6-(2-cyano-3-((3,5-dimethoxybenzyl)amino)-3-oxoprop-1-en-1-yl)-2-oxo-2H-benzo [h]chromene-3-carboxylate | C, 67.96; H, 4.72; N, 5.47; O, 21.80 | C, 67.97; H, 4.71; N, 5.46; O, 21.16 |
Table 1: Physiochemical Characterization of Synthesized Compounds
Infrared Spectroscopy (IR) spectrum of compound Va (ethyl 6-((Z)-2-cyano-3-oxo-3-(pyrrolidin-1-yl)prop- 1-enyl)-2-oxo-2H-benzo (h) chromene-3-carboxylate) shows strong C=O stretching vibration band at 1742.23 cm-1. Vibration band at 1596.84 cm-1 and 1645.38 cm-1 is arising due to C=C and cyclic -C-O and stretching, respectively. The emergence of the peak due to -C-N. Stretching at 2246.52 cm-1 confirmed the attachment of compound IVa to compound III.
The 1H NMR spectrum of compound Va has shown the signal at 8.35 due to exocyclic CH=C proton, multiplate in region 7.68-8.00 and 6.23-7.5 due to 4H naphthalene ring proton and 3H coumarin ring proton respectively. The signal at 3.25 and 1.72 is confirmed the presence of a pyrrolidine ring. While the signal at 45.7 and 25.0 in 13C NMR spectra also confirmed the presence of pyrrolidine ring in final compound Va. The compound Va mass spectrum revealed the peak at m/z 416.14+ in line with molecular formula C24H20N2O5. Likewise, spectral results of the remaining derivatives are represented in Table 2.
| Compound Id | IR Data | 1H NMR Data | 13C NMR Data | Mass Data |
|---|---|---|---|---|
| (KBr Cm-1) | ppm (DMSO-d6) | ppm (DMSO-d6) | m/z (M++1) | |
| Va | 2246.52 (-CN), 1742.23 (-CO), 1645.38 (cyclic-CO), 1596.84 (-C=C) | 8.29 (s, H, exocyclic CH=C), 7.68-8.00 (m, 4H naphthalene ring), 6.19-7.6 (3H, coumarin), 3.18 (t, 4H, pyrrolidine), 1.81 (t, 4H, pyrrolidine) | d169.8 (-CO), 159.9, (C-2), 156.2 (C-7), 154.4 (C, expcyclic ethylene), 145.8 (C-10), 124.9 (C-5), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 119.1 (C-8), 114.8 (-CN), 113.5 (C-6), 111.9 (C-3), 111.4 (C-9), 105.7 (C, ethylene), 46.2 (2C, of pyrrolidine), 24.9 (2C, of pyrrolidine) | 416.14 |
| Vb | 1742.34 (-CO), 2240.35 (-CN), 1648.28 (cyclic -CO), 1595.63 (-C=C) | 8.27 (s, H, exocyclic CH=C), 7.68-8.00 (m, 4H naphthalene ring), 6.2-7.44 (m, 3H, coumarinring), 3.68 (m, 4H, piperidine), 1.59 (m, 2H, piperidine),1.49 (m, 4H, piperidine) | d169.2 (CO), 159.4 (C-2), 154.6 (C-7),152.8 (C,ethylene), 151.9 (C-4),145.9 (C-10), 124.7 (C-5), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 117.5 (C-8), 114.3 (CN), 113.5 (C-6), 111.8 (C-3), 111.4 (C-9), 105.7 (C, ethylene), 46.2 (2C, piperidine), 24.6 (2C, piperidine), 23.4 (C, piperidine) | 430.15 |
| Vc | 3277.34 (-NH), 2230.43 (-CN), 1730.48 (-CO), 1674.81 (cyclic -CO), 1546.22 (-C=C) | 8.35 (s, H, exocyclic CH=C), 8.14 (s,H,–NH), 7.68-8.00 (m, 4H naphthalene ring), 6.0 (d,H), 6.5 (d, H), 7.5 (d, H), 7.8 (d, H), 3.49 (m, H), 1.11-1.3 (m, 4H), 1.45-1.53 (m, 2H), 1.62–1.82 (m, 4H) | d161.8 (CO), 159.9 (C-2), 149.7 (C-7), 152.8 (C,ethylene), 151.9 (C-4), 145.1 (C-10), 124.7 (C-5), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 117.4 (C-8), 114.9 (CN), 113.6 (C-6), 111.8 (C-3), 111.1 (C-9), 105.3 (C,C–CN), 46.5 (C, C-10), 32.7 (2C, C-20, C-60), 25.6 (C, C-40), 23.8 (2C, C-30, C-50) | 444.17 |
| Vd | 2240.45 (-CN), 3276.59 (-NH), 1729.24 (-CO, cyclic), 1624.47 (-CO), 1557.81 (-C=C-) | 8.85 (s, H, -NH)), 8.42 (s, H exocyclic CH=C), 7.68-8.00 (m, 4H naphthalene ring), 7.46 (d, H, coumarin ring), 6.72 (d, H, coumarin ring), 7.16-7.39 (m, 5H, phenyl), 6.11 (s, H, endocyclic), 4.52 (s, 2H, -CH
|
d162.7 (C-2),160.3, (-CO), 155.5 (C-7), 153.5 (C, ethylene),152.5 (C-4), 146.1 (C-10), 134.1 (C-40), 128.5 (C-20,C-60), 126.5 (C-30,C-50), 126 (C-10), 124.9 (C-5), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 117.8 (C-7), 114.6 (C, -CN), 113.7 (C-6),111.9 (C-3), 111.3 (C-9), 102.6 (C–CN), 42.5 (C, -CH2) | 452.14 |
| Ve | 2257.72 (-CN), 3273.35 (-NH), 1745.58 (-CO), 1646.62 (cyclic -CO), 1597.51 (-C=C) | 8.84 (s, H -NH), 8.46 (s, H, exocyclic CH=C), 8.49 (d, H), 7.68-8.00 (m, 4H naphthalene ring), 6.90 (d, 2H), 7.06 (t, 2H), 7.49 (d, H), 7.44 (d, H), 6.88 (d, H), 6.26 (s, H), 4.13 (t, 2H, -CH2-O-), 3.68 (s, 3H, -OCH3), 3.17 (q, 2H, -NH-CH2) | d 163.9 (C-40), 159.8 (C-2), 158.4 (-CO), 154.3 (C-7), 152.8 (C, exocyclic ethylene), 151.2 (C-4), 145.9 (C-10), 127.8 (C-20, C-60), 126.9 (C-30, C-50), 125.2 (C-10), 124.6 (C-5), 126.5 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 117.4 (C-8), 115.5 (-CN), 113.8 (C-6), 111.8 (C-3), 111.2 (C-9), 102.5 (C-exocyclic), 48.2 (-CH-NH), 20.9 (-CH3) | 512.16 |
| Vf | 2240.34 (-CN), 3275.53 (-NH),1742.79 (-CO), 1643.87 (cyclic -CO), 1584.56 (-C=C) | 8.35 (s, H, exocyclic ethylene), 8.0 (s, H, -NH), 7.68-8.00 (m, 4H naphthalene ring), 6.23-7.55 (m, 3H coumarin ring), 7.22-7.44 (m, 10H, benzhydril), 6.16 (s, H,-CH) | d 160.8 (C-2), 160 (C=O), 158.9 (C-10), 155 (C-7), 153.4 (C- exocyclic), 153 (C-4), 146 (C-10), 133 (C-40), 126.5 (C-30, C-50), 125.2 (C-5), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 118.2 (C-8), 115 (–CN), 114.0 (C-20, C-60, C-6), 112.5 (C-3), 112.0 (C-9), 103 (C- exocyclic), 56.1 (–OCH3), 49.0 (C, –CH–NH), 19.5 (–CH3 of C-4) | 528.17 |
| Vg | 3276.11 (-NH), 2229.18 (-CN), 1729.48 (-CO), 1673.65 (cyclic -CO), 1549.94 (-C=C) | 8.00 (d,H), 6.66 (d,H), 6.65 (d,H), 8.39 (s,H,exocyclic,CH=C), 8.09 (s,H,–NH), 7.68-8.00 (m, 4H naphthalene ring), 6.26 (s,H), 6.88 (d,H), 7.44 (d,H), 2.42 (s, 3H, -CH3) | d163.7 (C-2),160.6, (-CO), 153.7 (C-7), 152.4 (C, ethylene), 151.6 (C-4), 116.3 (C-60),144.7 (C-10), 125.7 (C-40), 141.6 (C-20), 131.7 (C-30), 129.4 (C-50), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 114.7 (C-5), 114.2 (C, -CN), 113.1 (C-6), 111.3 (C-3), 110.5 (C-9), 107.6 (C-CN) | 453. 13 |
| Vh | 2254.54 (-CN), 1741.62 (-CO), 1643.49 (cyclic-CO), 1593.27 (-C=C), 1547.31 (-C=C), 749.23 (C-S) | 8.34 (s, Hexocyclic ethylene), 7.94 (s, H, -NH), 7.68-8.00 (m, 4H naphthalene ring), 6.16-7.46 (m, 3H coumarin ring), 3.75 (t, 2H,-N-CH2-), 3.19 (t, 2H,-S-CH2) | d 168.7 (C-10), 63.56 (C, -CO), 161.1 (C-2),154.3 (C-7), 154.71 (C, exocyclic ethylene), 153.31 (C-4, C-50), 145.9 (C-10), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 126.1 (C-5), 122.8 (C-30), 119.0 (C-8), 118.1 (C-40), 116.7 (-CN), 115.7 (C-6), 113.6 (C-3), 113.4 (C-9), 105.3 (C-20), 105.2 (C, ethylene), 101.3 (C-60), 57.1 (2C, -OCH3) | 447.09 |
| Vi | 3278.15 (-NH), 2230.35 (-CN), 1732.47 (-CO), 1673.42 (cyclic -CO), 1556.67 (-C=C) | 8.81 (s, H, -NH), 8.49 (s, H, exocyclic CH=C), 7.68-8.00 (m, 4H naphthalene ring), 6.43-7.37 (m, 5H, phenyl), 5.91-6.38 (m, 2H, coumarin ring), 6.21 (d, 1H, endocyclic), 4.97 (q, H, -CH-NH), 1.47 (d, 3H, CH3) | d 161.7 (C-2), 159.9 (C=O), 151.2 (C-10), 156.1 (C-7), 154.6 (C-exocyclic), 154.2 (C-4), 152.5 (C-10), 147.3 (C-10), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 122.9 (C-40), 121.8 (C-30), 112.2 (C-50), 126.1 (C-5), 119.4 (C-8), 114.8 (-CN), 113.9 (C-60), 113.1 (C-3), 111.9 (C-9), 104.2 (C-exocyclic), 71.4 (C,-O-CH2-), 57.2 (-OCH3), 38.7 (C, -CH-NH) | 466.15 |
| Vj | 2254.41 (-CN), 1745.62 (-CO), 1643.37 (cyclic -CO), 1584.72 (-C=C), 1548.12 (-C=C) | 8.87 (s, H-NH), 8.49 (s, H, exocyclic CH=C), 7.68-8.00 (m, 4H naphthalene ring),6.86-7.31 (m, 4H, phenyl), 6.13-6.62 (m, 2H, coumarin ring), 5.11 (q, H,-CH-NH), 3.92 (s, 3H, -OCH3 1.61 (d, 3H, -CH3) | d 162.2 (C-2), 161.1 (C, exocyclic ethylene), 160.3 (CO),156.2 (C-7), 154.4 (C-4), 147.1 (C-10), 141.0 (C-40, C-400), 127.9 (C-50, C-500, C-30,C-300), 130.3 (C-60, C-600, C-20, C-200), 127.4 (C-10,C-100), 126.1 (C-5), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring),119.3 (C-8), 116.2 (-CN), 115.4 (C-6), 113.9 (C-3), 113.1 (C-9), 105.1 (C, ethylene), 53.6 (C, -CH of benzhydril) | 496.16 |
| Vk | 3274.24 (-NH), 2241.71(-CN), 1730.66 (-CO, cyclic), 1625.38 (-CO), 1558.15 (-C=C-) | 10.21 (s, H, -NH), 8.57 (d, H), 8.42 (s, H, exocyclic CH=C), 8.31 (d, H), 7.8 (t, 2H), 7.67 (d, H), 7.44 (d, H), 6.88 (d, H), 6.26 (s, H), 7.68-8.00 (m, 4H naphthalene ring). | d160.8 (C-2),164.5, (-CO), 154.1 (C-7), 153.2 (C,ethylene), 151.9 (C-4), 147.4 (C-10), 159.2 (C-40), 164.2 (C-20), 146.7 (C-60), 116.1 (C-30), 126.6 (2C, of naphthalene ring), 123.5 (2C, of naphthalene ring), 124.9 (C-5), 123.4 (C-50), 119.1 (C-8), 116.3 (C, -CN), 113.9 (C-6), 112.2 (C-3), 111.5 (C-9), 107.8 (C-CN), 22.0 (C,-CH3 of pyridine) | 483.11 |
| Vl | 2254.33 (-CN), 1,743.49 (-CO), 1643.81 (cyclic-CO), 1596.47 (-C=C), 3272,31 (-NH) | 10.62 (s, -NH), 8.41 (s, H exocyclic ethylene), 8.22 (d, H), 7.68-8.00 (m, 4H naphthalene ring), 6.14-7.46 (m, 3H coumarin ring), 6.62-6.79 (2H), 3.8 (s, 6H of -OCH3). | d 170.2 (CO), 164.4 (-N-C=N), 163.1 (C, exocyclic ethylene), 161.3 (C-2), 155 (C-7), 153 (C-4), 146 (C-10),126.6 (2C, of naphthalene ring), 125.2 (C-5), 123.5 (2C, of naphthalene ring),119.1 (C-8), 116.2 (-CN), 115.5 (C-6), 113.1 (C-3), 113.2 (C-9), 105.7 (C,ethylene), 54 (=N-CH2), 25.3 (-S- CH2) | 512.16 |
Table 2: Characterization of Synthesized Compounds By Spectral Analysis
In this work, rat fed with cholesterol reported expected rises in total cholesterol, TG, LDL and Aluminium (Al) with decreases in HDL and VLDL level after 30 d in the experimental group relative to the usual control group. In this study, it was observed that all synthesized compounds (Va-l) were able to decreases total cholesterol, LDL, Al and TG and increased HDL and VLDL in hyperlipidemic rats at the dose of 05 mg/kg (Table 3). Compound Vd, Vf, and Vk showed a highly significant reduction in total serum cholesterol, TG, LDL and Al. These compounds also significantly increase the HDL and VLDL levels with respect to the cholesterol control group. Especially compounds with diphenylmethanamine group (Vf) show promising antihyperlipidemic activity as compared to standard atorvastatin.
| Treatment groups | Total cholesterol (mg/dl) | Tri glycerides (mg/dl) | HDL (mg/dl) | LDL (mg/dl) | VLDL (mg/dl) | Al (mg/dl) | LDL/ HDL |
|---|---|---|---|---|---|---|---|
| Group-I | 51.59±3.12 | 57.10±2.89 | 27.38±2.41 | 15.12±1.11 | 32.71±2.21 | 0.95±0.89 | 0.55 |
| Group-II | 119.35±6.41**(a) | 160.4±5.68**(a) | 12.27±1.15**(a) | 32.78±2.27**(a) | 14.21±1.34**(a) | 9.03±4.56**(a) | 2.67 |
| Group-III | 78.51±4.31**(b) | 80.8±3.78**(b) | 22.47±2.06**(b) | 22.12±2.13**(b) | 27.42±1.15**(b) | 2.60±1.45**(b) | 0.98 |
| Group-IV | 94.47±2.87**(b) | 118.38±4.90**(b) | 16.78±1.11**(b) | 26.25±2.21**(b) | 20.12±1.26**(b) | 2.53±1.12**(b) | 1.56 |
| Group-V | 91.41±3.21**(b) | 95.68±3.80**(b) | 17.65±2.05**(b) | 21.46±2.48**(b) | 24.51±1.24**(b) | 3.19±2.14**(b) | 1.22 |
| Group-VI | 87.79±2.94**(b) | 90.10±3.77**(b) | 15.32±2.08**(b) | 21.52±2.12**(b) | 21.20±1.52**(b) | 2.99±1.45**(b) | 1.4 |
| Group-VII | 79.98±3.12**(b) | 82.83±3.77**(b) | 21.67±2.11**(b) | 21.12±2.04**(b) | 22.35±1.26**(b) | 2.20±2.11**(b) | 0.97 |
| Group-VIII | 82.93±3.25**(b) | 85.49±3.07**(b) | 19.34±2.27**(b) | 22.94±2.35**(b) | 24.32±1.52**(b) | 2.69±2.21**(b) | 1.13 |
| Group-IX | 78.89±3.71**(b) | 79.92±3.92**(b) | 22.45±2.33**(b) | 22.25±2.34**(b) | 23.14±1.54**(b) | 1.62±3.02**(b) | 0.99 |
| Group-X | 92.57±2.33**(b) | 105.94±4.90**(b) | 19.24±1.28**(b) | 20.86±2.24**(b) | 20.42±1.32**(b) | 3.38±2.02**(b) | 1.08 |
| Group-XI | 90.84±3.27**(b) | 93.45±3.07**(b) | 18.57±2.15**(b) | 30.58±2.57**(b) | 24.31±1.73**(b) | 2.09±2.11**(b) | 1.65 |
| Group-XII | 85.97±2.88l**(b) | 88.84±4.90**(b) | 17.58±1.21**(b) | 21.75±2.34**(b) | 20.45±1.12**(b) | 3.18±2.89**(b) | 1.24 |
| Group-XIII | 84.35±3.29**(b) | 87.72±3.80**(b) | 18.98±2.17**(b) | 22.65±2.21**(b) | 24.28±1.54**(b) | 2.72±2.11**(b) | 1.19 |
| Group-XIV | 80.59±3.15**(b) | 83.26±3.12**(b) | 22.86±2.41**(b) | 21.56±2.31**(b) | 23.74±1.48**(b) | 2.63±2.21**(b) | 0.94 |
| Group-XV | 89.83±3.24**(b) | 92.39±3.12**(b) | 18.14±2.24**(b) | 24.10±2.22**(b) | 23.42±1.54**(b) | 2.64±2.14**(b) | 1.33 |
Note: Values are expressed as Mean±SEM, Values were found out by using one way ANOVA followed by Newman Keul’s multiple range tests, **(a) values were significantly different from normal control at p<0.01, **(b) values were significantly different from hyperlipidemic control at p<0.01
Table 3: In Vivo Antihyperlipidmic Activity of Prapered Compounds and Standard Atorvastatin
Furthermore, the LDL/HDL risk ratios of compound Vd, Vf and Vk were found to be 0.97, 0.99 and 0.94, respectively, while that of the population treated with atorvastatin was 0.96. This indicates that the cardiac risk factor was slightly less in compound Vk. None of the rats displayed some morbidity or mortality after 15 d of treatment, which also encourages that all synthesized substances are well handled and are relatively safe. Histological variability in the liver parts of the population caused by hyperlipidemia and freshly synthesized compounds treated rats has been calculated and subsequent findings were seen in fig. 3 to help us consider the prevalence and extent of hyperlipidemia.
Some histological differences in the liver tissue of the cholesterol control and the compounds treated rat have been identified in contrast to normal controls. In the hepatocytes of cholesterol control rats (fig. 3b), enlargement of nucleus and membrane disruption (necrosis) was observed. This might be due to the localization of fatty acid. Administration of compound Va and Vc showed micro fatty modification in a large array of lymphoid cells had been shown to suggest very little necrosis or degeneration (fig. 3c and fig. 3d). Hepatocytes of atorvastatin treated rats and compound Vd, Vf, and Vk treated rats showing good fatty infiltration and granular degeneration.
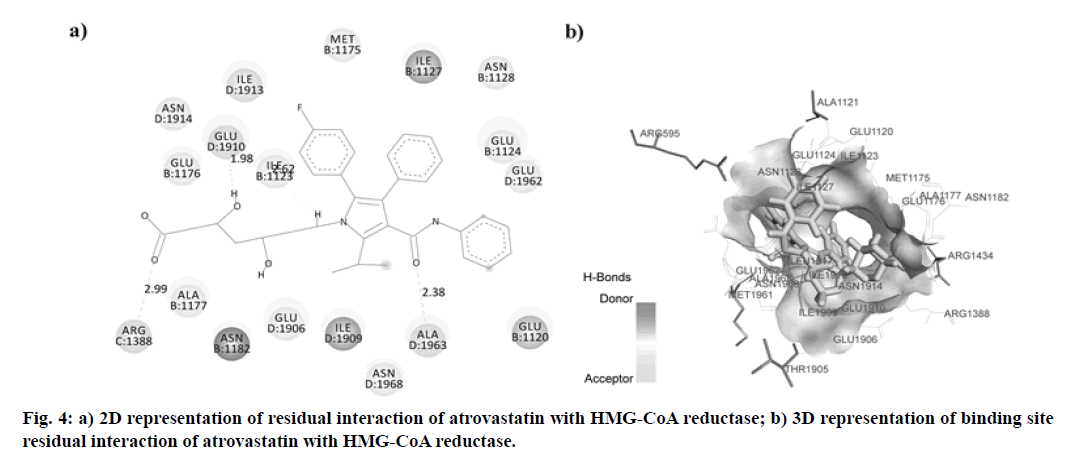
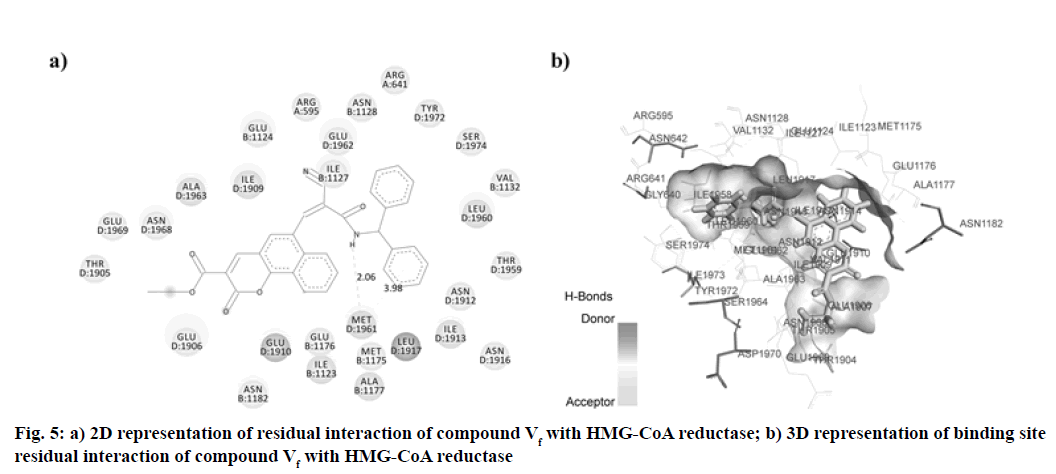
In this research, the synthesized molecule produced improved effects, which led us to consider its mechanism of action. We have, therefore, chosen the docking studies as an instrument to support our results. HMG- GOA reductase inhibition has been the most powerful method for managing hyperlipidemic disorders to date. This enzyme catalyzes HMG-CoA transformation into mevalonic acid, producing cholesterol eventually. In this analysis, we contrasted the findings with atorvastatin for the docking of our synthesized compounds. The binding data obtained from Argus Lab revealed, synthesized compounds have a good affinity towards HMG-CoA reductase enzymes with binding energy ranges in between -9.980 to -12.898 kcal/mol. Analysis of residual interaction between the inhibitor and target enzyme at the active binding site revealed that the atorvastatin and synthesized compounds bind with HMG-CoA reductase by van der Waals interaction, conventional hydrogen bond, carbon-hydrogen bond, Pi-anion interaction, Pi-sigma interaction, alkyl and pi-alkyl interaction. Atorvastatin bind with chain B, chain C and chain D of the receptor. It forms three hydrogen bonds with three residues of HMG-CoA reductase. The carboxylic group of atorvastatin forms hydrogen bonds with Arginine (ARG) 1388 residue of chain C at a distance of 2.991 Å. The Hydroxy (OH) group of 2, 4-dihydroxy pentanoic acid chain interacts with chain D residue Glutamic Acid (GLU) 1910 (at a distance of 1.976 Å). While residue Alanine (ALA) 1963 of chain D forms a hydrogen bond with distance 2.378 Å with C=O group of phenyl carbamoyl chain of atorvastatin (fig. 4). The compound Vf showed even better binding affinity (-12.898 kcal/mol) energy values compared with atorvastatin (-11.8605 kcal/mol) (Table 4). Out of 18 amino acid residue which interacts with atorvastatin, 13 were found common with synthesized compound Vf. The Amino (NH) group of diphenylethyl amino chain of compound Vf forms a hydrogen bond with D chain residue (Methionine (MET) 1961) of HMG-CoA reductase enzyme at bond length 2.067 Å. These interaction patterns give a strong impression that synthesized compounds possess good inhibitory potential against HMG-CoA reductase (fig. 5).
| Compound ID(s) | Binding Energy (kcal/mol) | Hydrogen Bonds | Enzyme's Binding Site residue | ||
|---|---|---|---|---|---|
| Quantity | Bond Length in Å | Receptor residue | |||
| Va | -10.4733 | 3 | 2.258 | 627 ARG | Chain A: 654 ALA (coil), 826 ALA (coil), 627 ARG (coil),830 ASN (coil), 653 ASP (coil), 827 CYS (coil), 652 GLY (coil), 806 GLY (coil), 832 GLY (coil), 828 LYS (coil), 659 MET (alpha helix), 628 PHE (coil), 831 PRP (coil), 651 SER (coil), 805 VAL (alpha helix) |
| 2.763 | 627 ARG | ||||
| 2.998 | 653 ASP | ||||
| Vb | -9.98073 | 1 | 2.938 | 483 THR | Chain A: 529 ASN (coil), 482 GLU (coil), 528 GLU (coil), 480 LYS (coil), 523 MET (coil), 477 PRO (coil), 483 THR (coil), 479 TYR (coil) Chain B: 958 ALA (alpha helix), 961 ASN (alpha helix), 957 VAL (alpha helix) |
| Vc | -10.7961 | 3 | 2.219 | 1527 ASN | Chain A: 783 ALA (beta strand), 732 ASN (alpha helix), 734 ASN (coil), 788 ASN (coil), 726 GLU (alpha helix), 730 GLU (alpha helix), 782 GLU (beta strand), 729 ILE (alpha helix), 733 ILE (alpha helix), 737 LEU (alpha helix), 780 LEU (beta strand), 781 MET (beta strand) Chain C: 1527 ASN (coil), 1523 GLU (alpha helix), 1575 GLU (beta strand), 1522 ILE (alpha helix), 1526 ILE (alpha helix), 1574 MET (beta strand), 1531 VAL (alpha helix) Chain D: 1775 ARG (beta strand) |
| 2.998 | 1527 ASN | ||||
| 2.697 | 730 GLU | ||||
| Vd | -11.578 | 1 | 2.888 | 483 THR | Chain A: 529 ASN (coil), 482 GLU (coil), 528 GLU (coil), 476 ILE (coil), 480 LYS (coil), 523 MET (coil), 477 PRO (coil), 483 THR (coil), 479 TYR (coil) Chain B: 958 ALA (alpha helix), 961 ASN (alpha helix), 957 VAL (alpha helix) |
| Ve | -11.0592 | 3 | 2.996 | 627 ARG | Chain A: 826 ALA (coil), 627 ARG (coil), 830 ASN (coil), 653 ASP (coil), 827 CYS (coil), 652 GLY (coil), 832 GLY (coil), 828 LYS (coil), 659 MET (alpha helix), 628 PHE (coil), 831 PRO (coil), 626 SER (coil) |
| 2.827 | 627 ARG | ||||
| 2.938 | 627 ARG | ||||
| Vf | -12.898 | 1 | 2.067 | 1961 MET | Chain A: 595 ARG (beta strand), 641 ARG (coil) Chain B: 1177 ALA (beta strand),1128 ASN (coil), 1124 GLU (alpha helix), 1176 GLU (beta strand), 1123 ILE (alpha helix), 1127 ILE (alpha helix), 1175 MET (beta strand), 1132 VAL (alpha helix) Chain D: 1963 ALA (beta strand), 1912 ASN (aplha helix), 1968 ASN (coil), 1906 GLU (alpha helix), 1962 GLU (beta strand), 1969 GLU (beta strand), 1090 ILE (alpha helix), 1913 ILE (alpha helix), 1917 LEU (alpha helix), 1960 LEU (beta strand), 1961 MET (beta strand), 1974 SER (beta strand), 1905 THR (alpha helix), 1959 THR (beta strand), 1972 TYR (beta strand) |
| Vg | -10.1287 | 5 | 2.617 | 1451 ASN | Chain C: 1447 ALA (coil), 1420 ARG (coil), 1451 ASN (alpha helix), 1449 GLY (coil), 1599 GLY (coil), 1600 GLY (coil), 1450 MET (alpha helix), 1452 MET (alpha helix), 1417 SER (coil), 1419 SER (coil), 1418 THR (coil), 1598 VAL (coil) |
| 2.576 | 1452 MET | ||||
| 2.468 | 1451 ASN | ||||
| 2.071 | 1419 SER | ||||
| 2.294 | 1420 ARG | ||||
| Vh | -10.3897 | 2 | 2.283 | 627 ARG | Chain A: 654 ALA (coil), 826 ALA (coil), 627 ARG (coil),653 ASP (coil), 827 CYS (coil), 652 GLY (coil), 832 GLY (coil), 659 MET (alpha helix), 628 PHE (coil), 831 PRP (coil), 826 SER (coil), 651 SER (coil) |
| 2.998 | 627 ARG | ||||
| Vj | -10.925 | 2 | 2.691 | 529 ASN | Chain A: 478 ALA (coil), 495 ARG (alpha helix), 496 ARG (alpha helix), 529 ASN (coil), 475 HIS (coil), 476 ILE (coil), 513 ILE (coil), 498 LEU (alpha helix), 499 LEU (alphahelix), 502 LYS (coil), 477 PRO (coil), 479 TYR (coil), 471 VAL (alpha helix) Chain B: 962 ARG (alpha helix), 965 ARG (alpha helix), 961 ASN (alpha helix), 946 GLN (beta strand), 936 GLY (coil), 934 VAL (beta strand) |
| 2.346 | 965 ARG | ||||
| Vj | -11.029 | 2 | 1.943 | 479 TYR | Chain A: 529 ASN (coil), 528 GLU (coil), 480 LYS (coil), 483 THR (coil), 479 TYR( coil) Chain B: 958 ALA (alpha helix), 965 ARG (alpha helix), 961 ASN (alpha helix), 1113 GLU (alpha helix), 969 LEU (coil), 957 VAL (alpha helix), 1114 VAL (alpha helix) |
| 2.999 | 965 ARG | ||||
| Vk | -11.3929 | 6 | 2.815 | 1451 ASN | Chain C: 1447 ALA (coil), 1619 ALA (coil), 1451 ASN (alpha helix), 1446 ASP (coil), 1620 CYS (coil), 1445 GLY (coil), 1449 GLY (alpha helix), 1599 GLY (coil), 1600 GLY (coil), 1625 GLY (coil), 1621 LYS (coil), 1448 MET (coil), 1450 MET (alpha helix), 1452 MET (alpha helix), 1421 PHE (coil), 1624 PRO (coil), 1419 SER (coil), 1444 SER (coil), 1418 THR (coil), 1598 VAL (coil) |
| 2.513 | 1452 MET | ||||
| 2.559 | 1449 GLY | ||||
| 2.677 | 1448 MET | ||||
| 2.987 | 1447 ALA | ||||
| 1.867 | 1446 ASP | ||||
| Vl | -10.559 | 4 | 2.9431 | 627 ARG | Chain A: 654 ALA (coil), 826 ALA (coil), 627 ARG (coil), 830 ASN (coil), 653 ASP (coil), 827 CYS (coil), 652 GLY (coil), 832 GLY (coil), 828 LYS (coil), 659 MET (alpha helix), 628 PHE (coil), 831 PRO (coil), 651 SER (coil), 805 VAL (alpha helix) |
| 2.256 | 627 ARG | ||||
| 2.853 | 627 ARG | ||||
| 2.906 | 653 ASP | ||||
| Atrovastatin | -11.8605 | 3 | 2.378 | 1963 ALA | Chain B: 1177 ALA (beta strand),1128 ASN (coil), 1182 ASN (coil), 1120 GLU (alpha helix), 1124 GLU (alpha helix), 1176 GLU (beta strand), 1123 ILE (alpha helix), 1127 ILE (alpha helix), 1175 MET (beta strand) Chain C: 1388 ARG (beta strand) Chain D: 1963 ALA (beta strand), 1914 ASN (coil), 1968 ASN (coil), 1906 GLU (alpha helix), 1910 GLU (alphahelix), 1962 GLU (beta strand), 1090 ILE (alpha helix), 1913 ILE (alpha helix) |
| 1.976 | 1910 GLU | ||||
| 2.991 | 1388 ARG | ||||
Table 4: Docking Result Of Ligands And Atrovastatin Against HMG-COA Reductase (PDB ID: 1HWK)
In summary, we have synthesized novel 12 substituted derivatives of benzo (h) chromene-3-carboxylate derivatives and screened their antihyperlipidemic activity. The molecular docking results revealed that compound Vf has better binding affinity (-12.898 kcal/mol) energy values compared with atorvastatin (-11.8605 kcal/mol) to HMG-CoA reductase enzyme. Interestingly, Vf also binds to the same binding site as that of atorvastatin with a similar amino acid binding residue of enzymes. Most of the synthesized compounds shown excellent in vivo antihyperlipidemic activity and found to be nontoxic at a given dose. Further, the values of LDL/HDL indicating cardiac risk factor was less in compound Vk, as compared to standard atorvastatin. Interestingly, high yield, good affinity, the nontoxic and low cardiac risk with potential antihyperlipidemic activity make these compounds the possible lead for future research.
Acknowledgements:
We gratefully acknowledge the management of Rungta College of Pharmaceutical Sciences and Research, Bhilai, for providing necessary infrastructural facilities.
Conflict of interests:
The authors have no conflict of interest to report.
References
- Kalalbandi VK, Seetharamappa J, Katrahalli U. Synthesis, crystal studies and in vivo anti-hyperlipidemic activities of indole derivatives containing fluvastatin nucleus. RSC Adv 2015;5(48):38748-59.
- Mendieta A, Jiménez F, Garduño-Siciliano L, Mojica-Villegas A, Rosales-Acosta B, Villa-Tanaca L, et al. Synthesis and highly potent hypolipidemic activity of alpha-asarone-and fibrate-based 2-acyl and 2-alkyl phenols as HMG-CoA reductase inhibitors. Bioorg Med Chem 2014;22(21):5871-82.
[Crossref] [Google Scholar] [Pub Med]
- Sashidhara KV, Kumar A, Kumar M, Srivastava A, Puri A. Synthesis and antihyperlipidemic activity of novel coumarin bisindole derivatives. Bioorg Med Chem Lett 2010;20(22):6504-7.
[Crossref] [Google Scholar] [Pub Med]
- El-Agrody AM, Halawa AH, Fouda AM, Al-Dies AA. The anti-proliferative activity of novel 4H-benzo [h] chromenes, 7H-benzo [h]-chromeno [2,3-d] pyrimidines and the structure–activity relationships of the 2-, 3-positions and fused rings at the 2, 3-positions. J Saudi Chem Soc 2017;21(1):82-90.
- Baral N, Mishra DR, Mishra NP, Mohapatra S, Raiguru BP, Panda P, et al. Microwave‐assisted rapid and efficient synthesis of chromene‐fused pyrrole derivatives through multicomponent reaction and evaluation of antibacterial activity with molecular docking investigation. J Heterocycl Chem 2020;57(2):575-89.
- Lakshmi Ranganatha V, Zameer F, Meghashri S, Rekha ND, Girish V, Gurupadaswamy HD, et al. Design, synthesis and anticancer properties of novel benzophenone‐conjugated coumarin analogs. Arch Pharm 2013;346(12):901-11.
[Crossref] [Google Scholar] [Pub Med]
- Vukovic N, Sukdolak S, Solujic S, Niciforovic N. Substituted imino and amino derivatives of 4-hydroxycoumarins as novel antioxidant, antibacterial and antifungal agents: Synthesis and in vitro assessments. Food Chem 2010;120(4):1011-8.
- Rapposelli S, Da Settimo F, Digiacomo M, La Motta C, Lapucci A, Sartini S, et al. Synthesis and biological evaluation of 2′‐oxo‐2, 3‐dihydro‐3′ h‐spiro [chromene‐4, 5′‐ [1, 3] oxazolidin]‐3′ yl] acetic acid derivatives as aldose reductase inhibitors. Arch Pharm 2011;344(6):372-85.
[Crossref] [Google Scholar] [Pub Med]
- Brühlmann C, Ooms F, Carrupt PA, Testa B, Catto M, Leonetti F, et al. Coumarins derivatives as dual inhibitors of acetylcholinesterase and monoamine oxidase. J Med Chem 2001;44(19):3195-8.
[Crossref] [Google Scholar] [Pub Med]
- Khan GA, Naikoo GA, War JA, Sheikh IA, Pandit UJ, Khan I, et al. An efficient green synthesis of some functionalized spiro chromene based scaffolds as potential antitubercular agents. J Heterocycl Chem 2018;55(3):699-708.
- Lee KS, Khil LY, Chae SH, Kim D, Lee BH, Hwang GS, et al. Effects of DK-002, a synthesized (6aS, cis)-9, 10-Dimethoxy-7, 11b-dihydro-indeno [2, 1-c] chromene-3, 6a-diol, on platelet activity. Life Sci 2006;78(10):1091-7.
[Crossref] [Google Scholar] [Pub Med]
- Abdella AM, Moatasim Y, Ali MA, Elwahy AH, Abdelhamid IA. Synthesis and anti‐influenza virus activity of novel bis (4H‐chromene‐3‐carbonitrile) Derivatives. J Heterocycl Chem 2017;54(3):1854-62.
- Venkatraman S, Meera R, Jagadeeswaran M, Devi P, Eruna A, PT Paramewari S et al. Anti hyperlipidemic activity of bisindole substituted coumarin ferivatives. J Biol Sci Opin 2014;2:24-32.
- Darla MM, Krishna BS, Umamaheswara Rao K, Reddy NB, Srivash MK, Adeppa K, et al. Synthesis and bio-evaluation of novel 7-hydroxy coumarin derivatives via Knoevenagel reaction. Res Chem Int 2015;41(2):1115-33.
- Dewangan D, Nakhate KT, Verma VS, Nagori K, Badwaik H, Nair N, et al. Synthesis and molecular docking study of novel hybrids of 1, 3, 4‐oxadiazoles and quinoxaline as a potential analgesic and anti‐inflammatory agents. J Heterocycl Chem 2018;55(12):2901-10.
- Ahirwar J, Ahirwar D, Lanjhiyana S, Jha AK, Dewangan D, Badwaik H. Analgesic and anti‐inflammatory potential of merged pharmacophore containing 1, 2, 4‐triazoles and substituted benzyl groups via thio linkage. J Heterocycl Chem 2018;55(9):2130-41.
- Ahirwar J, Ahirwar D, Lanjhiyana S, Jha AK, Dewangan D, Badwaik H. Synthesis, characterization, molecular modeling, and biological evaluation of 1, 2, 4‐triazole‐pyridine hybrids as potential antimicrobial agents. J Heterocycl Chem 2018;55(11):2598-609.